Abstract
Retinal vascular damages are the cardinal hallmarks of retinopathy of prematurity (ROP), a leading cause of vision impairment and blindness in childhood. Both angiogenesis and vasculogenesis are disrupted in the hyperoxia-induced vaso-obliteration phase, and recapitulated, although aberrantly, in the subsequent ischemia-induced neovessel formation phase of ROP. Yet, whereas the histopathological features of ROP are well characterized, many key modulators with a therapeutic potential remain unknown. The CCN1 protein also known as cysteine-rich protein 61 (Cyr61) is a dynamically expressed, matricellular protein required for proper angiogenesis and vasculogenesis during development. The expression of CCN1 becomes abnormally reduced during the hyperoxic and ischemic phases of ROP modeled in the mouse eye with oxygen-induced retinopathy (OIR). Lentivirus-mediated re-expression of CCN1 enhanced physiological adaptation of the retinal vasculature to hyperoxia and reduced pathological angiogenesis following ischemia. Remarkably, injection into the vitreous of OIR mice of hematopoietic stem cells (HSCs) engineered to express CCN1 harnessed ischemia-induced neovessel outgrowth without adversely affecting the physiological adaptation of retinal vessels to hyperoxia. In vitro exposure of HSCs to recombinant CCN1 induced integrin-dependent cell adhesion, migration, and expression of specific endothelial cell markers as well as many components of the Wnt signaling pathway including Wnt ligands, their receptors, inhibitors, and downstream targets. CCN1-induced Wnt signaling mediated, at least in part, adhesion and endothelial differentiation of cultured HSCs, and inhibition of Wnt signaling interfered with normalization of the retinal vasculature induced by CCN1-primed HSCs in OIR mice. These newly identified functions of CCN1 suggest its possible therapeutic utility in ischemic retinopathy.
Keywords: Endothelium, Extracellular Matrix Proteins, Ischemia, Retina, Stem Cell, CCN Proteins, Angiogenesis, Cysteine-rich Protein 61, Endothelial Progenitor Cells, Oxygen-induced Retinopathy
Introduction
Retinopathy of prematurity (ROP),2 a leading cause of visual impairment in low birth weight infants, is initiated by delayed retinal vascular growth and insufficient vascularization after premature birth (1). A disrupted oxygen environment in the retina of severely premature neonates is a key factor in the onset of retinal vaso-obliteration and the development of ROP. Retinal ischemia is the underlying cause triggering the release of angiogenic factors that promote the formation of aberrant vessels, which then break through the inner limiting membrane into the vitreous causing vitreous hemorrhage, tractional retinal detachment, and vision loss.
The normal retinal vasculature, which is planar in nature, is well organized into three capillary plexuses generated during retinal development in a well coordinated manner. Conversely, pathological neovascularization generates disorganized, leaky, and tortuous vessels prone to exudation, hemorrhage, and fibrosis that damage the retina ultimately causing blindness. The collective work of many investigators has resulted in a greater appreciation of the pathogenic factors involved (2–5). The balance between proangiogenic and antiangiogenic factors and the activation state of progenitor and inflammatory cells recruited to the retina are considered to be the determinant factors for the development of ROP (6). Current therapeutic strategies are designed to eliminate/reduce the actual causes of retinal neovascularization while normalizing simultaneously the retinal vasculature.
Development of a normal retinal vasculature involves various sources of endothelial cells, such as circulating stem cells and redeployment of mural cells from regressing vessel segments. Grant et al. (7) has pioneered cell-based therapy studies demonstrating that a subset of transplanted bone marrow-derived hematopoietic stem cells (HSCs) can function as blood vessel progenitors during retinal regeneration/repair. Other groups further reported that HSCs may be used either directly or as a delivery vehicle of specific drugs to selectively prevent blood vessel loss and/or inhibit abnormal neovascularization (8, 9). Not only do progenitor cells incorporate and stabilize nascent blood vessels, but also they elicit a paracrine role by secreting factors that contribute to normalization of resident endothelial cells (10). However, the identity of the chemical and/or environmental signals directing HSCs to sites of angiogenesis and enhancing their commitment to and efficacy in vascular regeneration and repair remain poorly understood. Elucidating these signals will open up new avenues of research in directed cell therapy for ischemic retinopathies.
Extracellular matrix (ECM) proteins are structural and informational entities supporting key signaling events involved in the regulation of endothelial cell differentiation and function during developmental morphogenesis, in response to injury and in pathological conditions (11). Within the ECM, the CCN proteins form an intriguing family of six molecules with distinct features for each member (12, 13). In particular, the cysteine-rich protein 61 (CCN1/Cyr61), an inducible immediate early gene-encoded protein secreted by many mesenchymally and ectodermally derived cells (14, 15), is pleiotropic in function and affects processes as disparate as bone formation and tumorigenesis (16). The CCN1 gene is exquisitely regulated during fetal development and in adult vascular tissue remodeling. During mouse development, CCN1 gene expression peaks as the chorioallantoic plate is invaded by fetal blood vessels from the allantois and most CCN1-deficient mouse embryos die prematurely due to severe vascular defects including placental vascular insufficiency, hemorrhaging blood vessels that appeared with disorganized vascular cells, and absence of discrete basement membrane (17, 18). Clearly, vascular development is impaired/destabilized in the absence of CCN1 despite the presence of other notable angiogenic factors of the vascular endothelial growth factor (VEGF) and fibroblast growth factor families. Recently, CCN1 activities have been expanded to include induction of stem cell self-renewal and differentiation (19), suggesting that CCN1 is potentially a novel therapeutic target in vascular diseases.
At the molecular level, CCN1 functions primarily through direct binding to specific integrin receptors, thereby activating signal transduction cascades that culminate in the regulation of cell adhesion, migration, proliferation, gene expression, differentiation, and survival (20, 21). CCN1 can also modulate the activities of several ECM, growth factors, and cytokines, including transforming growth factor (TGF)-β, tumor necrosis factor (TNF)-α, VEGF, and bone morphogenic proteins through direct physical interaction with either these ligands or their receptors (13, 22). Of particular interest, CCN1 was shown to enhance cytotoxicity of TNF-α, an inflammatory cytokine that influences neovessel growth (23). Here, we focused on the function and therapeutic potential of CCN1 in pathological vaso-obliteration and destructive angiogenesis associated with oxygen-induced retinopathy (OIR). We asked whether CCN1, expressed ectopically in situ or in HSCs that are subsequently transplanted in the retina, could affect vascular repair and/or inhibit pathological angiogenesis associated with OIR.
EXPERIMENTAL PROCEDURES
Reagents, Antibodies, and Proteins
All chemicals used were of reagent grade. Bovine serum albumin (BSA) and heparin were from Sigma. Polyclonal anti-CCN1 antibody was from Biovision Inc. (Mountain View, CA). Antibodies against phospho-extracellular signal-regulated kinases (ERK1/2), phospho-c-Jun-N-terminal kinase (JNK) 1/2, β-catenin, phospho-Pyk2, and integrin subtypes were from Millipore (Billerica, MA). Antibodies against phosphoglycogen synthase kinase-3β (GSK-3β) and GSK-3β were from Cell Signaling Technology (Danvers, MA). Ulex europaeus agglutinin-1 (UEA-1), acetylated low density lipoprotein, labeled with 1,1′-dioctadecyl-3,3,3′,3′-tetramethyl indocarbocyanine perchlorate (DiI-acLDL), 4′,6-diamidino-2-phenylindole (DAPI), and fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse, and TRITC-conjugated goat anti-rabbit IgG antibodies were from Vector Laboratories (Burlingame, CA). Recombinant (r)CCN1 protein was synthesized in a Baculovirus expression system using Sf9 cells and purified from serum-free insect cell-conditioned medium on Sepharose-S columns as previously described (24). Recombinant CCN2 was obtained from Abnova (Walnut, CA).
Adenoviral and Lentiviral Vectors
Mouse CCN1 and CCN2 cDNAs were isolated by PCR amplification using DNA templates obtained from ATCC (Manassas, VA) and cloned into a shuttle vector. The recombinant adenoviruses, Ad-CCN1 and Ad-CCN2, were produced by cotransfecting an adenoviral shuttle vector with a viral backbone in which the recombinant cDNA is driven by the CMV promoter. The adenovirus encoding green fluorescent protein, Ad-GFP, was used as a control for infection. All adenoviruses were replication deficient and used at 20 multiplicity of infection to infect rat retinal endothelial cells (REC) only. Because transduction of bone marrow-derived HSCs with adenoviral vectors is inefficient, lentiviral vectors expressing the CCN1 (Lnv-CCN1), luciferase (Lnv-Luc), and GFP (Lnv-GFP) genes were constructed. Lentiviruses were produced by transient CaPO4-mediated transfection of 293T cells with lentiviral backbone plasmids, a vsv.g-envelope plasmid, and a gag/pol plasmid. Titers of 5 × 106 IU/ml were achieved and concentrated by ultracentrifugation to 1 × 1012 IU/ml. Production of both adenoviral and lentiviral vectors was performed under the auspice of the National Heart, Lung, and Blood Institute Gene Therapy Resource Program Vector Core Laboratory at the University of Pennsylvania.
