Abstract
Two-pore channels (TPCs) localize to the endolysosomal system and have recently emerged as targets for the Ca2+-mobilizing messenger, nicotinic acid adenine dinucleotide phosphate (NAADP). However, their membrane topology is unknown. Using fluorescence protease protection assays, we show that human TPC1 and TPC2 possess cytosolic N and C termini and therefore an even number of transmembrane regions. Fluorophores placed at position 225 or 347 in TPC1, or 339 in TPC2 were also cytosolic, whereas a fluorophore at position 628 in TPC1 was luminal. These data together with sequence similarity to voltage-gated Ca2+ and Na+ channels, and unbiased in silico predictions are consistent with a topology in which two homologous domains are present, each comprising 6 transmembrane regions and a re-entrant pore loop. Immunocytochemical analysis of selectively permeabilized cells using antipeptide antibodies confirmed that the C-terminal tails of recombinant TPCs are cytosolic and that residues 240–254 of TPC2 prior to putative pore 1 are luminal. Both TPC1 and TPC2 are N-glycosylated with residues 599, 611, and 616 contributing to glycosylation of TPC1. This confirms the luminal position of these residues, which immediately precede the putative pore loop of the second domain. Mutation of all three glycosylation sites in TPC1 enhances NAADP-evoked cytosolic Ca2+ signals. Our data establish essential features of the topology of two-pore channels.
Keywords: Calcium, Calcium Imaging, Calcium Intracellular Release, Calcium Transport, Glycosylation, Ion Channels, Lysosomes, Site-directed Mutagenesis, NAADP, Two-pore Channels
Introduction
Regulated increases in the concentration of cytosolic Ca2+ form the basis of a ubiquitous signal transduction pathway critical for controlling a host of cellular events ranging from fertilization to cell death (1). Rapid changes in cytosolic Ca2+ concentration are achieved through opening of Ca2+-permeable channels located within the plasma membrane and the membranes of intracellular Ca2+ stores (2). Numerous plasma membrane Ca2+-permeable channels have been described including voltage-gated Ca2+ channels and ligand-gated ion channels activated by diverse neurotransmitters (1, 2). The portfolio of intracellular Ca2+ channels, however, is much more limited. The main intracellular Ca2+ channels are those activated by second messengers such as inositol 1,4,5-trisphosphate (IP3),4 cyclic ADP-ribose, and nicotinic acid adenine dinucleotide phosphate (NAADP) (3–5). These channels thus represent a point of convergence in transducing a multitude of extracellular cues into cellular responses. IP3 and cyclic ADP-ribose release Ca2+ from the endoplasmic reticulum via activation of the well characterized family of IP3 (6) and ryanodine receptors (7), respectively. In contrast, in most cells studied, NAADP appears to target acidic stores of Ca2+ (8) such as lysosomes or lysosome-like organelles (9, 10), endosomes (11) and secretory vesicles (12). The molecular target for NAADP has remained elusive since the first description of the Ca2+-mobilizing properties of NAADP in sea urchin eggs over 15 years ago (13, 14), but recent independent studies have converged on a poorly characterized family of ion channels, the two-pore channels (TPCs), as credible candidates (15–18). Consistent with their identity as NAADP receptors, TPCs localize to the endolysosomal system and potentiate NAADP-mediated Ca2+ signals when overexpressed (15–17, 19, 20). Additionally, interfering with endogenous TPC function by siRNA-mediated silencing (15), overexpression of an inactive TPC (15) or genetic ablation (16), inhibits NAADP-mediated Ca2+ signals. Biophysical analyses confirm that TPC2 is an NAADP-gated Ca2+-permeable channel (21–23) targeted to lysosomes by an N-terminal dileucine motif (23). Transcripts for TPCs are present in a range of mammalian tissues consistent with the widespread actions of NAADP (15–17).
Three TPC isoforms (TPC1–3) are present in the sea urchin and most other deuterostome animals (19). Unusually, however, the gene encoding TPC3 has been lost in some closely related mammals, including humans (19, 24). TPCs share modest sequence identity with voltage-gated Ca2+ channels (25). The latter, like most members of the voltage-gated ion channel family, are assembled from domains with 6 trans-membrane (TM) regions and a re-entrant pore loop between TM regions 5 and 6 (26). Voltage-gated Ca2+ (and Na+) channels possess four such domains, whereas TPCs have only two. The predicted topology of TPCs is therefore unique and may represent an evolutionary intermediate between four-repeat channels and one-repeat (single domain) channels (25), such as voltage-gated K+ channels and TRP channels. At present, however, no experimental evidence is available relating to the molecular architecture of TPCs.
N-Linked glycosylation is the process whereby carbohydrate residues are added to asparagine residues of proteins (27). This modification can affect protein folding, targeting, stability, and function (28). The effect of N-glycosylation on plasma membrane ion channels has been extensively studied, but corresponding information for intracellular channels is scant. IP3 receptors are N-glycosylated at two residues within the variable region preceding the pore (29), but the functional consequences are unknown. TRP mucolipins (TRMLs) are N-glycosylated (30, 31), and glycosylation of TRPML3 is altered in the A419P mutant that underlies the varitint-waddler phenotype (31). TPCs were recently been shown to be N-glycosylated (16, 17, 23), but the functional impact is unknown.
In the present study, we use fluorescence protease protection assays with mapping of antibody epitopes and mutation of putative N-glycosylation sites to define the topology of human TPCs. We also show that N-glycosylation regulates NAADP-mediated Ca2+ release by TPC1.
MATERIALS AND METHODS
Bioinformatics
TM regions were predicted using the algorithms listed in supplemental Table S1. N-Linked glycosylation sites were predicted using Prosite.
