Abstract
The holoenzyme of transcription integrates the positive and negative signals from the promoters of eukaryotic genes. We demonstrate that the essential holoenzyme component Srb7p is a physiologically relevant target of the global repressor Tup1p in Saccharomyces cerevisiae. Tup1p binds Srb7p in vivo and in vitro, and all genes tested that are repressed by Tup1p are derepressed when wild-type Srb7p is replaced by a mutant derivative of Srb7p that is no longer capable of interacting with Tup1p. Therefore, Srb7p is the first holoenzyme component essential for repression by Tup1p for which a physical interaction with Tup1p has been demonstrated. Furthermore, we find that Srb7p also binds Med6p and that this interaction is necessary for full transcriptional activation by different activators. Our finding that Med6p and Tup1p compete for the interaction with Srb7p suggests a model for Tup1p-mediated repression.
Keywords: repression/split-ubiquitin system/Srb7p/transcription/Tup1p
Introduction
The holoenzyme of transcription integrates the positive and negative signals from the promoters of eukaryotic genes (Koleske and Young, 1995). Positive signals emanate from activators bound to enhancers and negative signals from repressors bound to silencers. Both activators and repressors can also influence transcription by modulating the chromatin structure (Struhl, 1999). Activators can be associated with histone acetylases, leading to a chromatin structure favourable for transcription (Peterson and Workman, 2000), and repressors can bind chromatin components (Edmondson et al., 1996; Lehming et al., 1998; Laser et al., 2000) or be associated with histone deacetylases, resulting in a condensed chromatin structure hindering transcription (Ng and Bird, 2000). But both activators and repressors can also influence the holoenzyme of transcription directly. For activators, physiologically relevant interactions with specific components of the holoenzyme have been demonstrated (Lee et al., 1999). Various signals from different activators to different subunits of the holoenzyme are thought to be transmitted to Med6p, a holoenzyme component required for transcriptional activation processes (Han et al., 1999). The Saccharomyces cerevisiae transcriptional activator Gal4p, for example, binds Srb4p, and mutations in Gal4p can be compensated for by mutations in Srb4p (Koh et al., 1998). Srb4p, on the other hand, binds Med6p, and this interaction is necessary for the transmission of the signal to the polymerase. Mutations in MED6 can be suppressed by mutations in SRB4 in an allele-specific manner (Lee and Kim, 1998). For transcriptional repressors, the situation is less clear. Deletions of specific components of the holoenzyme result in derepression (Lee et al., 2000); for example, deletion of SRB10 leads to derepression of MFA2, a gene repressed by α2 with the help of the global repressor complex Ssn6p–Tup1p (Wahi and Johnson, 1995). Ssn6p is thought to interact directly with various repressor proteins bound to different promoters and to recruit the global repressor Tup1p to those promoters (Keleher et al., 1992). However, a physical association between Tup1p and Srb10p or any other Srb subunit has not yet been reported. Tup1p binds to the holoenzyme component Med3p, but deletion of MED3 does not result in upregulation of any of the genes repressed by Tup1p, demonstrating that Med3p does not mediate repression by Tup1p (Papamichos-Chronakis et al., 2000).
Here we show that the essential holoenzyme component Srb7p is a physiologically relevant target of Tup1p. Tup1p binds Srb7p in vivo and in vitro, and all genes tested that are repressed by Tup1p are derepressed when wild-type Srb7p is replaced by a mutant derivative of Srb7p that is no longer capable of interacting with Tup1p. Furthermore, we find that Srb7p also binds Med6p, and that this interaction is necessary for full transcriptional activation by different activators. Our finding that Med6p and Tup1p compete for the interaction with Srb7p suggests a model for Tup1p-mediated repression.