Oxygen-induced Retinopathy
Animal studies were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Committee on the Ethics of Animal Experiments of the State University of New York-Downstate Medical Center (approval number 416-10-6). Ischemic retinopathy was produced in C57BL/6J mice as described by Smith et al. (25). Neonatal mice and their nursing dams were exposed to 75% oxygen in a PRO-OX 110 chamber oxygen controller from Biospherix Ltd. (Redfield, NY) between postnatal day 7 (P7) and P12 producing vaso-obliteration and cessation of vascular development in the capillary beds of the central retina. The mice were exposed to 12-h cyclical broad spectrum light. The room temperature was maintained at 28 °C. On P12, the mice were placed at room air until P17 when the retinas were assessed for maximum neovascular response. For developmental studies, room air mice were raised under normal light and temperature conditions. Mice were sacrificed at the indicated time periods after birth by CO2 euthanasia and cervical dislocation or by decapitation. Eyes were enucleated and processed for histological and molecular analyses as described below. For gene and cell therapy studies, animals were anesthetized by intraperitoneal injection of ketamine (65 mg/kg) and xylazine (35 mg/kg). Lnv-CCN1 (∼1 μl) was injected into the vitreous in one eye of each animal using a 33-gauge syringe. In the contralateral eye, an equal volume of Lnv-Luc was injected. For cell injection studies, bone marrow CD34+ cells from pathogen-free C57BL/6 mice were used. For this purpose, mice were killed by cervical dislocation and femora were excised and transferred into sterile phosphate-buffered saline (PBS). Bone marrow cells were flushed out of bones with PBS containing 1% fetal bovine serum (FBS). CD34+ cells were obtained through separation by magnetic cell sorting (MACS) with an Auto MACS system using the Lineage Cell Depletion Kit from Miltenyi Biotec (Bergisch Gladbach, Germany). Cells were further incubated with streptavidin-conjugated Texas Red and sorted by flow cytometry. Sorted cells were cultured in the StemSpan serum-free medium supplemented with 100 ng/ml of stem cell factor, 20 ng/ml of thrombopoietin, 20 ng/ml of insulin-like growth factor-2, and 10 ng/ml of human fibroblast growth factor, all obtained from StemCell Technologies (Vancouver, BC). Cells were transduced with 10 multiplicity of infection of either Lnv-CCN1 or Lnv-Luc lentiviral vector for 24 h. Cells infected with Lnv-GFP at 5–10 multiplicity of infection achieved transduction efficiency of 70 to 90% as determined by counting the number of GFP-positive cells. Transduced cells were injected intravitreally in a 1-μl volume (∼1 × 105).
Visualization of the Retinal Vasculature and Image Analyses
Quantification of vascular obliteration, retinal vascularization, and preretinal neovascular tufts was performed at P12 and P17. Eyes were enucleated and fixed in 4% paraformaldehyde for 1 h. Retinas were then dissected, flat mounted through four incisions dividing it into 4 quadrants and incubated overnight in 10 μg/ml of fluorescently labeled UEA-1, a plant lectin with high affinity for the endothelium. Images were acquired using a Nikon Eclipse E400 fluorescence microscope. The area of vascular obliteration was measured by delineating the avascular zone in the central retina and calculating the total area using Photoshop CS5 (Adobe). Similarly, the area of preretinal neovascularization was calculated by selecting tufts, which appear more brightly stained that normal vasculature, based on pixel intensities. Selected regions were then summed to generate the total area of neovascularization. The avascular and neovascularization areas were expressed as a percentage of the total retinal area.
Cell Culture and Treatments
In vitro studies were performed with bone marrow-derived HSCs prepared to 99% purity from healthy human donors (StemCell Technologies). HSCs were first expanded in Poietics HPGM medium supplemented with cytokines, e.g. Flt-3 ligand, stem cell factor, and thrombopoietin. In this medium, HSCs undergo a limited amount of proliferation and still maintain an undifferentiated phenotype. Rat RECs were obtained from Cellpro (San Pedro, CA) and maintained in culture according the manufacturer's instructions. Cells were characterized by immunostaining using endothelial cell markers, e.g. von Willebrand Factor and CD31 obtained from Millipore and Miltenyi Biotec, respectively. The cells were propagated in 35-mm dishes in predefined endothelial growth medium containing 10% of FBS obtained from Atlanta Biological Inc. (San Diego, CA). Cells at 80% confluence were treated as described in the text and further processed for various analyses.
Proliferation Assay
Proliferation rate of RECs and HSCs was determined using the CyQUANT® Direct Cell Proliferation Assay obtained from Invitrogen according to the manufacturer's protocol. The fluorescence intensity was measured with a fluorescence microplate reader using an FITC filter set.
Adhesion Assay
Cell adhesion was examined in 96-well plates uncoated or coated overnight (4 °C) with various substrates. Cells were plated at a concentration of 5 × 104 cells/well in 100 μl of serum free-medium containing 0.1% BSA. Adhesion was carried out for 4 h at 37 °C. Removal of non-adherent cells was achieved by 2 washing steps with serum-free medium containing 0.1% BSA at room temperature. Adhesion was quantified in duplicate by counting adherent cells using an inverted cell culture microscope.
Cell Migration
Proteins tested for their chemotactic activity were diluted, each, in serum-free medium containing 0.5% BSA and loaded in the lower well of Transwell 12-well plates. The lower well was then covered with a polycarbonate filter (5 μm pore diameter, Nucleopore), which was treated with 2.9% (v/v) glacial acetic acid overnight, and rinsed three times in PBS for 1 h immediately before use. Cells were re-suspended in 1.0 ml of pre-equilibrated serum-free medium containing BSA (0.5%) and added into each Transwell insert. Where indicated, cells were treated with either pharmacological inhibitors or function-blocking antibodies for 1 h before chamber loading. After incubation for 6 h at 37 °C, the membrane was removed and stained with Diff-Quik. The chemotactic response was determined by counting the total number of cells migrated in 10 randomly selected microscope fields at ×100 magnification.
RNA Isolation and Quantitative Analysis of mRNA
Total RNA was extracted from cells using RNA Easy column purification protocol (Qiagen). Quantitative real time PCR (qPCR) assay was performed to determine the levels of a specific mRNA using TaqMan technology on an ABI 7000 sequence detection system from Applied Biosystems (Carlsbad, CA). Highly specific primers were designed using Primer3, a Web-based primer design program. Primers were designed to span exon-exon junctions to prevent amplification of genomic DNA. The cycling parameters for qPCR amplification reactions were: AmpliTaq activation at 95 °C for 10 min, denaturation at 95 °C for 15 s, and annealing/extension at 60 °C for 1 min (40 cycles). Triplicate Ct values were analyzed with Microsoft Excel using the comparative Ct (ΔΔCt) method as described by the manufacturer (Applied Biosystems). The transcript amount (2−ΔΔCt) was obtained by normalizing to an endogenous reference (18S rRNA) relative to a calibrator (one experimental sample). Samples with a high starting copy number showed an increase in the fluorescence early in the PCR process resulting in a low Ct number, whereas a lower starting copy number resulted in a higher Ct number.
Western Immunoblotting and Fluorescence Microscopy
For protein analysis from retinas, mouse eyes were enucleated and retinas were carefully dissected and homogenized in lysis buffer containing 10 mm NaF, 300 mm NaCl, 50 mm Tris, pH 7.4, 1% Triton X-100, 10% glycerol, and 1 mm EDTA with a 1% volume of phosphatase and protease inhibitor mixture. Protein samples (50 μg) were fractioned in a 10% SDS-polyacrylamide gel, transferred to nitrocellulose membrane, and Western blot analysis was performed with each of the indicated primary antibodies. Immunodetection was performed using enhanced chemiluminescence (ECL) from Pierce. Protein bands were quantified by densitometric scanning. To analyze proteins from cell cultures, cells were homogenized in lysis buffer fractioned by electrophoresis and analyzed as described above.
For immunohistochemical analyses, eyes were fixed in 4% paraformaldehyde for 2 h followed by an overnight incubation in sucrose solution. A set of eyes were frozen in Tissue-Tek Optimal Cutting temperature compound and 10–30-μm thick cryostat sections were prepared. Retinas were dissected from another set of eyes and laid flat on SuperFrost® Plus-coated slides to obtain whole mount preparations. Sections and flat mount preparations were then permeabilized in 0.1% Triton X-100 at room temperature for 20 min and incubated for 24 h with the indicated primary antibody. Immunodetection was performed with either rhodamine- or fluorescein-conjugated anti-rabbit IgG. Retina mounts and sections were washed several times in PBS between incubations.
To localize proteins in cultured cells, cells were fixed in paraformaldehyde for 10 min and permeabilized in 0.1% Triton X-100 at room temperature for 5 min. Cells were then incubated with the indicated primary antibodies overnight at 4 °C and treated with TRITC- or FITC-conjugated secondary antibodies. Images were acquired using Bio-Rad 1024 MDC laser scanning fluorescence imaging system.
Pathway-specific Microarray Gene Expression Profiling
Expression of the Wnt pathway genes was performed using the real time PCR-based RT2 Profiler PCR Array from SABiosciences (Frederick, MD). The array used analyzed 84 genes related to the Wnt-mediated signal transduction pathway. Side-by-side amplification experiments determine differential gene expression between the two samples. First, total RNA from Lnv-Luc- and Lnv-CCN1-infected cells was subjected to DNase digestion to eliminate the genomic DNA contaminant and used as a template for reverse transcription and real time PCR. The cDNA template was then mixed with the ready-to-use RT2 qPCR Master Mix. The mixture was aliquoted into each well of the same plate containing pre-dispensed gene-specific primer sets. Real time PCR was performed and amplification data were collected using ABI software. The relative gene expression was determined using the ΔΔCT method. The threshold value was set by using the Log View of the amplification plots and placing it above the background signal but within the lower one-third to one-half of the linear phase of the amplification plot.