Plasmids
pCS2+ plasmids into which the coding sequence of mRFP (pCS2+ N mRFP) or GFP (pCS2+ C GFP; see Ref. 15) were introduced at the BamHI/EcoRI or Xho/XbaI sites, respectively, were kindly provided by Dr. Masa Tada (UCL). Full-length TPC1 and TPC2 constructs tagged at their C termini with GFP (TPC1-GFP, TPC2-GFP) or mRFP (TPC1-mRFP, TPC2-mRFP) were described previously (15). To generate TPC1 tagged at its N terminus with mRFP (mRFP-TPC1), the full-length coding sequence of TPC1 including the stop codon was amplified by PCR using I.M.A.G.E. clone 40148827 (accession number: BC150203) as a template and primers TPC1 1F and 1R (supplemental Table S2). The product was then cloned into pCS2+ N mRFP at the EcoRI and XbaI sites. Truncated TPC1 constructs tagged at their C termini with GFP were generated by PCR using forward primer TPC1 2F and reverse primers TPC1 2R, 3R, and 4R followed by cloning of the products into the ClaI and EcoRI sites of pCS2+ C GFP. These plasmids (TPC11–225GFP, TPC11–347GFP, TPC11–628GFP) encoded residues 1–225, 1–347, and 1–628 of TPC1, respectively. Constructs in which Asn-599, Asn-611, or Asn-616 of TPC1 GFP were substituted by Gln were generated using the Stratagene QuikChange® Site-directed Mutagenesis kit and mutagenic primers TPC1 5F and 5R (TPC1N599QGFP), TPC1 6F and 6R (TPC1N611QGFP), and TPC1 7F and 7R (TPC1N616QGFP), respectively. A triple mutant (TPC13QGFP) in which all three Asn residues were substituted was generated by first creating a double mutant (TPC1N599Q/N616QGFP) using primer 7F and 7R with pCS2+ TPC1N599QGFP as the template. Primers 8F and 8R were then used to introduce the N611Q mutation. To generate TPC2 tagged at its N terminus with mRFP (mRFP-TPC2), the full-length coding sequence of TPC2 including the stop codon was amplified by PCR using I.M.A.G.E. clone 5214862 (accession number: BC063008) as a template and primers TPC2 1F and 1R. The product was cloned in to pCS2+ N mRFP at the EcoRI and XbaI sites. A truncated TPC2 construct encoding residues 1–339 tagged with GFP at the C terminus (TPC11–339GFP) was generated using primers TPC2 2F and 2R followed by cloning into pCS2+ C GFP at the EcoRI and XhoI sites. The coding sequences of all plasmids were fully sequenced to confirm that only the desired mutations were introduced.
Cell Culture and Transfection
HEK cells and SKBR3 human breast cancer cells were cultured and plated as described (15). They were transiently transfected with plasmids using LipofectamineTM 2000 transfection reagent (Invitrogen) according to the manufacturer's instructions. Cells were used 1–2 days after transfection.
Fluorescence Protease Protection Assays
The methods are based on those described by Lorenz et al. (32). HEK cells attached to glass coverslips were washed in HEPES-buffered saline and the plasma membrane was then selectively permeabilized by addition of 50 μm digitonin for 7 min at 20 °C in a cytosol-like medium containing 110 mm K+ acetate, 2 mm MgCl2, and 20 mm NaHEPES (pH 7.2). The cells were washed twice in cytosol-like medium and the coverslips mounted on the stage of an inverted epifluorescence microscope (Olympus IX71) equipped with a ×20 objective, a cooled CCD camera and a monochromotor light source (T.I.L.L. Photonics). Fluorescence of GFP (emission >500 nm) and mRFP (emission >570 nm) was captured at 3-s intervals following excitation at 488 nm and 543 nm, respectively. Signals in regions of interest corresponding to individual cells (typically 10–25 cells/imaging field) were corrected for background and normalized to that recorded prior to addition of trypsin (4 μm) (Sigma). Results are presented as means ± S.E. of 4–5 fields from at least two independent transfections.
Antibodies
Affinity-purified goat polyclonal antibody raised to the C terminus of human TPC1 was from Santa Cruz Biotechnology. Affinity-purified rabbit polyclonal antibody raised to the C terminus of human TPC2 (residues 695–752) was from Sigma. Affinity-purified rabbit polyclonal antibody raised to residues 240–254 (GGKQDDGQDRERLTY) of human TPC2 was produced by Eurogentec.
Immunocytochemistry
Transfected HEK cells were fixed in paraformaldehyde (4% w/v) at 25 °C for 10 min and washed three times in phosphate-buffered saline. They were then permeabilized with either Triton X-100 (0.1% v/v, 10 min) to permeabilize all membranes and so expose cytosolic and luminal epitopes, or with digitonin (50 μm, 6 min) to selectively permeabilize the plasma membrane thus exposing only cytosolic epitopes. Cells were washed again (three times with phosphate-buffered saline) and then incubated in blocking buffer consisting of phosphate-buffered saline supplemented with fetal bovine serum (5% v/v) and bovine serum albumin (1% w/v) for 1 h at 25 °C. Incubation of primary and secondary antibodies was performed sequentially in blocking buffer at 37 °C for 1 h. Unbound antibody was removed after each incubation by washing coverslips three times in phosphate-buffered saline supplemented with 0.1% v/v Tween-20. Primary antibodies were used at a dilution of 1:50 (C-terminal TPC1), 1:40 (C-terminal TPC2), or 1:20 (residues 240–254 in TPC2). Antibody binding was visualized using secondary antibodies (1: 100) conjugated to either ALEXA Fluor® 488 or 568 (Invitrogen). The specificity of the labeling was verified by omitting the primary antibody. Coverslips were mounted on slides with 1,4-diazabicyclo[2,2,2]octane (DABCO) and sealed immediately. Confocal microscopy was performed as described (15).