Results
Tup1p physically interacts with Srb7p in vivo
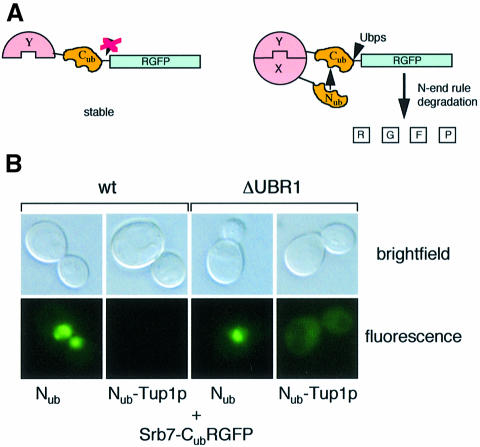
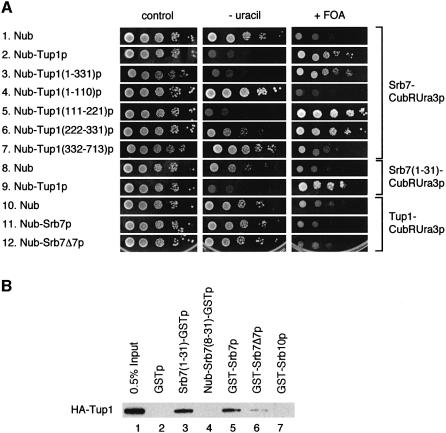
In order to identify potential targets of Tup1p in the holoenzyme of transcription, we tested 15 of its known components in the split-ubiquitin assay. The split-ubiquitin assay is an alternative two-hybrid system suited to analysing transcription factors (Johnsson and Varshavsky, 1994; Wellhausen and Lehming, 1999; Rojo-Niersbach et al., 2000). One of the variants of the split-ubiquitin system used here is outlined in Figure 1A. Ubiquitin fused to a green fluorescent protein (GFP) reporter with an arginine in position 1 (RGFP) is cleaved off by the ubiquitin-specific proteases (Ubps). Arginine is a destabilizing residue in the N-end rule pathway of protein degradation (Varshavsky, 1996), and therefore RGFP is rapidly degraded. No cleavage occurs if a fusion contains only the C-terminal half of ubiquitin (Cub, Figure 1A, left). However, two interacting proteins, X and Y, can be fused to the N-terminal half of ubiquitin (Nub) and the Cub-RGFP reporter, respectively. If X and Y form a complex inside the cell, the resulting increase in local concentration leads to the re-association of the two halves of ubiquitin. This native-like ubiquitin is now recognized by the Ubps and cleaved. Subsequently, the RGFP reporter is rapidly degraded by the enzymes of the N-end rule (Figure 1A, right). The following holoenzyme components were tested for interaction with Tup1p in the split-ubiquitin system: Rpb1p, Srb2p, Srb4p, Srb5p, Srb6p, Srb7p, Srb8p, Srb9p, Srb10p, Med6p, Gal11p, Toa1p, Toa2p, Sua7p and Spt15p. Out of these 15 proteins, only Srb7p interacted with Tup1p in vivo. Figure 1B shows that the expression of an Srb7–Cub-RGFP fusion resulted in localized nuclear staining in the presence of Nub. As a consequence of the interaction between Tup1p and Srb7p, the staining disappeared when Nub was replaced by Nub–Tup1p. Srb7p is essential, and the Srb7–Cub-RGFP fusion was able to complement an SRB7 deletion (data not shown). Apparently, Srb7–Cub-RGFP was able to replace Srb7p in the holoenzyme, making it possible for Nub–Tup1p to interact with Srb7–Cub-RGFP in the context of the holoenzyme. Figure 1B further shows that coexpression of Nub–Tup1p and Srb7–Cub-RGFP resulted in delocalization of the staining when an S.cerevisiae strain deficient for the recognition component of the N-end rule pathway (ΔUBR1) was used. After cleavage, RGFP was no longer fused to the nuclear localized Srb7p. In the absence of a functional N-end rule pathway, RGFP was stable and evenly distributed throughout the cytosol of the cell. For Srb10p, a holoenzyme component necessary for transcriptional repression by Tup1p, no interaction was observed in the split-ubiquitin system (data not shown). The interaction between Srb7p and Tup1p was analysed in more detail with the help of the Cub-RUra3p reporter (Wittke et al., 1999). Here, the RUra3p moiety is subject to rapid degradation upon release from the Cub part of the fusion. Therefore, interaction between the two proteins is indicated by uracil auxotrophy and resistance to the drug 5-fluoroorotic acid (FOA). Cells coexpressing Nub–Tup1p and Srb7–Cub-RUra3p do not grow on plates lacking uracil but on plates containing FOA, confirming the interaction observed with the Cub-RGFP reporter (Figure 2A, compare lines 1 and 2). A deletion analysis of Tup1p revealed that the N-terminal half of the protein contains two non-overlapping Srb7p-interacting regions, located between residues 111 and 221, and between residues 222 and 331, respectively (Figure 2A, compare lines 3–6). The C-terminal half of Tup1p, containing the WD repeats, did not interact with Srb7p (Figure 2A, line 7). The same results were obtained when the fusions were expressed in a strain lacking endogenous Tup1p (data not shown). The Nub part of the