Statistical Analyses
Data were expressed as mean ± S.E. To test differences among several means for significance, a one-way analysis of variance with the Newman-Keuls multiple comparison test was used. Where appropriate, post hoc unpaired t test was used to compare two means/groups, and p values < 0.05 or <0.01 were considered significant. Statistical analyses were performed using the Prism software for Windows version 4 from GraphPad Inc. (San Diego, CA).
RESULTS
Regulation of CCN1 Gene Expression in the Retinal Vasculature during Development and in Response to Hyperoxia
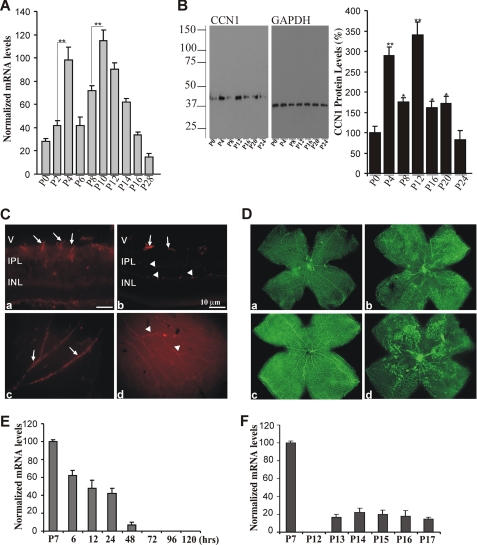
Expression of the CCN1 gene is limited to developmental and pathological events only (12). Because development of mouse retinal vessels occurs during the first 2–3 weeks after birth, we examined the expression pattern and localization of CCN1 in the developing mouse retina. As shown in Fig. 1A, CCN1 mRNA levels peaked between P2 and P4, when the budding of superficial vessel begins and between P8 and P12 when the secondary deep layer of the retinal vasculature starts extending and growing radially outward toward the inner nuclear and outer plexiform layers. The CCN1 mRNA levels then progressively declined and became barely detectable as the adult vasculature was established. These changes correlated well with those seen at the protein level (Fig. 1B). Immunoreactivity to the CCN1 protein at P4 appeared in superficial retinal vessels, which grow radially from the optic nerve, and in the deeper retinal vasculature, which forms during the second week (P10) through branching of the superficial layer (Fig. 1C). The protein localized primarily in large and moderately sized vessels. There was no evidence of association of CCN1 with the microvasculature.
FIGURE 1.
Expression profile of the CCN1 gene during normal retinal vessel development and in response to OIR. A, room air mice pups were raised under normal light and temperature conditions and sacrificed at the indicated time periods. Retinas were dissected and used for quantitative real time PCR analysis. CCN1 mRNA levels were normalized to those of 18S rRNA. **, p < 0.001 (n = 4). B, CCN1 protein expression in retinal tissue homogenates was detected by Western blot with an anti-CCN1 antibody. The same blot was probed with an anti-GAPDH antibody to control for total protein loading. Densitometric values of the CCN1 protein band (39-kDa) were normalized to those of GAPDH. Data are mean ± S.E. of three experiments. **, p < 0.01 versus P0. *, p < 0.05 versus P4 or P12. C, immunohistochemical localization of the CCN1 protein in cross-sections (a and b) and flat mount preparations (c and d) of retinas at P4 (a and c) and P10 (b and d). All retinal preparations were fixed in 4% paraformaldehyde and permeabilized prior to immunostaining with anti-CCN1 antibody. Arrows and arrowheads indicate immunoreactivity to CCN1 in the superficial and deeper retinal vessels, respectively. V, vitreous; IPL, inner plexiform layer; INL, inner nuclear layer. Bars, 10 μm (a and b). Magnification, ×20 (c and d). D, representative flat mount preparations of retinas from mice pups subjected to OIR. Exposure of mice pups to hyperoxia (from day P7 to P12) led to vessel obliteration in the central retina (b) as compared with normaxia (a). Upon return to room air, preretinal neovascular tufts formed in the central retina at P17 (d) as compared with normal retinal vasculature in eyes under normaxia at the same age (c). Magnification, ×4. E and F, temporal expression of CCN1 mRNA during the hyperoxic (P7 to P12) and ischemic (P12 to P17) phases of OIR as determined by real time PCR. CCN1 mRNA levels were normalized to those of 18S rRNA. The values represent the average determinations from tissue samples obtained from 4 animals each measured in triplicate.
The expression pattern of CCN1 was also examined in the mouse model of OIR. In this model, hyperoxia-induced regression of superficial vessels along the arteries in the central retina and development of the deep plexus was suppressed (Fig. 1D). Upon return to room air for 5 days, neovascular tufts can be seen in the midperiphery halfway between the central retina and the more vascularized peripheral retina. Remarkably, CCN1 mRNA levels were significantly reduced after 6 h of hyperoxia, reached basal levels after 24 h, and remained low throughout the 5-day course of hyperoxia (Fig. 1E). CCN1 mRNA levels were not significantly increased during the subsequent hypoxic phase associated with both vessel regrowth and retinal neovascularization (Fig. 1F). Similarly, the CCN1 protein was undetectable in ischemic retina (at P17) and most neovascular tufts examined in retinal whole mounts did not express the CCN1 protein (data not shown).
Effect of CCN1 on Hyperoxia-induced Retinal Vessel Obliteration and Neovessel Formation
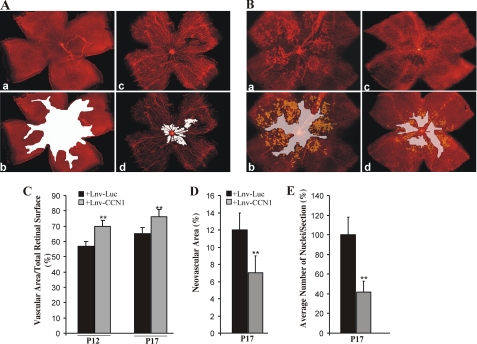
To determine whether ectopic expression of the CCN1 gene affects hyperoxia-induced vaso-obliteration and recovery, we first performed intravitreal injection of a lentiviral vector expressing CCN1 (Lnv-CCN1) in mice pups subjected to OIR. Injection of the control vector, Lnv-Luc, and measurement of luciferase activity in retinal homogenates demonstrated a 3-fold increase of luciferase activity as early as 24 h after injection suggesting an efficient gene transfer into retinal tissue (data not shown). At P12, an extensive vaso-obliteration was clearly evident in retinas from Lnv-Luc-injected control eyes, whereas retinas from Lnv-CCN1-injected eyes showed normally formed blood vessels over a considerably wider area of the central region (Fig. 2A). Quantitative analyses showed that more than 70% of the retinal surface was vascularized in Lnv-CCN1-injected eyes but less than 60% was vascularized in Lnv-Luc-injected eyes (Fig. 2C). At P17, the retinal vasculature in eyes injected with Lnv-CCN1 exhibited normal morphology and branching in the midperipheral and peripheral regions except for spares areas around the central retina (Fig. 2B). Quantitative analyses showed that retinal vessels extended over 68 and 78% of the retinal surface in Lnv-Luc- and Lnv-CCN1-injected eyes, respectively (Fig. 2C). Preretinal neovascular tufts, which can be seen abundantly in the central and midperipheral retina in Lnv-Luc-injected eyes, were minimally present in eyes injected with the Lnv-CCN1 vector (Fig. 2D). Similarly, the number of preretinal nuclei significantly decreased in eyes injected with CCN1 vector as compared with those injected with a control vector (Fig. 2E).
FIGURE 2.
Effects of lentivirus-mediated expression of CCN1 on vaso-obliteration and retinal neovascularization following OIR. A and B, representative flat mount preparations of rhodamine-UEA-1-stained retinas from OIR eyes at P12 (A) and P17 (B) injected with either Lnv-Luc (a) or Lnv-CCN1 (c). Areas of vaso-obliteration and preretinal neovascular tufts as determined by computer-assisted image analyses are shown in white and yellow, respectively (b for a and d for c). Magnification, ×4. C and D, compiled data showing percentage of vascularized and neovascular tuft areas in Lnv-Luc- and Lnv-CCN1-injected eyes. **, p < 0.01 versus Lnv-Luc (n = 6–8). E, extent of neovascularization as determined by counting the number of nuclei in random mid-peripheral areas of retinas. **, p < 0.05 (n = 4).