Western Blotting
HEK cells expressing GFP-tagged TPC were harvested by scraping, washed by centrifugation (500 × g, 5 min) and resuspended in solubilization medium (10 μl/cm2 adherent cells) comprising 20 mm Tris (pH 7.2), 50 mm NaCl, 10 mm MgAcetate, 1% Surfact-Amps X-100 (Pierce) and an EDTA-free protease inhibitor mixture (Roche). The suspension was rotated for 60 min at 4 °C, then centrifuged (90,000 × g, 60 min, 4 °C). The supernatant was incubated with or without PNGaseF (50,000 units/ml, New England Biolabs) for 60 min at 37 °C according to the manufacturer's instructions, except that the denaturation step was omitted. Samples (10 μg) were resuspended in NuPAGE® LDS-sample buffer (Invitrogen) supplemented with 100 mm DTT and incubated at 20–50 °C for 15–30 min prior to SDS-PAGE using NuPAGE® Novex 4–12% Bis-Tris gels (Invitrogen). Proteins were transferred to nitrocellulose and Western blotting was performed as described (33) using a mouse anti-GFP primary antibody (Roche, 1:1000) and an anti-mouse horseradish peroxidase-conjugated secondary antibody (Invitrogen, 1:1000).
Microinjection
Injection of NAADP into SKBR3 cells was performed using Femtotips I and the InjectMan N12 Femtojet system (Eppendorf). Pipettes were back-filled with an intracellular solution composed of 110 mm KCl, 10 mm NaCl, and 20 mm Hepes, pH 7.2, and supplemented with or without 10 nm NAADP (Sigma). The injection time was 0.3 s at 75 hPa with a compensation pressure of 25 hPa. Only cells that maintained a resting basal fura-2 fluorescence ratio (see following paragraph) of <0.7 and that expressed similar levels of the transfection reporter were injected.
Ca2+ Imaging
Cells were incubated with 5 μm fura-2 AM (Invitrogen) in Hanks' balanced salt solution at 20 °C for 45 min in the dark, washed three times with dye-free buffer, and incubated for an additional 45 min to allow de-esterification of the dye. Coverslips were then mounted in a custom-designed bath on the stage of an inverted microscope (Eclipse Ti; Nikon) equipped with a “Perfect Focus System,” a 40× oil immersion objective and a CCD camera (CoolSnap HQ2, Roper Scientific). Cells were superfused with Hanks' balanced salt solution at a flow rate of 0.5 ml/min. Fura-2 fluorescence (510-nm emission), after alternate excitation at 340 and 380 nm, was acquired at a frequency of 0.25 Hz. Captured images were analyzed using NIS-Elements AR 3.1 software (Nikon). Calibration was performed as described in (34) Autofluorescence was negligible.
RESULTS AND DISCUSSION
Domain Architecture of TPCs
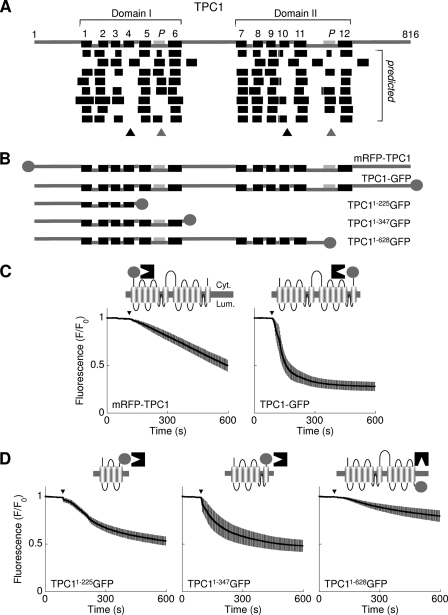
Fig. 1A (top) shows a schematic representation of the amino acid sequence of human TPC1 highlighting the positions of the 12 putative TM regions (black) and the 2 pore (pale gray) regions. The boundaries were determined based on alignments with voltage-sensitive Ca2+ and Na+ channels (25). We compared this topology with topologies predicted by 8 different algorithms (Fig. 1A, bottom). The number of predicted TM regions ranged from 10–14 (supplemental Table S1). Whereas there was convergence on TM regions 1, 2, 3, 5, and 6 of domain I and the corresponding regions in domain II, TM region 4 of domain I and the equivalent region of domain II (TM region 10) were not predicted by most algorithms (Fig. 1A, black triangles). Additionally, several programs predicted TM regions corresponding to the pore regions (Fig. 1A, gray triangles). To resolve these discrepancies, we generated the constructs depicted in Fig. 1B and performed fluorescence protease protection assays (see “Materials and Methods”) to examine directly the membrane topology.
FIGURE 1.
Domain architecture of TPC1. A, top, schematic depiction of human TPC1 based on alignment to voltage-sensitive Ca2+ and Na+ channels showing the positions of the predicted TM regions (numbered, black) and pores (P, pale gray). Two repeated domains (I and II) are identified. Linker regions predicted to be cytosolic (upper connectors) or luminal (lower connectors) are shown. Bottom, unbiased predictions of the TM regions derived using (from top to bottom) Das, SOSUI, TMHMM, HMMTOP, TMPred, Split Server, TMMOD, and Phobius (see supplemental Table S1). Ambiguous predictions of TM4/10 and the pores are highlighted by the black and gray triangles, respectively. B, schematic depiction of the human TPC1 fusion proteins used, where circles represent the fluorescent tags. C and D, fluorescence protease protection assays using cells expressing the indicated TPC1 fusion protein. Trypsin was added at the arrowhead. Fluorescence signals are normalized to initial fluorescence and are presented as means ± S.E. from >40 cells derived from at least 4 fields from two independent transfections. Cartoons show a schematic of the channel with the assumed position of the tag (circle), which faces either the cytoplasm (Cyt.), where it is accessible to trypsin (black structure); or the lumen (Lum.), where it is protected.