fusions contained the haemagglutinin (HA) epitope, which was used to confirm correct protein sizes and equal expression by western analysis (data not shown). Fusions to the red fluorescent protein DsRed1 (Clontech) revealed that all Nub–Tup1p derivatives were located in the nucleus (data not shown). Surprisingly, when we switched the fusions, coexpressing Nub–Srb7p and Tup1–Cub-RUra3p, the interaction was abolished (Figure 2A, compare lines 10 and 11). A deletion mutant lacking the first seven amino acids of Srb7p (Nub–Srb7Δ7p) also showed no interaction with Tup1p (Figure 2A, line 12). The integrity of the Tup1–Cub-RUra3p fusion was demonstrated in a control experiment, coexpressing Nub–Ssn6p and Tup1–Cub-RUra3p (data not shown), and Nub–Srb7p and Nub–Srb7Δ7p were both able to complement an SRB7 deletion. One possible explanation is that the interaction between Tup1p and Srb7p involves the N-terminus of Srb7p, and that the Nub part of the Nub–Srb7p fusion interferes with this interaction. Indeed, we found that the first 31 amino acids of Srb7p were sufficient for the interaction with Tup1p, as shown in Figure 2A, lines 8 and 9. When we tested the Nub–Tup1p deletion derivatives, we found that both Srb7-interacting regions in Tup1p interacted independently with the N-terminal 31 amino acids of Srb7p (data not shown).
Fig. 1. Tup1p interacts with Srb7p in vivo. (A) The split-ubiquitin system. A fusion containing a protein Y, Cub and the RGFP reporteris not subject to proteolysis by the Ubps. Co-expression of a fusion containing Nub and a protein X capable of interacting with protein Y leads to the formation of a native-like ubiquitin, to subsequent cleavage by the Ubps and to degradation of the RGFP reporter by the enzymes of the N-end rule. (B) The interaction between Tup1p and Srb7p leads to the destruction of the nuclear localized GFP signal in wild-type cells (left), or to the delocalization in cells without a functional N-end rule pathway (ΔUBR1, right). Cells expressing the depicted fusions were grown in liquid medium, spotted onto slides and analysed with the help of a fluorescent microscope (Leitz).
Fig. 2. The N-terminus of Srb7p is necessary and sufficient for the interaction with Tup1p. (A) Tup1p interacts with Srb7p in vivo. Serial dilutions of cells expressing the depicted fusions were spotted onto control plates selecting for the presence of these fusions, onto the same plates lacking uracil and onto plates also containing FOA. Interactions are revealed by the lack of growth on the plates without uracil and by growth on the plates containing FOA. (B) Tup1p interacts with Srb7p in vitro. GST-pulldown assays with the depicted proteins purified from E.coli were loaded onto an SDS gel, and HA-Tup1p was detected in a western blot with the help of an α-HA antibody.
In order to demonstrate that the observed in vivo interaction between Tup1p and Srb7p was direct, both proteins were expressed in Escherichia coli. Tup1p was fused to a tag containing His6 and the HA epitope, and Srb7p and its derivatives were fused either N- or C-terminally to glutathione S-transferase (GST). Figure 2B shows that Srb7p and Tup1p interacted in a GST-pulldown assay in vitro, and that the N-terminus of Srb7p was necessary and sufficient for this interaction (compare lanes 2 and 3). The figure further shows that Nub seemed to play an active role in the destruction of the Srb7p–Tup1p interaction. The addition of GST to the N-terminus of Srb7p had only a small effect on the interaction with Tup1p (Figure 2B, compare lanes 3 and 5), and the deletion of the first seven amino acids of Srb7p reduced the interaction with Tup1p ∼3-fold (Figure 2B, compare lanes 5 and 6). However, substitution of the first seven amino acids of Srb7p for Nub eliminated the interaction (Figure 2B, compare lanes 3 and 4). For Srb10p, no interaction with Tup1p was observed (Figure 2B, lane 7).