Because HSCs play a critical role in the repair process of the vasculature, we examined whether HSCs transduced in vitro with Lnv-CCN1, injected intravitreally between P2 and P4, modulate the effects of hyperoxia and ischemia on the retinal vasculature. At P12, less than 15% of the retinal surface was vaso-obliterated upon injection of Lnv-CCN1-transduced HSCs, whereas more that 30% of the retinal surface remained avascular upon injection of HSCs transduced with Lnv-Luc (Fig. 3, A and C). Clearly, Lnv-CCN1-transduced HSCs provided a greater vascular protection against hyperoxia. At P17, there was clear evidence of reduced avascular areas (<5%) in the central retina upon injection of Lnv-CCN1-transduced HSCs (Fig. 3, B and C). Similarly, Lnv-CCN1-transduced HSCs induced a significant reduction of preretinal neovascular tufts that were minimally present and in many cases undetectable as compared with eyes injected with Lnv-Luc-transduced HSCs (Fig. 3D). Interestingly, immunohistochemical analyses showed that immunoreactivity to CCN1 was localized in sparse areas in the superficial retinal layer and within and around blood vessel walls (Fig. 3E). Such a diffused localization is likely due to the extracellular secretion of the CCN1 protein by the intravitreally injected Lnv-CCN1-transduced HSCs. The precise localization of HSCs within the retina was further examined upon injection of HSCs co-transduced with both Lnv-CCN1 and Lnv-GFP vectors to facilitate tracing of cell fate in vivo. As shown in Fig. 3F, fluorescently labeled cells were localized in the retinal vessel wall in the superficial layer and secondary plexus. Flat mount preparations of the retina further confirmed that HSC localization was confined to the inner wall of medium and small size vessels (Fig. 3G).
FIGURE 3.
Effects of CCN1-primed HSCs on vaso-obliteration and preretinal neovascularization following OIR. A and B, representative flat mount preparations of rhodamine-UEA-1-stained retinas from OIR eyes at P12 (A) and P17 (B). Retinas were from eyes injected with HSCs transduced with either the Lnv-Luc (a) or Lnv-CCN1 (c) vector. Areas of vaso-obliteration and preretinal neovascular tufts are shown in white and yellow, respectively (b for a and d for c). Magnification, ×4. C and D, compiled data showing percentage of vascularized and neovascular tuft areas in eyes injected with HSCs transduced with either Lnv-Luc or Lnv-CCN1 vector. **, p < 0.01 versus +Lnv-Luc-HSCs (n = 6). E, immunolocalization of CCN1 and CD31, a marker of retinal vessels, in a retinal cross-section at P17 after intravitreal injection of Lnv-CCN1-transduced HSCs at P4 and OIR at P7. DAPI shows nuclear staining. IPL, inner plexiform layer; ONL, outer nuclear layer. Bars, 20 μm. F, autofluorescence of a retinal cross-section at P17 after intravitreal injection of Lnv-CCN1- and Lnv-GFP-transduced HSCs at P4 and OIR at P7. Co-transduction with Lnv-GFP facilitates cell fate tracing in vivo. Arrows indicate areas in the blood vessel wall where labeled HSCs were integrated. V, vitreous. G, localization at P17 of HSCs within a blood vessel wall in a flat mount preparation of a retina from an eye injected with Lnv-CCN1- and Lnv-GFP-transduced HSCs at P4 and subjected to OIR at P7. Rhodamine UEA-1-stained retina whole mount is shown in a. Retinal autofluorescence carried by Lnv-GFP-transduced HSCs is shown in b. Merged image of a and b is shown in c. Magnification, ×40.
Effects of CCN1 Protein on Retinal Endothelial Cells and Their Progenitors
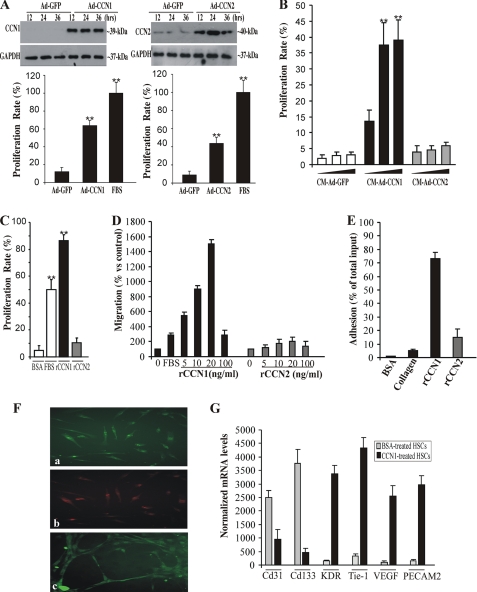
We determined what cellular and molecular effects CCN1 exerts on retinal endothelial cells and their progenitors. We first examined the effects of CCN1 on the proliferation rate of RECs and HSCs. As shown in Fig. 4A, ectopic expression of CCN1 in RECs via adenovirus gene transfer increased by 65% their proliferation rate as compared with a 100% increase by FBS. Ectopic expression of CCN2, a protein structurally related to CCN1, induced a 45% increase of the proliferation rate of RECs as well. This is consistent with the previously reported proangiogenic activity of CCN1 and CCN2 in vivo (26). Both CCN1 and CCN2 are secreted in the medium and their ectopic expression in RECs via adenoviruses gene transfer ensures proper folding of the full-length protein. Therefore, we examined the effects of conditioned media (CM) from RECs transduced with either Ad-CCN1 or Ad-CCN2 on HSCs. Fig. 4B shows that CM-Ad-CCN1 stimulated HSC growth in a dose-dependent manner, whereas CM-Ad-CCN2 had no significant effect on their proliferation rate. Concordantly, incubation with rCCN1 increased the proliferation rate of HSCs, whereas incubation with either rCCN2 or BSA had no effect (Fig. 4C).
FIGURE 4.
Effects of CCN1 on RECs and their progenitors. A, RECs were transduced with adenoviral vector expressing GFP (Ad-GFP), CCN1 (Ad-CCN1), or CCN2 (Ad-CCN2) for the indicated periods of time. A representative autoradiogram of the ectopically expressed CCN1 and CCN2 proteins in the medium as determined by Western blot and immunodetection analyses is shown. The proliferation rate of the cells cultured under these conditions was measured as described under “Experimental Procedures.” B, effects of CCN1 and CCN2 on the growth of HSCs. HSCs were incubated with increasing concentrations of CM-Ad-GFP, CM-Ad-CCN1, or CM-Ad-CCN2. **, p < 0.001 versus CM-Ad-GFP of three separate experiments. C, proliferation rate of HSCs upon their exposure to either rCCN1 or rCCN2. Data are the mean ± S.E. of three separate experiments. **, p < 0.001 versus BSA. D, chemotactic activity of rCCN1 vis à vis HSCs. HSC migration was analyzed by chemotaxis assay as described under “Experimental Procedures.” The data shown are the mean ± S.E. of three determinations. The experiments were repeated twice with similar results. E, rCCN1-induced HSC adhesion. HSCs were plated in microtiter plates precoated with BSA (1%), type I collagen (1 μg/ml), rCCN1 (1 μg/ml), or CCN2 (1 μg/ml). Data shown are the mean ± S.E. of three determinations. F, rCCN1-induced HSC differentiation into endothelial cells. HSCs were plated in CCN1-coated wells. Adherent cells were stained with either rhodamine-labeled UEA-1 (a), DiI-acLDL (b), or an anti-von Willebrand factor antibody (c). Magnification, ×40. G, steady state mRNA levels of Cd31, Cd133, KDR, Tie-1, VEGF, and PECAM2 after incubation with rCCN1. Data are the mean ± S.E. of three measurements, each performed in triplicate.
The CCN1 protein functions primarily through direct binding to integrin receptors and heparan sulfate proteoglycans, thereby activating multiple signaling molecules (e.g. focal adhesion kinases and mitogen-activated protein (MAP) kinases). These activities culminate in the regulation of cell migration, adhesion, proliferation, gene expression, differentiation, and survival. In agreement with this background information, we found that phospho-Erk1/2, phospho-JNK1/2, and Pyk2 levels were increased upon incubation of HSCs with rCCN1 (supplemental Fig. S1A). Pyk2 is a focal adhesion kinase-like factor commonly involved in integrin signaling. The p38 MAP kinase was not phosphorylated in response to rCCN1 stimulation (data not shown). Therefore, we examined the effects of rCCN1 on HSC migration and adhesion. As shown in Fig. 4D, rCCN1 enhanced transwell migration of HSCs in a dose-dependent manner. The response of HSCs to rCCN1 at a concentration of 20 ng/ml was more than 5 times higher than that of 10% FBS. HSCs were responsive even to the lowest rCCN1 concentration of 5 ng/ml but a concentration of 100 ng/ml was less effective in promoting cell migration leading to a bell-shaped curve characteristic of most chemoattractant factors. Similarly, incubation of HSCs in rCCN1-coated wells significantly increased their adhesion (Fig. 4E). We further established that the effects of CCN1 on HSC adhesion and migration were mediated through the integrin pathway and that heparin-binding activity is required for cell adhesion as well (supplemental Fig. S1, B and C). Exposure to rCCN1 increased the expression of α4, αv, and α5 integrin chain subunits, whereas that of αM and β3 chains was down-regulated (supplemental Fig. S1D). Using function blocking antibodies, we found that CCN1-induced adhesion is mediated through both integrin α4β1 and HSPGs and that α4β1 and α6β1 integrins were required for cell migration (supplemental Fig. S1, E and F). Furthermore, more than 90% of rCCN1-treated cells were positively stained with either UEA-1 or DiI-acLDL, a characteristic feature of cells in the endothelial lineage (Fig. 4F). CCN1-treated HSCs aligned and connected into cord-like structures forming hollow lumens, a typical feature of the early stages of capillary morphogenesis. Conversely, rCCN2 induced neither migration nor adhesion and differentiation of HSCs (Fig. 4, D and E), suggesting specificity of the effects of CCN1 on HSCs. The expression pattern of several marker genes confirmed the endothelial cell lineage features of CCN1-treated HSCs. As shown in Fig. 4G, stem cell markers including CD31 and CD133 were minimally expressed, whereas endothelial cell marker genes like the KDR, Tie-1, VEGF-A, and PECAM-2 were highly expressed in CCN-1-treated HSCs. Markers of smooth muscle cell lineage such as smooth muscle α-actin and caldesmon were expressed, although minimally in rCCN1-treated cells (data not shown), suggesting that a minor fraction of attached cells may exhibit a smooth muscle-like phenotype.