In the first experiments we compared the susceptibility of fluorescent tags introduced at either the N terminus or C terminus of TPC1 to trypsin. According to the 12-TM model, both fluorophores should be cytosolic and therefore degraded after selective permeabilization of the plasma membrane with digitonin (32). As shown in Fig. 1C, trypsin caused a marked decrease in fluorescence in cells expressing mRFP-TPC1 or TPC1-GFP. This indicates that both termini are cytosolic and that TPC1 therefore contains an even number of TM regions.
To address the status of the putative TM regions 4 and 10 and the pore regions, we performed fluorescence protease protection analysis of cells expressing truncated C-terminally tagged TPC1 constructs. The first construct (TPC11–225GFP) corresponded to residues 1–225 of TPC1. According to the 12-TM region model, the fluorophore proceeds TM region 4 and should therefore be cytosolic. In contrast, if the first 224 amino acids harbor only three TM regions, as predicted by most algorithms, then the fluorophore should be luminal. As shown in Fig. 1D, fluorescence was significantly reduced by addition of trypsin to cells expressing TPC11–225GFP indicating that the fluorophore is cytosolic. These data suggest that the first 225 amino acids of TPC1 contain an even number of TM regions, likely 4. Failure of some algorithms to predict TM region 4 may relate to the presence of positively charged residues within this region which in voltage-gated Ca2+ channels contribute to voltage-sensing.
We next analyzed a construct corresponding to the first 347 residues of TPC1 (TPC11–347GFP). The region between residues 225 and 347 harbors TM region 5, a pore region and TM region 6. However, some algorithms predicted an additional TM region in place of the pore (Fig. 1A). The fluorophore would thus be cytosolic in the 12-TM region model, but luminal if the pore region includes an additional TM region. The results of the fluorescence protease protection assays showed that trypsin caused a substantial loss of fluorescence from cells expressing TPC11–347GFP. The fluorophore at position 347 is thus cyostolic. With both residues 225 and 347 being cytosolic, the intervening region must contain an even number of TM regions corresponding to TM regions 5 and 6 in the 12-TM region model. The additional predicted TM region likely corresponds instead to the pore.
The accessibility of a fluorophore placed at position 628 was also analyzed. This residue within domain II proceeds a reasonably well predicted TM region. In the 12 TM region model, this would place the fluorophore after TM region 11 and thus in a luminal location. As with TM region 4 in domain I, TM region 10 which lies upstream is not well predicted (Fig. 1A, black triangle). Thus, the fluorophore would be cytosolic if TM region 10 were absent. As shown in Fig. 1D, trypsin had little effect on the fluorescence of permeabilized cells expressing TPC11–628GFP. These results place residue 628 within the lumen and confirms the specificity of action of trypsin.
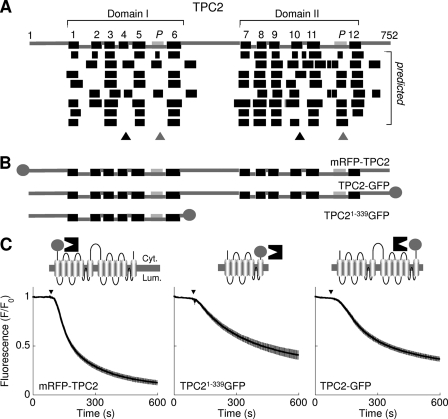
Finally, in a more limited analysis, we examined the topology of TPC2 both in silico and in vitro. As with TPC1, TM regions 4 and 10 in TPC2 were not well predicted (Fig. 2A, black triangles). There was loss of fluorescence after trypsin addition to cells expressing TPC2 with the fluorophore at the N terminus of full-length protein (mRFP-TPC2) or the C terminus of TPC2 truncated at residue 339 (TPC21–339GFP; Fig. 2, B and C). The region between the two fluorophores must therefore contain an even number of TM regions (probably 6). The fluorophore in cells expressing C-terminally-tagged full-length TPC2 (TPC2-GFP) was also accessible to trypsin (Fig. 2, B and C). Thus, residues 340–813 also contain an even number of TM regions, again likely to be 6. The overall architecture of TPC2 is therefore comparable to TPC1.
FIGURE 2.
Domain architecture of TPC2. A–C, schematic depictions of human TPC2 topology (A), and fusion proteins (B) used for fluorescence protease protection assays (C). Refer to legend of Fig. 1 for further details.
A quantitative summary of the fluorescence protease protection analyses is provided in Table 1. For some constructs where the rate of release of the tag is consistent with it being cytosolic, there is almost complete loss of fluorescence (e.g. mRFP-TPC2), while for others the loss is incomplete (50–75%), though still significantly greater than for the only construct (TPC1–628GFP) where our results suggest a luminal location of the tag (Table 1). These observations may indicate heterogeneous populations of protein, but we note that both TPC1-GFP and TPC2-GFP, which showed only partial loss of fluorescence (Table 1), are functional (15, 19, 23).
TABLE 1.