Disrupting the Tup1p–Srb7p interaction causes derepression
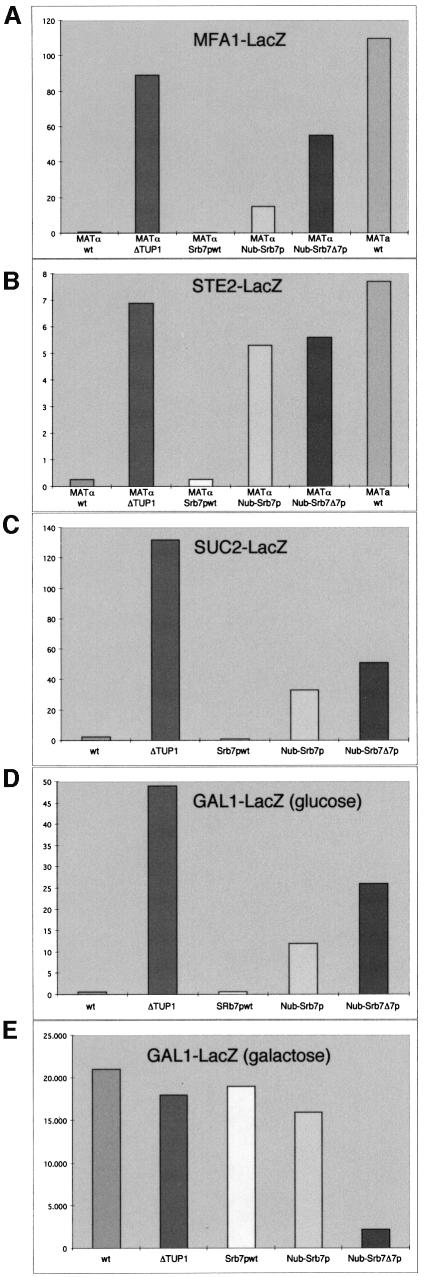
SRB7 is an essential gene and cannot be deleted. However, as mentioned above, Nub–Srb7Δ7p was able to complement an SRB7 deletion. This enabled us to test whether the disruption of the interaction between Srb7p and Tup1p had physiological consequences. We noticed that independent S.cerevisiae strains expressing Nub–Srb7Δ7p instead of wild-type Srb7p grew clumpy, as did strains in which TUP1 had been deleted. Consistently, the FLO11 gene was derepressed in both TUP1-deleted and Nub–Srb7Δ7p strains, as revealed by northern blot analysis (Figure 3A, compare lanes 1 and 4 with 5). Deletion of the holoenzyme component SRB10 also resulted in a clumpy growth phenotype and derepression of the FLO11 gene (Figure 3A, lane 2). Re-integration of a PCR fragment containing the entire wild-type SRB7 gene into the disrupted SRB7 locus (Srb7pwt) complemented the clumpy phenotype and the loss of FLO11 repression (Figure 3A, lane 3). We performed mating assays and observed that the mating efficiency of the Nub–Srb7Δ7p MATα strain was decreased, as it was for the isogenic strain deleted for TUP1 (Figure 3B). We tested four other genes known to be repressed by Tup1p (Trumbly, 1986; Mukai et al., 1991; Zhang et al., 1991; DeRisi et al., 1997) by fusing their promoters to the LacZ gene and integrating these promoter fusions into different strain backgrounds. Figure 4 shows that MFA1, STE2, SUC2 and GAL1, which were derepressed in strains deleted for TUP1 (columns 2), were also derepressed in the Nub–Srb7Δ7p strains (columns 5). The deletion of SRB10 also resulted in derepression, but to a lesser extent (data not shown). Disruption of the Tup1p–Srb7p interaction had a weaker effect than deletion of the entire TUP1 gene. This probably reflected the repressive effect Tup1p has on chromatin (Huang et al., 1997). The Nub–Srb7p strains also showed derepression, but with an intermediate phenotype (columns 4), consistent with the data presented in Figure 2B, where deletion of the first seven amino acids of Srb7p decreased the interaction between Srb7p and Tup1p in vitro. The derepression was readily complemented upon re-integration of wild-type SRB7 into the disrupted SRB7 locus (columns 3). Northern blot analysis with a LacZ probe revealed that the derepression took place at the level of transcription (data not shown).