Role of the Wnt Signaling Pathway in CCN1 Regulation of HSC Differentiation
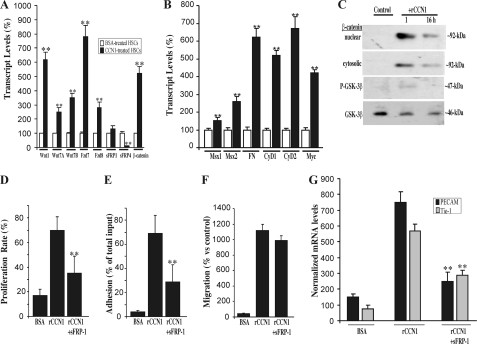
Increasing evidence suggests that the Wnt signaling pathway, which plays an important role in expansion of vascular progenitor populations and vascular cell differentiation, contributes to the activity of CCN1 (27, 28). To investigate the potential regulation of the Wnt pathway-related genes by CCN1, we used a real time PCR-based array to examine the expression pattern of Wnt molecules as well as a focused panel of genes related to Wnt-mediated signal transduction in rCCN1-treated and non-treated HSCs. An exhaustive list of genes differentially expressed in treated and non-treated cells is shown in Table 1. Interestingly, several components of the Frizzled-1 and -2 and Wnt receptor signaling pathways, such as DVL1, DVL2, Fzd-7, and Fzd-8, were strongly up-regulated in rCCN1-treated cells. In contrast, secreted frizzled-related protein (sFRP)-1 and sFRP-4, which are naturally occurring soluble decoy receptors that prevent binding of Wnt proteins to the Fzd/lipoprotein receptor-related protein receptor complex, were down-regulated in rCCN1-treated cells. The expression of selected Wnt signaling pathway genes was further independently validated by real time PCR. As shown in Fig. 5A, Wnt1 and Wnt7A of the Fzd2 signaling pathway were up-regulated in rCCN1-treated HSCs. The mRNA levels of Fzd7 and Fzd8 increased 5- and 4-fold, respectively, whereas those of sFRP-4 decreased below basal levels in rCCN1-treated cells, which corroborated the microarray data. Similarly, expression of downstream gene targets of the Wnt pathway such as Msx1, Msx2, fibronectin, cyclin D1, cyclin D2, and Myc were strongly up-regulated in rCCN1-treated cells (Fig. 5B).
TABLE 1.
Pathway-focused expression profiling of the Wnt signaling pathway genes in rCCN1-treated HSCs
| Pathway/gene name | Unigene | GenBankTM | Description | Fold-change |
|---|---|---|---|---|
| Frizzled-2 signaling pathway | ||||
| WNT1 | Hs.248164 | NM_005430 | Wingless-type MMTV integration site 1 family, member 1 | 5.62 |
| WNT5A | Hs.696364 | NM_003392 | Wingless-type MMTV integration site family 5A | 1.83 |
| WNT7A | Hs.72290 | NM_004625 | Wingless-type MMTV integration site family 7A | 1.83 |
| WNT7B | Hs.512714 | NM_058238 | Wingless-type MMTV integration site family 7B | 3.68 |
| Wnt receptor signaling pathway | ||||
| NLK | Hs.208759 | NM_016231 | Nemo-like kinase | 1.96 |
| SENP2 | Hs.401388 | NM_021627 | SUMO1/sentrin/SMT3 specific peptidase 2 | 6.45 |
| TCF7L1 | Hs.516297 | NM_031283 | Transcription factor 7-like 1 (T-cell specific, HMG-box) | 2.20 |
| Protein modification activities | ||||
| FBXW2 | Hs.494985 | NM_012164 | F-box and WD repeat domain containing 2 | 43.71 |
| WIF1 | Hs.284122 | NM_007191 | WNT inhibitory factor 1 | 16.11 |
| Frizzled-1 signaling pathway | ||||
| DVL1 | Hs.74375 | NM_004421 | Dishevelled, dsh homolog1(Drosophila) | 2.27 |
| DVL2 | Hs.118640 | NM_004422 | Dishevelled, dsh homolog 2 (Drosophila) | 4.96 |
| FZD7 | Hs.173859 | NM_003507 | Frizzled homolog 7 (Drosophila) | 6.82 |
| FZD8 | Hs.302634 | NM_031866 | Frizzled homolog 8 (Drosophila) | 3.68 |
| Regulation of growth and proliferation | ||||
| TCF7L1 | Hs.516297 | NM_031283 | Transcription factor 7-like 1 (T-cell specific, HMG-box) | 2.20 |
| SRFP1 | Hs.713546 | NM_003012 | Secreted frizzled- related protein 1 | 1.32 |
| SRFP4 | Hs.658169 | NM_003014 | Secreted frizzled-related protein 4 | −28.70 |
| Regulation of cell cycle | ||||
| CCND1 | Hs.523852 | NM_053056 | Cyclin D1 | 9.51 |
| CCND2 CCND3 | Hs.376071Hs.534307 | NM_001760 | Cyclin D3 | 5.17 |
| PPP2CA | Hs.483408 | NM_002715 | Protein phosphatase 2 (formerly 2A), catalytic subunit α | −3.41 |
| Regulation of transcription | ||||
| CTNNB1 | Hs.476018 | NM_001904 | Catenin (cadherin- associated protein), β1 | 6.77 |
| PITX2 | Hs.643588 | NM_000325 | Paired-like homeodomain 2 | −5.66 |
| SOX17 | Hs.98367 | NM_022454 | SRY (sex determining region Y)-box 17 | −8.22 |
| TCF7L1 | Hs.516297 | NM_031283 | Transcription factor 7-like 1(T-cell specific, HMG-box) | 2.20 |
| TLE1 | Hs.197320 | NM_005077 | Transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) | 2.87 |
FIGURE 5.
Involvement of the Wnt signaling pathway in CCN1-induced HSC differentiation. A, HSCs were incubated with either BSA or rCCN-1. Gene expression of Wnt1, Wnt7A, Wnt7B, Fzd7, Fzd8, sFRP-1, sFRP-4, and β-catenin was quantified by real time PCR. The data shown are the mean ± S.E. of three determinations each performed in triplicate. RNA levels in BSA-treated cells were set to 100% to facilitate comparison among experiments. **, p < 0.05 versus BSA-treated HSCs. B, the expression of gene targets of the Wnt pathway including Msx1, Msx2, fibronectin (FN), cyclin D1 (CyD1), cyclin D2 (CyD2), and Myc was measured by real time PCR. Data shown are the mean ± S.E. of three determinations each performed in triplicate. **, p < 0.05 versus BSA-treated HSCs. C, cell lysates from rCCN1-treated cells were analyzed by Western immunoblotting using antibodies against β-catenin, phospho (P)-GSK-3β, and GSK-3β. A representative profile is shown. Both cytoplasmic and nuclear fraction contents of β-catenin were analyzed. D, effects of sFRP-1 on HSC behavior. Cells were incubated for 24 h in rCCN1-coated plates in the presence and absence of sFRP-1 (10 μg/ml) and their proliferation rate was measured. **, p < 0.05 versus rCCN1. E, cells were incubated in rCCN1-coated wells with and without sFRP-1 (10 μg/ml). The effect on cell adhesion was determined. **, p < 0.05 versus rCCN1. F, migration of cells exposed to rCCN1 and sFRP-1 was determined using chemotaxis assay. Data shown are the mean ± S.E. of three determinations. G, cells were incubated with rCCN1 in the presence and absence of sFRP-1 (10 μg/ml) and the steady state mRNA levels of PECAM2 and Tie-1 were determined by real time PCR. The mRNA levels were normalized to those of 18S rRNA. Data are the mean ± S.E. of three determinations each performed in triplicate. **, p < 0.01 versus rCCN1.
Functionally, Wnt proteins bind to their Fzd receptors, thereby preventing proteosomal degradation of the Wnt mediator, β-catenin. Subsequently, β-catenin translocates to the nucleus where it forms an active transcription complex with transcription factors downstream of the Wnt pathway (29). In the absence of Wnt ligands, cytosolic β-catenin is phosphorylated by a protein complex containing GSK-3β and is degraded rapidly via the proteosomal pathway. As shown in Fig. 5C, increased levels of β-catenin were detected in nuclear fractions of rCCN1-treated HSCs as compared with non-treated cells. Additionally, GSK-3β was only weakly activated in rCCN1-treated cells indicating enhanced Wnt activity in these cells.
To determine whether inhibition of Wnt signaling interferes with CCN1 effects on HSC differentiation into the endothelial lineage, cells were incubated with rCCN1 in the presence and absence of exogenous sFRP-1, which blocks Wnt signaling by interfering with Wnt ligand binding to their receptors. As shown in Fig. 5, D–F, sFRP-1 significantly reduced rCCN1-induced HSC proliferation and adhesion but had no effect on HSC migration. No evidence of cell death was observed when HSCs were exposed to sFRP-1 (data not shown). Moreover, sFRP-1 significantly reduced the expression of molecular markers of endothelial cell lineage such as in Tie-1, and PECAM-2 in rCCN1-treated HSCs (Fig. 5G).