Fluorescence protease protection analysis of TPC1 and TPC2
Summary data (from n experiments) quantifying the extent (at 10 min) and initial rate of fluorescence loss for each of the indicated constructs.
| Construct | Extent | Initial rate | n |
|---|---|---|---|
| % | %/min | ||
| mRFP-TPC1 | 50 ± 6 | 7 ± 1 | 5 |
| TPC1-GFP | 75 ± 5 | 88 ± 21 | 5 |
| TPC11–225GFP | 46 ± 5 | 11 ± 1 | 5 |
| TPC11–347GFP | 52 ± 6 | 21 ± 6 | 5 |
| TPC11–628GFP | 21 ± 5 | 3 ± 1 | 5 |
| mRFP-TPC2 | 88 ± 2 | 40 ± 5 | 6 |
| TPC21–339GFP | 59 ± 5 | 13 ± 2 | 5 |
| TPC2-GFP | 60 ± 2 | 15 ± 1 | 9 |
Topological Mapping of Residues Using Anti-TPC Antibodies
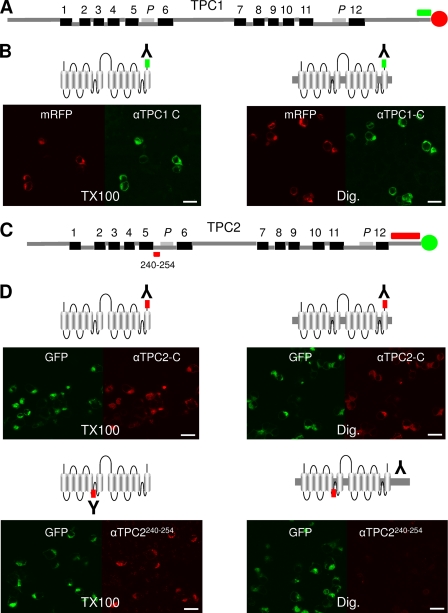
In an independent approach to probe the topology of the TPCs, we used anti-peptide antibodies for immunolocalization of TPCs. Fig. 3, A and C show the location of the epitopes for the three antibodies used. Immunostaining was performed using cells that over expressed TPC1-mRFP or TPC2-GFP and which were permeabilized with either Triton X-100 (to expose both cytosolic and luminal epitopes) or digitonin (to expose only cytosolic epitopes). In the first experiments, we used an antibody raised to the extreme C terminus of TPC1. This antibody recognized TPC1 as evinced by confocal fluorescence images of Triton X-100-permeabilized cells expressing TPC1-mRFP showing signals in both the red channel (corresponding to mRFP fluorescence of the tagged protein) and green channel (indicative of antibody binding) (Fig. 3B, left). Similar results were obtained when only the plasma membrane was permeabilized with digitonin (Fig. 3B, right). The latter indicates that the epitope (residues 803–816) is cytosolic. For this and the other TPC antibodies used, no signals were obtained in neighboring untransfected cells or in the absence of primary antibody confirming the specificity of the antibody labeling (supplemental Fig. S1). We performed similar experiments with TPC2 using an antibody raised to residues corresponding to the C-terminal 58 residues (Fig. 3C). This antibody again detected its epitope in TPC2-GFP-expressing cells permeabilized with either Triton X-100 or digitonin (Fig. 3D). These results place residues 694–752 in the cytosol. Accessibility of C-terminal tails to the anti-TPC1 antibodies in digitonin-permeabilized cells is consistent with the accessibility of the C-terminal fluorophores in fluorescence protease protection assays (Figs. 1C and 2C). They thereby provide independent evidence that the C termini of TPCs are cytosolic. Finally, we performed similar experiments using an antibody to an epitope of TPC2 which according to the 12-TM region model is luminal and precedes pore 1 (Fig. 3C). Whereas this antibody labeled cells expressing TPC2-GFP when Triton X-100 was used for permeabilization, no labeling was observed when cells were permeabilized with digitonin (Fig. 3D). These results suggest that residues 240–254 are luminal. The inaccessibility of these residues until intracellular membranes are permeabilized also establishes that incomplete removal of the C-terminal GFP tag during trypsin treatment (Table 1) is not due to some of the protein adopting a reversed topology. Together, the immunolocalization results further support the proposed 12-TM region topology of the TPCs.
FIGURE 3.
Mapping the topology of TPCs with anti-peptide antibodies. A, schematic depiction of TPC1 showing the position of a C-terminal sequence (green) to which an anti-TPC1 antibody was raised. B, confocal fluorescence images of cell populations transfected with TPC1-mRFP showing fluorescence of the tag (red) or the bound anti-TPC1 antibody using an ALEXA Fluor® 488-labeled secondary antibody (green). Cells were fixed and then permeabilized with either Triton X-100 (TX100; left) or digitonin (Dig; right). C, schematic depiction of TPC2 showing the positions of sequences within domain I (residues 240–254) and the C terminus to which anti-TPC2 antibodies were raised (red). D, similar to B, except cells were transfected with TPC2-GFP (green) and probed with the indicated antibodies to TPC2 and ALEXA Fluor® 568-labeled secondary antibody (red). Results are representative of at least 2 fields of view from two independent transfections. Scale bars, 20 μm.