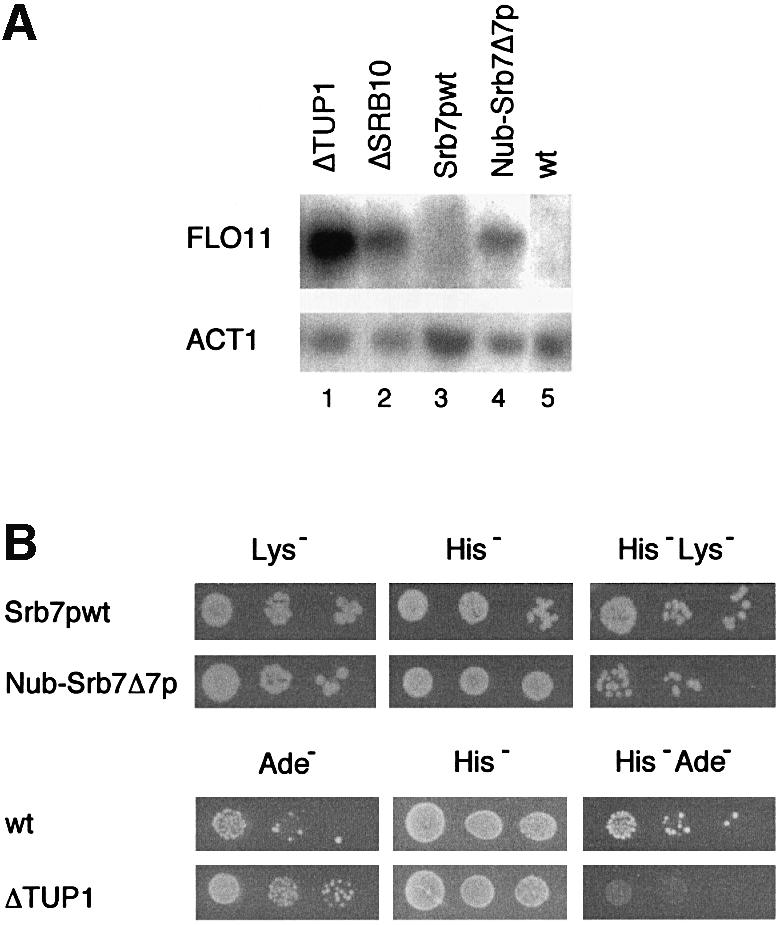
Fig. 3. Srb7p is a physiological target of Tup1p. (A) Disruption of the Tup1p–Srb7p interaction leads to derepression of FLO11. Total RNA was isolated from JD53ΔTUP1 (ΔTUP1), JD53ΔSRB10 (ΔSRB10), JD53ΔSRB7 + Srb7p (Srb7pwt), JD53ΔSRB7 + Nub–Srb7Δ7p (Nub–Srb7Δ7p) and JD53 (wt), and the northern blot was probed with probes derived from the FLO11 and ACT1 genes. (B) Disruption of the Tup1p–Srb7p interaction leads to a decreased mating efficiency. JD53ΔSRB7 + Srb7p (Srb7pwt) and JD53ΔSRB7 + Nub–Srb7Δ7p (Nub–Srb7Δ7p), which are prototrophic for lysine, as well as JD53 (wt) and JD53ΔTUP1 (ΔTUP1), which are prototrophic for adenine, were mated with JD52 + HIS3, which is prototrophic for histidine. Serial dilutions of the crosses were spotted onto the indicated plates and only diploids were able to grow on the double selection plates.
Fig. 4. Srb7p affects transcriptional repression and activation. (A) An MFA1-LacZ fusion was integrated into the MFA1 locus of the strains JD53 (MATα; wt), JD53ΔTUP1 (MATα; ΔTUP1), JD53ΔSRB7 + Srb7p (MATα; Srb7pwt), JD53ΔSRB7 + Nub–Srb7p (MATα; Nub–Srb7p), JD53ΔSRB7 + Nub–Srb7Δ7p (MATα; Nub–Srb7Δ7p) and JD52 (MATa; wt). The cells were grown in liquid culture and β-galactosidase activity was determined. (B) A STE2-LacZ fusion was integrated into the URA3 locus of the same strains as in (A). The cells were grown in liquid culture and β-galactosidase activity was determined. (C) A SUC2-LacZ fusion was integrated into the URA3 locus of the strains described in (A), with the exception of JD52. The cells were grown in liquid culture and β-galactosidase activity was determined. (D) A GAL1-LacZ fusion was integrated into the GAL1 locus of the strains described in (A), with the exception of JD52. The cells were grown on YPDA plates, washed off the plates and β-galactosidase activity was determined. (E) The strains from (D) were grown in liquid medium with 2% galactose, and β-galactosidase activity was determined. All experiments were performed in triplicate and the standard deviation was <20%.