Intravitreal Injection of sFRP-1 Compromises CCN1-induced Retinal Vessel Normalization in OIR Mice
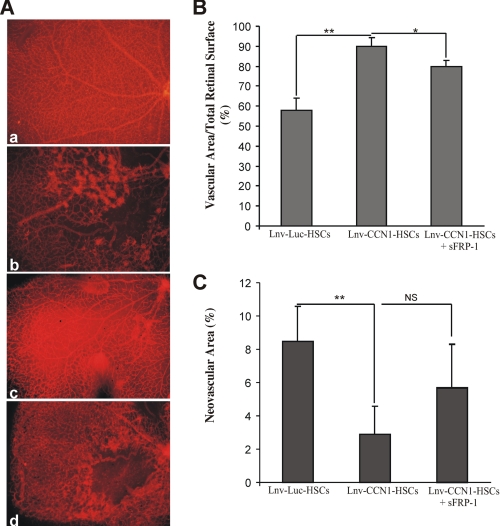
The involvement of CCN1-induced Wnt signaling in retinal vessel normalization was examined in OIR mice through intravitreal injection of sFRP-1. As shown in Fig. 6, injection of Lnv-CCN1-transduced HSCs between P2 and P4 increased resistance to retinopathy at P17. Conversely, retinas from mice co-injected with Lnv-CCN1-transduced HSCs at P2 and sFRP-1 at P7 showed persistence of areas of vaso-obliteration. Such areas were often confined to specific portions of the retina, possibly where sFRP-1 was concentrated the most. In most sFRP-1-treated eyes, neovascular tufts formed but were rare in sFRP-1-untreated eyes. Similar results were observed in retinas from eyes co-injected with Lnv-CCN1 and sFRP-1 (data not shown).
FIGURE 6.
The activity CCN1-primed HSCs during OIR is dependent on the Wnt signaling pathway. A, representative retina quadrants at P17 from a normal untreated eye (a) and OIR eyes injected with Lnv-Luc-transduced HSCs (b), Lnv-CCN1-transduced HSCs (c), and Lnv-CCN1-transduced HSCs at P2 and sFRP-1 at P7 (d). Magnification, ×10. B and C, compiled data showing percentage of vascularized and neovascular tuft areas in retinas from the eyes subjected to treatments described in A. **, p < 0.05 (n = 5).
DISCUSSION
The present study identified previously unrecognized functions of the matricellular CCN1 to induce either directly or more efficiently via HSCs, a rapid development of stable retinal blood vessels following vaso-obliteration in the OIR model. CCN1 activities reduced ischemia and subsequently, suppressed preretinal neovascularization.
Several new findings emerged through examination of the vascular implications of CCN1 expression during development and in response to hyperoxic injury. First, our data showed a close correlation in topography and timing between CCN1 gene expression and retinal vessel growth and spacial distribution during development of the retinal vasculature. The expression of the CCN1 gene increased concomitantly with blood vessel budding. This suggests that CCN1 functions as a vasoformative molecule that promotes the proper formation and stabilization of blood vessels and/or affects vascular precursor cell function and behavior. Because CCN1 expression is transient during normal retinal vessel development, we speculate that it may be regulated by the function of the newly formed vessels in transporting blood-borne oxygen, although further studies are needed to test this hypothesis. Furthermore, hyperoxia-induced vaso-obliteration is associated with a concomitant decrease of CCN1 gene expression, which plummeted below normal levels within 24 h after the onset of hyperoxia suggesting, (i) a direct inhibitory effect of CCN1 gene expression by hyperoxia, and (ii) a cause-and-effect relationship between CCN1 levels and growth of endothelial cells and their precursors within the retina. These hypotheses are supported by three observations: 1) hyperoxia causes developing retinal blood vessels to stop growing and retinal capillaries to drop-out in the central retina as a result of a decreased proliferation and migration of both endothelial cells and their precursors (30), processes that are potently stimulated by CCN1 as shown in Fig. 1. 2) Hyperoxia-induced vaso-obliteration is triggered, at least in part, by oxidative stress-induced endothelial cell apoptosis (31, 32), whereas CCN1 is known to elicit a protective effect against oxidative stress (33, 34). However, contribution of cell apoptosis to hyperoxia-induced vaso-obliteration was reported to be much lower than the decrease in cell number due to inhibition of proliferation. Thus, the antioxidative effects of CCN1 may not play a major role in this process. 3) Hyperoxia was reported to inhibit key signaling events by inducing disassembly of the cell cytoskeleton (30, 35), a major event commonly known to modulate CCN1 gene expression (14, 36). The cytoskeleton of endothelial progenitors, in particular, is more susceptible to the damaging effects of hyperoxia than that of endothelial cells and this contributes largely to retinal vaso-obliteration. Thus, cytoskeletally based mechanisms cause both hyperoxia-induced endothelial progenitor damage and repression of the CCN1 gene. Meanwhile, hyperoxia represses the CCN1 gene and further decreases its responsiveness to hypoxia even though the relative ischemia after hyperoxia-induced vaso-obliteration is commonly known to stimulate proangiogenic factor synthesis (e.g. oxygen-dependent VEGF and oxygen-independent insulin-like growth factor-I). Such fluctuations of angiogenic gene expression between hyperoxia and hypoxia are considered to be important risk factors for the development of ROP in humans.
Ectopic expression of the CCN1 gene before the onset of hyperoxia resulted in a reduced vascular obliteration during hyperoxic injury. As such, the presence of CCN1 is protective to resident cells against the damaging effects of hyperoxia including vascular progenitor cells, which have been reported to localize in the retina based on the identification of CXCR4 and ADPase positive cells (37). Other vascular progenitor cells may be recruited from within the circulating blood. We contend that CCN1 exerts chemotactic effects by stimulating HSC migration and recruitment from the circulation into avascular areas, thus facilitating vascular repair. Our in vitro data, shown in Fig. 4, support this hypothesis because CCN1 exerted, not only a mitogenic effect on retinal endothelial cells, but it also promoted migration, tube formation, and differentiation of HSCs into endothelial cells. These effects are further supported by previously reported data in other model systems. In particular, Shintani et al. (38) have shown that the levels of CCN1 protein correlated well with increased circulating HSCs in response to cardiac ischemia further supporting a role of CCN1 as a chemoattractant factor vis à vis HSCs in vivo. Similarly, Estrada et al. (39) have shown that CCN1 was secreted by mesenchymal stem cells, which differentiate into endothelial cells as well and integrate into nascent blood vessels in vivo. Immunodepletion of CCN1 abrogated the angiogenesis-promoting capability of mesenchymal stem cell secretome, whereas addition of human recombinant CCN1 to the depleted secretome restored their angiogenic activity both in vitro and in vivo.
Interestingly, when HSCs were used as delivery vehicle of CCN1, an even greater protective and regenerative effect of normal retinal vasculature was observed. CCN1-primed HSCs exerted their function using mainly two strategies: 1) differentiating into mature endothelial cells that integrate into the vessels, and 2) producing new intermediates that act in an autocrine/paracrine fashion to provide additional protection against hyperoxia and resistance to ischemia.
First, it is well established that HSCs have a long-term capacity for multilineage differentiation including the ability to give rise to functional endothelial cells, after either a single-cell transplantation or serial transplantations (8). However, their effectiveness in the repair process relies on their activation state. Concordantly, unstimulated HSCs were less effective that CCN1-primed HSCs in the normalization of the retinal vasculture in OIR mice. Previous studies have shown that unstimulated HSCs were, indeed unable to stimulate angiogenesis upon their transplantation in a Matrigel plug subcutaneously inserted in mice (40). Recently identified factors involved in the process of HSC commitment to the endothelial lineage include the nitric oxide pathway (41), Jagged-1-dependent Notch signaling (42), and the Lnk adaptor protein associated with the SCF-c-Kit axis. However, these molecules are ubiquitously and constitutively expressed and regulate multiple functions in various types of mature and immature cells. Unlike the latter molecules, CCN1 is functionally important primarily in developmental and pathological vascular events. Our finding that CCN1 controls endothelial lineage commitment of HSCs suggests its particular importance in developmental and adult vasculogenesis.