TPCs are N-Glycosylated
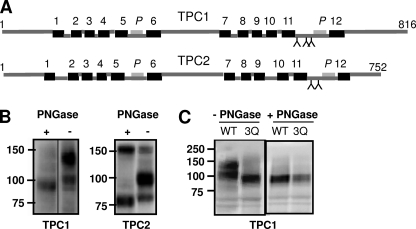
N-Glycosylation of proteins occurs within the secretory pathway and results in modification of only luminal asparagine residues (27). Mapping of such sites therefore provides another means of defining topology. Fig. 4A shows the positions of putative N-glycosylation sites within TPC1 and TPC2 conforming to the consensus N-X-T/S (where X is any residue except proline). Notably, there is a conserved cluster of sites in domain II within a putative luminal loop preceding pore 2. Additional sites were predicted in the C-terminal tail of TPC1 and within TM region 4 for TPC2 (not shown). To determine whether TPCs were glycosylated, we compared the mobility of recombinant TPCs after SDS-PAGE and Western blot analysis. Analysis was performed with or without treatment with PNGase F which removes N-glycan chains from asparagine residues. In the absence of PNGase, two bands were observed for TPC1-GFP with apparent molecular masses of 130 ± 5 kDa and 100 ± 5 kDa (n = 8) (Fig. 4B). After pretreatment with PNGase F, a single band of 95 ± 2 kDa (n = 4) was observed. This decrease in apparent molecular mass indicates that TPC1 is glycosylated. The higher molecular mass band likely corresponds to the mature fully glycosylated protein and the lower band to the core glycosylated protein. Two prominent bands (100 ± 5 kDa and 82 ± 5 kDa, n = 7) were also detected for TPC2-GFP, and a single band of lower molecular mass (77 ± 2 kDa, n = 3) after PNGaseF treatment (Fig. 4B). These results indicate that TPC2, like TPC1, is N-glycosylated.
FIGURE 4.
TPCs are N-glycosylated. A, schematic depictions of TPC1 and TPC2 showing the positions of the predicted N-glycosylation sites (Y). B, Western blot analysis of extracts prepared from cells expressing TPC1-GFP (left) or TPC2-GFP (right). Extracts were incubated with (+) or without (−) PNGase-F. C, Western blot analysis of extracts prepared from cells expressing a GFP-tagged TPC1 triple mutant in which Asn-599, Asn-611, and Asn-616 were substituted with Gln (TPC13QGFP). The migration of TPC1-GFP (WT) is shown for comparison. Results are representative of at least three independent transfections. Mr markers (kDa) are shown.
A previous study showed that mutation of Asn-594 and Asn-601 in mouse TPC2 (conserved as Asn-611 and Asn-618 in human TPC2) abolished glycosylation (17). To determine which residues are responsible for glycosylation of TPC1, we focused on asparagines 599, 611, and 616 because they are clustered in the same region as the glycosylated residues in TPC2. All three residues were substituted for glutamine and the resulting mutant (TPC13QGFP) was analyzed by Western blotting with and without PNGaseF treatment. TPC13QGFP migrated substantially faster than the wild-type protein and as a single band with an apparent mass (95 ± 1 kDa, n = 3) similar to that of the deglycosylated wild-type protein (Fig. 4C). Migration of TPC13QGFP was unaffected by PNGase F treatment. These data suggest that asparagines 599, 611 and 616 of TPC1 include glycosylation sites and are therefore luminal. We noted a modest decrease in the apparent molecular mass of the upper and lower band for TPC1 proteins individually mutated in the three potential glycosylation sites (TPC1N599QGFP, TPC1N611QGFP and TPC1N616QGFP) compared with wild-type protein (supplemental Fig. S2), consistent with N-glycosylation of each of the three sites. The luminal location of the region that includes these residues is consistent with the fluorescence protease protection analysis of TPC11–628GFP (Fig. 1C).
N-Glycosylation of TPCs Regulates NAADP-mediated Ca2+ Release
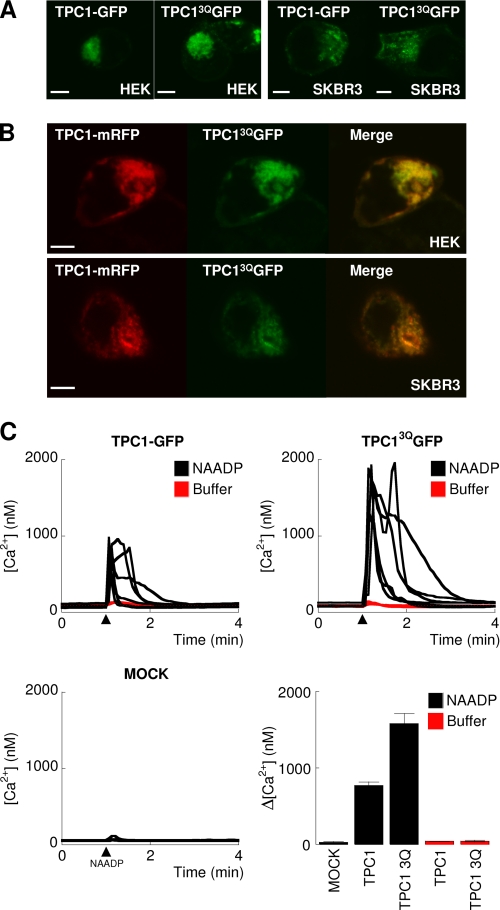
N-Glycosylation can regulate protein trafficking (28) and impaired N-linked glycosylation is associated with disease (35, 36). We therefore compared the sub-cellular distribution of TPC1-GFP and TPC13QGFP. As shown in Fig. 5A, both proteins showed a similar punctate intracellular distribution in HEK and SKBR3 cells. Additionally, co-expression of TPC1-mRFP with TPC13QGFP showed substantial overlap in fluorescence (Fig. 5B). Moreover, TPC13QGFP showed partial colocalization with markers for endosomes and lysosomes (supplemental Fig. S3) as reported previously for TPC1 (15). Thus, glycosylation appeared not to affect the subcellular localization of TPC1.
FIGURE 5.
N-Glycosylation of TPC1 regulates NAADP-mediated Ca2+ release. A, confocal fluorescence images of HEK (left) or SKBR3 (right) cells expressing wild-type TPC1-GFP or TPC13QGFP. In B, cells were contransfected with wild-type TPC1-mRFP and TPC13QGFP. Merged images are shown to the right. Scale bars, 5 μm. C, cytosolic Ca2+ signals from individual fura-2-loaded SKBR3 cells microinjected (arrowheads) with 10 nm NAADP (black lines) or buffer (red lines). Cells were either mock-transfected or transiently transfected with TPC1-GFP or TPC13QGFP. Summary results quantifying amplitudes of the Ca2+ signals in individual cells (means ± S.E. of 4–6 cells) from the indicated cells are shown in the last panel.