Srb7p is a physiological target of Med6p
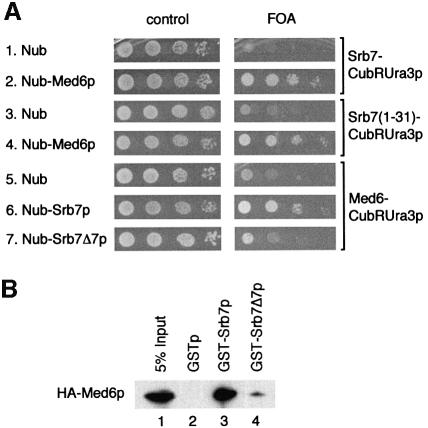
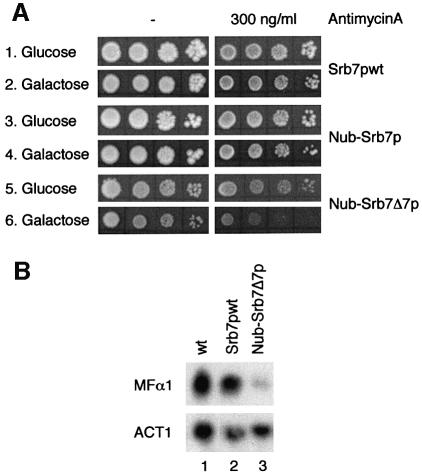
When we tested the strains with the integrated GAL1-LacZ fusion in galactose, we noticed that the Nub–Srb7Δ7p strain was not only defective for transcriptional repression but also for transcriptional activation (Figure 4E, column 5). But whereas the Nub–Srb7Δ7p strain showed a 10-fold decrease in transcriptional activation by Gal4p, the Nub–Srb7p strain gave results similar to wild type (Figure 4E, column 4). We wanted to know how Srb7p might be involved in transcriptional activation, and so we tested the 14 other holoenzyme components mentioned above for interaction with Srb7p in the split-ubiquitin assay. Only Med6p gave the expected result. Figure 5A shows that Srb7p interacted with Med6p in vivo, as revealed by the FOA resistance of cells coexpressing Nub–Med6p and Srb7–Cub-RUra3p (Figure 5A, lines 1 and 2). The figure further shows that the N-terminal 31 amino acids of Srb7p were sufficient for the interaction (Figure 5A, lines 3 and 4). And whereas Nub–Srb7p interacted with Med6–Cub-RUra3p, Nub–Srb7Δ7p showed no interaction (Figure 5A, lines 5–7). GST-pulldown assays with proteins expressed in E.coli demonstrated that the interaction between Srb7p and Med6p was direct (Figure 5B, compare lanes 2 and 3), and that an intact N-terminus of Srb7p was needed for the interaction (Figure 5B, compare lanes 3 and 4). The decrease in transcriptional activation by Gal4p that had been seen in Figure 4E was reflected by a GAL– phenotype of the Nub–Srb7Δ7p strain when the cells were grown on galactose plates in the presence of the respiration inhibitor antimycin A (Figure 6A, line 6). Consistently, the Nub–Srb7p strain grew like wild type on galactose even in the presence of antimycin A (Figure 6A, compare lines 2 and 4). The activation of the MFα1 gene in α cells expressing Nub–Srb7Δ7p instead of wild-type Srb7p was decreased too, as can be seen in the northern blot presented in Figure 6B (compare lanes 1 and 2 with 3).
Fig. 5. Med6p interacts with Srb7p in vivo and in vitro. (A) Med6p interacts with Srb7p in vivo. Serial dilutions of cells expressing the depicted fusions were spotted onto control plates selecting for the presence of these fusions and onto plates also containing FOA. Interactions are revealed by growth on the FOA plates. (B) Med6p interacts with Srb7p in vitro. GST-pulldown assays with the depicted proteins were loaded onto an SDS gel and HA-Med6p was detected in a western blot with the help of an α-HA antibody.