Second, differentiation of HSCs into fully differentiated endothelial cells is commonly viewed as a multistep process in which effector molecules bind to receptors, in turn activating, via a number of signaling pathways, key transcription factors that transactivate differentiation associated genes. These include structural (e.g. cytoskeletal and extracellular matrix proteins) and regulatory molecules (e.g. enzymes, channels, and transporters). These molecules activate other cell types and/or modules via positive or negative feedbacks. Herein, we showed that, as an effector molecule, CCN1 binds to integrin receptors (e.g. α4β1 and α6β1), activates signaling cascades involving focal adhesion kinases and MAP kinases, and stimulates the expression of paracrine factors of the Wnt signaling pathway and this contributes, at least in part to the vascular activities of CCN1. In total, 19 different Wnt proteins have been identified in both human and mouse genomes and their patterns of expression overlap both spatially and temporally during development, raising the possibility of functional redundancy among these proteins (28, 43). Remarkably, we found that a number of genes within the canonical Wnt signaling pathway are preferentially expressed in CCN1-treated HSCs. In particular, candidate components of the Frizzled-1 (e.g. DVL1, DVL2, Fzd7, and Fzd8) and Frizzled-2 (e.g. Wnt1, -5, and -7) signaling pathways are up-regulated. Moreover, negative transcriptional regulators of Wnt signaling such a SOX17 and naturally occurring soluble decoy receptors such as sFRP-1 and sFRP-4 were down-regulated. Thus, both positive and negative regulators of Wnt signaling were either similarly or inversely modulated suggesting that this signaling pathway requires tight control. The up-regulation of known Wnt target genes including Msx1, Msx2, fibronectin, cyclin D1, cyclin D2, and Myc in CCN1-primed HSCs is consistent with an enhanced Wnt activity in CCN1-treated HSCs. Functionally, Wnt ligands have been implicated in regulating angiogenesis in the placenta, gonads, and central nervous system in vivo (44, 45). In particular, Wnt5 and Wnt7b proteins regulate endothelial cell survival, proliferation, and gene expression (46). Wnt2-deficient mice exhibit defects in placental angiogenesis. Similarly, inactivation of the Wnt receptor Fzd5 during mouse development severely impairs yolk sac vascularization and causes intrauterine death (47). Other reports demonstrated that Wnt proteins have direct effects on endothelial cell differentiation from progenitor cells although the exact Wnt proteins involved and their respective roles have not been delineated (48). Additional studies of the candidates identified through our gene expression profiles will likely develop a clearer picture of the role of Wnts and their downstream targets in developmental and postnatal angiogenesis. Taken together with our data, these observations suggest that CCN1-induced Wnt activation is both necessary and sufficient for normal blood vessel formation and regeneration, at least in part through regulation of differentiation and proliferation of endothelial progenitors.
Another interesting outcome of our study design is that CCN1 delivery either directly or indirectly via HSCs, reduced the formation of abnormal preretinal neovessels. These effects are likely to be mediated through a paracrine mode of action on resident vascular cells because the contribution of endothelial progenitors to the formation of neovessels was shown to be very low (49). There are a number of possible explanations to account for the ability of CCN1 to reduce the formation of neovessels. First, the neovascular tufts, which form at the transition zone between vascular and avascular regions of the retina are due to hypoxia-induced up-regulation of VEGF resulting in an over-proliferation of endothelial cells (50). Interestingly, most neovascular tufts examined in retinal whole mounts did not express CCN1 (data not shown). Thus, it is plausible that CCN1 induces a rapid vascular regeneration of the central retina and such activity overrides that of VEGF on neovessel outgrowth. It is reasonable to speculate that if the equilibrium between CCN1 and VEGF is shifted toward a certain threshold ratio, CCN1 activity outweighs that of VEGF resulting in more physiological angiogenesis and vasculogenesis and less pathological angiogenesis. Indeed, previous studies from our group and others have shown that CCN1 stimulates an angiogenic gene program characteristic of earlier developmental stages, which is synonymous of normal blood vessel growth (51, 52). Other reports have shown that CCN1 improves angiogenesis and collateral blood flow in rabbit ischemic hind limb and rat cornea models to an even larger extent than VEGF (53, 54). This role of CCN1 does not preclude a direct physical and/or functional interaction between CCN1 and either VEGF or VEGF receptors to modulate angiogenesis in an antagonistic, additive, or synergistic manner. In particular, VEGF164 was shown to be required for pathological but not physiological angiogenesis (55). Not only is the expression of CCN1 and VEGF164 mutually regulated but also they may physically interact with one another. Indeed, CCN2, a protein structurally and functionally related to CCN1, directly interacts with VEGF164 both in vitro and in vivo via its modular domain, TSP1, which is highly conserved between CCN1 and CCN2 (56). Such interaction hinders VEGF164 binding to its receptor and reduced VEGF-mediated angiogenesis. Future studies in mice with conditional inactivation of CCN1 and/or VEGF164 will further address potential physical and functional interactions between CCN1 and VEGF isoforms.
Second, many lines of evidence indicate that macrophages presumably recruited from the circulation play a major role in vascular tuft regression (50, 57). Depending on the microenvironment, these cells are also capable of mediating endothelial cell death via initiation of caspase-mediated apoptosis, which prevents neovessel outgrowth (58). Interestingly, CCN1 regulates macrophage function during inflammation (33). Essentially, CCN1 supports macrophage recruitment and activation, induces NF-κB-dependent gene expression of multiple proinflammatory cytokines and chemokines (e.g. TNF-α, IL-1a, IL-6, and IL-12b), and further enables TNF-α to be cytotoxic without blocking NF-κB activity. The activity of TNF-α is particularly important in vascular tuft regression. TNF receptor-deficient mice exhibit a prolonged neovascular response in the model of OIR (59), with an associated decrease in endothelial cell apoptosis in the neovascular tufts. Other studies showed that apoptotic bodies derived from neovessel mediate release of cytokines and chemokines that facilitate progenitor cell recruitment, further supporting a role for TNF-α in vascular tuft regression (60). Additionally, CCN1 enhances the apoptotic activity of TRAIL and Fas ligand, which has been shown to be involved in endothelial cell death associated with vascular tuft regression (61). Thus, CCN1 induces a selective modulation of TNF-α activity that may translate into a more efficient and rapid vascular recovery. Current studies in our laboratory are comparing the vascular phenotype in mice with conditional inactivation of CCN1 and/or TNF-α to clarify the interaction between these factors in ischemic retinopathies.
Findings reported here highlight the influence of a novel inducible ECM protein, CCN1, on physiological and pathological angiogenesis in the retina. Our studies have shown that CCN1 has direct effects on HSC migration, adhesion, differentiation, and tube formation. These are critical processes required for vascular repair/rescue and adaptability to hyperoxia. CCN1 exerts a vaso-stabilizing effect as shown by its ability to reduce both hyperoxia-induced vaso-obliteration and pathological neovascularization associated with ischemia in the mouse model of OIR. We postulate that hyperoxia- and post-hyperoxia-induced decreases in CCN1 expression may deprive the retina of an important angiogenic/vasculogenic factor critical for the physiological response to hyperoxia and proper revascularization and repair after ischemic injury. Further understanding of CCN1 signaling pathways may hold promise as a strategy of rational therapeutic design to treat/prevent inadequate vascular repair and/or destructive angiogenesis.
Supplementary Material
Acknowledgments
We thank all members of the laboratory for technical assistance and critical discussion and review of this work. Our thanks to Dr. N. Schutze and his group for technical help with the preparation of rCCN1.
This work was supported, in whole or in part, by National Institutes of Health Grant EY019387-01A1 (to B. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- ROP
- retinopathy of prematurity
- CCN1/Cyr61
- cysteine-rich protein 61
- ECM
- extracellular matrix
- OIR
- oxygen-induced retinopathy
- HSC
- hematopoietic stem cell
- REC
- retinal endothelial cell
- Lnv
- lentivirus
- UEA-1
- U. europaeus agglutinin-1
- sFRP-1
- secreted frizzled-related protein-1
- CM
- conditioned medium
- TRITC
- tetramethylrhodamine isothiocyanate
- VEGF
- vascular endothelial growth factor
- P0
- postnatal day 0.