In several ion channels including NaV1.1 (37), Kv12.2 (38), and TRPV1 (39), N-glycosylation sites have been mapped to pore-forming regions and shown to regulate channel activity. Given the proximity of the glycosylation sites to the putative pore in domain II of TPCs, we examined the effect of glycosylation on NAADP-mediated Ca2+ release. We compared NAADP-mediated cytosolic Ca2+ signals in SKBR3 cells overexpressing TPC1-GFP or TPC13QGFP. As shown in Fig. 5C, injection of NAADP evoked robust Ca2+ signals in TPC1-GFP-expressing cells, but not in mock-transfected cells consistent with our previous analyses (15, 19, 23). NAADP-evoked Ca2+ signals were substantially larger in cells expressing TPC13QGFP (Fig. 5C). Injection of buffer in place of NAADP failed to affect the cytosolic Ca2+ concentration (Fig. 5C, red lines). These results show that N-linked glycosylation inhibits NAADP-evoked Ca2+ release.
CONCLUSIONS
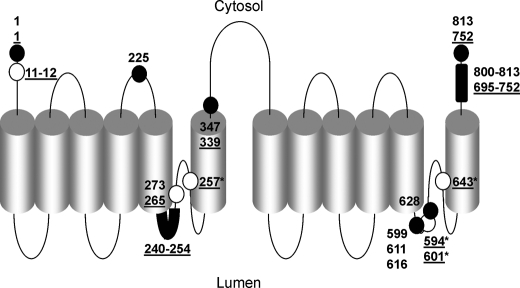
We applied several complementary techniques to determine the topology of the TPCs including mapping of regions prior to the putative pores. A schematic summary which incorporates previous mutagenesis studies (15, 17, 21, 23) is shown in Fig. 6. Our results are consistent with the structural assignment of TPCs to the family of voltage-gated ion channels and provides a framework for informed assessment of consensus regulatory sites. We show that TPCs are N-glycosylated, through identified residues and that this functions to dampen channel activity perhaps by conformational changes close to the pore. Alternatively, the negatively charged oligosaccharide chains may influence binding of luminal H+ or Ca2+, both of which are proposed to regulate TPC2 activity (21, 22). N-Glycosylation is a dynamic process (40) raising the possibility that triggering of cytosolic Ca2+ signals by TPCs may be regulated by physiological or patho-physiological changes in N-linked glycosylation.
FIGURE 6.
Current topological model for TPCs. Schematic summarizing the positions of the numbered residues in human TPCs identified in this study (filled circles). Underlined residues correspond to TPC2. Open circles show the positions of residues identified in previous studies (from left to right): TPC2 Leu-11–12 (23), TPC1 Leu-273 (15), TPC2 Leu-265 (23), TPC2 Asn-257 (21), TPC2 Asn 594, 601 (17), Glu-643 (21). *, numbers refer to amino acid residues of mouse TPC isoforms.
Supplementary Material
Acknowledgment
We thank Chi Li for useful discussion.
This work was supported, in whole or in part, by Grants from the National Institutes of Health (HL90804, to E. B.), the Biotechnology and Biological Sciences Research Council (BB/G013721/1, to S. P., and BB/H009736/1, to C. W. T.) and the Wellcome Trust (085295, to C. W. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S3.
- IP3
- inositol 1,4,5-trisphosphate
- mRFP
- monomeric red fluorescent protein
- NAADP
- nicotinic acid adenine dinucleotide phosphate
- TM
- transmembrane
- TPC
- two-pore channel.
REFERENCES
- 1. Berridge M. J., Lipp P., Bootman M. D. (2000) Nat. Rev. Mol. Cell Biol. 1, 11–21 [DOI] [PubMed] [Google Scholar]
- 2. Clapham D. E. (2007) Cell 131, 1047–1058 [DOI] [PubMed] [Google Scholar]
- 3. Berridge M. J. (1993) Nature 361, 315–325 [DOI] [PubMed] [Google Scholar]
- 4. Lee H. C. (1997) Physiol. Rev. 77, 1133–1164 [DOI] [PubMed] [Google Scholar]
- 5. Guse A. H., Lee H. C. (2008) Sci. Signal. 1, re10. [DOI] [PubMed] [Google Scholar]
- 6. Foskett J. K., White C., Cheung K. H., Mak D. O. (2007) Physiol. Rev. 87, 593–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fill M., Copello J. A. (2002) Physiol. Rev. 82, 893–922 [DOI] [PubMed] [Google Scholar]
- 8. Patel S., Docampo R. (2010) Trends Cell Biol. 20, 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Churchill G. C., Okada Y., Thomas J. M., Genazzani A. A., Patel S., Galione A. (2002) Cell 111, 703–708 [DOI] [PubMed] [Google Scholar]
- 10. Yamasaki M., Masgrau R., Morgan A. J., Churchill G. C., Patel S., Ashcroft S. J. H., Galione A. (2004) J. Biol. Chem. 279, 7234–7240 [DOI] [PubMed] [Google Scholar]
- 11. Menteyne A., Burdakov A., Charpentier G., Petersen O. H., Cancela J. M. (2006) Curr. Biol. 16, 1931–1937 [DOI] [PubMed] [Google Scholar]
- 12. Mitchell K. J., Lai F. A., Rutter G. A. (2003) J. Biol. Chem. 278, 11057–11064 [DOI] [PubMed] [Google Scholar]
- 13. Clapper D. L., Walseth T. F., Dargie P. J., Lee H. C. (1987) J. Biol. Chem. 262, 9561–9568 [PubMed] [Google Scholar]
- 14. Lee H. C., Aarhus R. (1995) J. Biol. Chem. 270, 2152–2157 [DOI] [PubMed] [Google Scholar]
- 15. Brailoiu E., Churamani D., Cai X., Schrlau M. G., Brailoiu G. C., Gao X., Hooper R., Boulware M. J., Dun N. J., Marchant J. S., Patel S. (2009) J. Cell Biol. 186, 201–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Calcraft P. J., Ruas M., Pan Z., Cheng X., Arredouani A., Hao X., Tang J., Rietdorf K., Teboul L., Chuang K. T., Lin P., Xiao R., Wang C., Zhu Y., Lin Y., Wyatt C. N., Parrington J., Ma J., Evans A. M., Galione A., Zhu M. X. (2009) Nature 459, 596–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zong X., Schieder M., Cuny H., Fenske S., Gruner C., Rötzer K., Griesbeck O., Harz H., Biel M., Wahl-Schott C. (2009) Pflugers Arch. 458, 891–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Patel S., Marchant J. S., Brailoiu E. (2010) Cell Calcium 47, 480–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brailoiu E., Hooper R., Cai X., Brailoiu G. C., Keebler M. V., Dun N. J., Marchant J. S., Patel S. (2010) J. Biol. Chem. 285, 2897–2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ruas M., Rietdorf K., Arredouani A., Davis L. C., Lloyd-Evans E., Koegel H., Funnell T. M., Morgan A. J., Ward J. A., Watanabe K., Cheng X., Churchill G. C., Zhu M. X., Platt F. M., Wessel G. M., Parrington J., Galione A. (2010) Curr. Biol. 20, 703–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schieder M., Rötzer K., Brüggemann A., Biel M., Wahl-Schott C. A. (2010) J. Biol. Chem. 285, 21219–21222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pitt S. J., Funnell T., Sitsapesan M., Venturi E., Rietdorf K., Ruas M., Ganesan A., Gosain R., Churchill G. C., Zhu M. X., Parrington J., Galione A., Sitsapesan R. (2010) J. Biol. Chem. 285, 35039–35046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brailoiu E., Rahman T., Churamani D., Prole D. L., Brailoiu G. C., Hooper R., Taylor C. W., Patel S. (2010) J. Biol. Chem. 285, 38511–38516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cai X., Patel S. (2010) Mol. Biol. Evol. 27, 2352–2359 [DOI] [PubMed] [Google Scholar]
- 25. Ishibashi K., Suzuki M., Imai M. (2000) Biochem. Biophys. Res. Commun. 270, 370–376 [DOI] [PubMed] [Google Scholar]
- 26. Yu F. H., Yarov-Yarovoy V., Gutman G. A., Catterall W. A. (2005) Pharmacol. Rev. 57, 387–395 [DOI] [PubMed] [Google Scholar]
- 27. Ohtsubo K., Marth J. D. (2006) Cell 126, 855–867 [DOI] [PubMed] [Google Scholar]
- 28. Helenius A., Aebi M. (2004) Annu. Rev. Biochem. 73, 1019–1049 [DOI] [PubMed] [Google Scholar]
- 29. Michikawa T., Hamanaka H., Otsu H., Yamamoto A., Miyawaki A., Furuichi T., Tashiro Y., Mikoshiba K. (1994) J. Biol. Chem. 269, 9184–9189 [PubMed] [Google Scholar]
- 30. Kiselyov K., Chen J., Rbaibi Y., Oberdick D., Tjon-Kon-Sang S., Shcheynikov N., Muallem S., Soyombo A. (2005) J. Biol. Chem. 280, 43218–43223 [DOI] [PubMed] [Google Scholar]
- 31. Kim H. J., Li Q., Tjon-Kon-Sang S., So I., Kiselyov K., Muallem S. (2007) J. Biol. Chem. 282, 36138–36142 [DOI] [PubMed] [Google Scholar]
- 32. Lorenz H., Hailey D. W., Wunder C., Lippincott-Schwartz J. (2006) Nat. Protoc. 1, 276–279 [DOI] [PubMed] [Google Scholar]
- 33. Churamani D., Boulware M. J., Geach T. J., Martin A. C., Moy G. W., Su Y. H., Vacquier V. D., Marchant J. S., Dale L., Patel S. (2007) PLoS. ONE. 2, e797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Grynkiewicz G., Poenie M., Tsien R. Y. (1985) J. Biol. Chem. 260, 3440–3450 [PubMed] [Google Scholar]
- 35. Petrecca K., Atanasiu R., Akhavan A., Shrier A. (1999) J. Physiol. 515, 41–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Satler C. A., Vesely M. R., Duggal P., Ginsburg G. S., Beggs A. H. (1998) Hum. Genet. 102, 265–272 [DOI] [PubMed] [Google Scholar]
- 37. Bennett E. S. (2002) J. Physiol. 538, 675–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Noma K., Kimura K., Minatohara K., Nakashima H., Nagao Y., Mizoguchi A., Fujiyoshi Y. (2009) J. Biol. Chem. 284, 33139–33150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wirkner K., Hognestad H., Jahnel R., Hucho F., Illes P. (2005) Neuroreport 16, 997–1001 [DOI] [PubMed] [Google Scholar]
- 40. Montpetit M. L., Stocker P. J., Schwetz T. A., Harper J. M., Norring S. A., Schaffer L., North S. J., Jang-Lee J., Gilmartin T., Head S. R., Haslam S. M., Dell A., Marth J. D., Bennett E. S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 16517–16522 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.