REFERENCES
- 1. Pollan C. (2009) Neonatal. Netw. 28, 93–101 [DOI] [PubMed] [Google Scholar]
- 2. Chang K. H., Chan-Ling T., McFarland E. L., Afzal A., Pan H., Baxter L. C., Shaw L. C., Caballero S., Sengupta N., Li Calzi S., Sullivan S. M., Grant M. B. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10595–10600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Connor K. M., SanGiovanni J. P., Lofqvist C., Aderman C. M., Chen J., Higuchi A., Hong S., Pravda E. A., Majchrzak S., Carper D., Hellstrom A., Kang J. X., Chew E. Y., Salem N., Jr., Serhan C. N., Smith L. E. (2007) Nat. Med. 13, 868–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Friedlander M., Dorrell M. I., Ritter M. R., Marchetti V., Moreno S. K., El-Kalay M., Bird A. C., Banin E., Aguilar E. (2007) Angiogenesis 10, 89–101 [DOI] [PubMed] [Google Scholar]
- 5. Penn J. S., Madan A., Caldwell R. B., Bartoli M., Caldwell R. W., Hartnett M. E. (2008) Prog. Retin. Eye Res. 27, 331–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Flynn J. T., Chan-Ling T. (2006) Am. J. Ophthalmol. 142, 46–59 [DOI] [PubMed] [Google Scholar]
- 7. Grant M. B., May W. S., Caballero S., Brown G. A., Guthrie S. M., Mames R. N., Byrne B. J., Vaught T., Spoerri P. E., Peck A. B., Scott E. W. (2002) Nat. Med. 8, 607–612 [DOI] [PubMed] [Google Scholar]
- 8. Asahara T. (2007) Yakugaku Zasshi 127, 841–845 [DOI] [PubMed] [Google Scholar]
- 9. Otani A., Slike B. M., Dorrell M. I., Hood J., Kinder K., Ewalt K. L., Cheresh D., Schimmel P., Friedlander M. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 178–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Asahara T. (2005) Ernst. Schering. Res. Found. Workshop 54, 111–129 [DOI] [PubMed] [Google Scholar]
- 11. Davis G. E., Senger D. R. (2005) Circ. Res. 97, 1093–1107 [DOI] [PubMed] [Google Scholar]
- 12. Chaqour B., Goppelt-Struebe M. (2006) FEBS J. 273, 3639–3649 [DOI] [PubMed] [Google Scholar]
- 13. Chen C. C., Lau L. F. (2009) Int. J. Biochem. Cell Biol. 41, 771–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hanna M., Liu H., Amir J., Sun Y., Morris S. W., Siddiqui M. A., Lau L. F., Chaqour B. (2009) J. Biol. Chem. 284, 23125–23136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Katsube K., Sakamoto K., Tamamura Y., Yamaguchi A. (2009) Dev. Growth Differ. 51, 55–67 [DOI] [PubMed] [Google Scholar]
- 16. Schütze N., Schenk R., Fiedler J., Mattes T., Jakob F., Brenner R. E. (2007) BMC Cell Biol. 8, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kireeva M. L., Latinkić B. V., Kolesnikova T. V., Chen C. C., Yang G. P., Abler A. S., Lau L. F. (1997) Exp. Cell Res. 233, 63–77 [DOI] [PubMed] [Google Scholar]
- 18. Mo F. E., Muntean A. G., Chen C. C., Stolz D. B., Watkins S. C., Lau L. F. (2002) Mol. Cell. Biol. 22, 8709–8720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bourillot P. Y., Aksoy I., Schreiber V., Wianny F., Schulz H., Hummel O., Hubner N., Savatier P. (2009) Stem Cells 27, 1760–1771 [DOI] [PubMed] [Google Scholar]
- 20. Liu H., Yang R., Tinner B., Choudhry A., Schutze N., Chaqour B. (2008) Endocrinology 149, 1666–1677 [DOI] [PubMed] [Google Scholar]
- 21. Perbal B. (2004) Lancet 363, 62–64 [DOI] [PubMed] [Google Scholar]
- 22. Francischetti I. M., Kotsyfakis M., Andersen J. F., Lukszo J. (2010) PLoS One 5, e9356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen C. C., Young J. L., Monzon R. I., Chen N., Todorović V., Lau L. F. (2007) EMBO J. 26, 1257–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schütze N., Kunzi-Rapp K., Wagemanns R., Nöth U., Jatzke S., Jakob F. (2005) Protein Expr. Purif. 42, 219–225 [DOI] [PubMed] [Google Scholar]
- 25. Smith L. E., Wesolowski E., McLellan A., Kostyk S. K., D'Amato R., Sullivan R., D'Amore P. A. (1994) Invest. Ophthalmol. Vis. Sci. 35, 101–111 [PubMed] [Google Scholar]
- 26. Suzuma K., Naruse K., Suzuma I., Takahara N., Ueki K., Aiello L. P., King G. L. (2000) J. Biol. Chem. 275, 40725–40731 [DOI] [PubMed] [Google Scholar]
- 27. Si W., Kang Q., Luu H. H., Park J. K., Luo Q., Song W. X., Jiang W., Luo X., Li X., Yin H., Montag A. G., Haydon R. C., He T. C. (2006) Mol. Cell. Biol. 26, 2955–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Franco C. A., Liebner S., Gerhardt H. (2009) Curr. Opin. Genet. Dev. 19, 476–483 [DOI] [PubMed] [Google Scholar]
- 29. Gordon M. D., Nusse R. (2006) J. Biol. Chem. 281, 22429–22433 [DOI] [PubMed] [Google Scholar]
- 30. Uno K., Merges C. A., Grebe R., Lutty G. A., Prow T. W. (2007) Dev. Dyn. 236, 981–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alon T., Hemo I., Itin A., Pe'er J., Stone J., Keshet E. (1995) Nat. Med. 1, 1024–1028 [DOI] [PubMed] [Google Scholar]
- 32. Zhang C., Rosenbaum D. M., Shaikh A. R., Li Q., Rosenbaum P. S., Pelham D. J., Roth S. (2002) Invest. Ophthalmol. Vis. Sci. 43, 3059–3066 [PubMed] [Google Scholar]
- 33. Bai T., Chen C. C., Lau L. F. (2010) J. Immunol. 184, 3223–3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yoshida Y., Togi K., Matsumae H., Nakashima Y., Kojima Y., Yamamoto H., Ono K., Nakamura T., Kita T., Tanaka M. (2007) Biochem. Biophys. Res. Commun. 355, 611–618 [DOI] [PubMed] [Google Scholar]
- 35. Brooks S. E., Gu X., Samuel S., Marcus D. M., Bartoli M., Huang P. L., Caldwell R. B. (2001) Invest. Ophthalmol. Vis. Sci. 42, 222–228 [PubMed] [Google Scholar]
- 36. Zhou D., Herrick D. J., Rosenbloom J., Chaqour B. (2005) J. Appl. Physiol. 98, 2344–2354 [DOI] [PubMed] [Google Scholar]
- 37. McLeod D. S., Hasegawa T., Prow T., Merges C., Lutty G. (2006) Dev. Dyn. 235, 3336–3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shintani S., Murohara T., Ikeda H., Ueno T., Sasaki K., Duan J., Imaizumi T. (2001) Circulation 103, 897–903 [DOI] [PubMed] [Google Scholar]
- 39. Estrada R., Li N., Sarojini H., An J., Lee M. J., Wang E. (2009) J. Cell. Physiol. 219, 563–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Grote K., Salguero G., Ballmaier M., Dangers M., Drexler H., Schieffer B. (2007) Blood 110, 877–885 [DOI] [PubMed] [Google Scholar]
- 41. Guthrie S. M., Curtis L. M., Mames R. N., Simon G. G., Grant M. B., Scott E. W. (2005) Blood 105, 1916–1922 [DOI] [PubMed] [Google Scholar]
- 42. Kwon S. M., Eguchi M., Wada M., Iwami Y., Hozumi K., Iwaguro H., Masuda H., Kawamoto A., Asahara T. (2008) Circulation 118, 157–165 [DOI] [PubMed] [Google Scholar]
- 43. Wang Z., Shu W., Lu M. M., Morrisey E. E. (2005) Mol. Cell. Biol. 25, 5022–5030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Daneman R., Agalliu D., Zhou L., Kuhnert F., Kuo C. J., Barres B. A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 641–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Monkley S. J., Delaney S. J., Pennisi D. J., Christiansen J. H., Wainwright B. J. (1996) Development 122, 3343–3353 [DOI] [PubMed] [Google Scholar]
- 46. Masckauchan T. N., Kitajewski J. (2006) Physiology 21, 181–188 [DOI] [PubMed] [Google Scholar]
- 47. Ishikawa T., Tamai Y., Zorn A. M., Yoshida H., Seldin M. F., Nishikawa S., Taketo M. M. (2001) Development 128, 25–33 [DOI] [PubMed] [Google Scholar]
- 48. Liu B. Y., Soloviev I., Chang P., Lee J., Huang X., Zhong C., Ferrara N., Polakis P., Sakanaka C. (2010) PLoS One 5, e8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Grunewald M., Avraham I., Dor Y., Bachar-Lustig E., Itin A., Jung S., Yung S., Chimenti S., Landsman L., Abramovitch R., Keshet E. (2006) Cell 124, 175–189 [DOI] [PubMed] [Google Scholar]
- 50. Dace D. S., Khan A. A., Kelly J., Apte R. S. (2008) PLoS One 3, e3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen C. C., Mo F. E., Lau L. F. (2001) J. Biol. Chem. 276, 47329–47337 [DOI] [PubMed] [Google Scholar]
- 52. Yang R., Amir J., Liu H., Chaqour B. (2008) Physiol. Genomics 36, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fataccioli V., Abergel V., Wingertsmann L., Neuville P., Spitz E., Adnot S., Calenda V., Teiger E. (2002) Hum. Gene Ther. 13, 1461–1470 [DOI] [PubMed] [Google Scholar]
- 54. Rayssac A., Neveu C., Pucelle M., Van den Berghe L., Prado-Lourenco L., Arnal J. F., Chaufour X., Prats A. C. (2009) Mol. Ther. 17, 2010–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ishida S., Usui T., Yamashiro K., Kaji Y., Amano S., Ogura Y., Hida T., Oguchi Y., Ambati J., Miller J. W., Gragoudas E. S., Ng Y. S., D'Amore P. A., Shima D. T., Adamis A. P. (2003) J. Exp. Med. 198, 483–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Inoki I., Shiomi T., Hashimoto G., Enomoto H., Nakamura H., Makino K., Ikeda E., Takata S., Kobayashi K., Okada Y. (2002) FASEB J. 16, 219–221 [DOI] [PubMed] [Google Scholar]
- 57. Davies M. H., Eubanks J. P., Powers M. R. (2006) Mol. Vis. 12, 467–477 [PubMed] [Google Scholar]
- 58. Lang R., Lustig M., Francois F., Sellinger M., Plesken H. (1994) Development 120, 3395–3403 [DOI] [PubMed] [Google Scholar]
- 59. Gardiner T. A., Gibson D. S., de Gooyer T. E., de la Cruz V. F., McDonald D. M., Stitt A. W. (2005) Am. J. Pathol. 166, 637–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bhatwadekar A. D., Glenn J. V., Curtis T. M., Grant M. B., Stitt A. W., Gardiner T. A. (2009) Invest. Ophthalmol. Vis. Sci. 50, 4967–4973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hubert K. E., Davies M. H., Stempel A. J., Griffith T. S., Powers M. R. (2009) Am. J. Pathol. 175, 2697–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.