Fig. 6. Srb7p is a physiological target of Med6p. (A) Disruption of the Med6p–Srb7p interaction results in a GAL– phenotype on plates containing the respiration inhibitor antimycin A. Serial dilutions of the strains JD53ΔSRB7 + Srb7p (Srb7pwt), JD53ΔSRB7 + Nub–Srb7p (Nub–Srb7p) and JD53ΔSRB7 + Nub–Srb7Δ7p (Nub–Srb7Δ7p) were spotted onto plates containing the depicted medium. (B) Disruption of the Med6p–Srb7p interaction leads to a reduction in the transcriptional activation of MFα1. Total RNA was isolated from JD53 (wt), JD53ΔSRB7 + Srb7p (Srb7pwt) and JD53ΔSRB7 + Nub–Srb7Δ7p (Nub–Srb7Δ7p), and the northern blot was probed with probes derived from the MFα1 and ACT1 genes.
Tup1p and Med6p compete for Srb7p
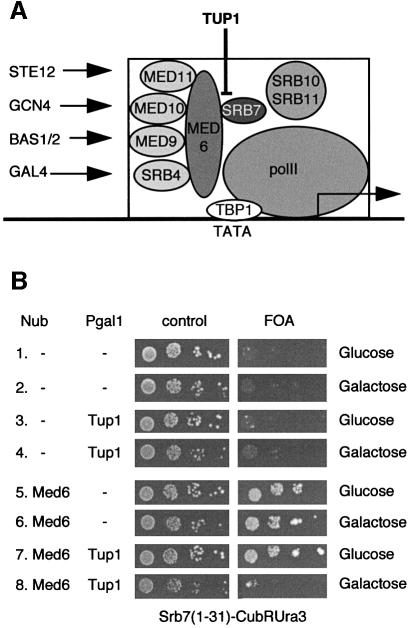
Med6p is involved in the transmission of positive signals from activators to the polymerase (Han et al., 1999). We propose that this process involves the interaction with the N-terminus of Srb7p. Apparently, Med6p and Tup1p bind the same part of Srb7p, suggesting a model for how Tup1p could inhibit the holoenzyme directly. Tup1p binds the N-terminus of Srb7p, hindering Med6p transmitting the signal (Figure 7A). In order to test this hypothesis, we performed a competition experiment. Cells coexpressing Nub–Med6p and Srb7(1-31)–Cub-RUra3p were FOA resistant due to the interaction between Med6p and Srb7p (Figure 7B, lines 5–7). Overexpression of Tup1p resulted in a decreased interaction between Med6p and Srb7p, consistent with our model (Figure 7B, line 8). Finally, we have observed that overexpression of Tup1p reduced activated levels of GAL1 transcription to 50% (data not shown).
Fig. 7. Tup1p works by blocking the access of Med6p to Srb7p. (A) A model for Tup1p action. The interaction between Med6p and Srb7p is needed to transmit positive signals emanating from transcriptional activators to the polymerase, and Tup1p interferes with this interaction. (B) Overexpression of Tup1p inhibits interaction between Med6p and Srb7p. Cells expressing Nub or Nub–Med6p together with the SRB7(1–31)–Cub-RUra3p fusion were transformed with a plasmid expressing Tup1p under the control of the GAL1 promoter, or the empty expression vector. Serial dilutions of the transformants were spotted onto plates containing the depicted medium. The presence of the interaction is revealed by the growth on the FOA plate.
Discussion
We have shown that Srb7p is involved in both transcriptional activation and repression. Our experiments suggest that Srb7p is a physiological target of the global repressor Tup1p, and that it also mediates positive signals obtained from Med6p. Tup1p and Med6p compete for binding to Srb7p, and we propose that this is how Tup1p exerts its direct effect on the holoenzyme of transcription.
The deletion analysis presented above revealed that Tup1p contains two independent Srb7p-interacting regions within its N-terminal half, both of them interacting with the N-terminal 31 amino acids of Srb7p. These two Srb7p-interacting regions map to the two non-overlapping transcriptional repression regions of Tup1p described previously (Tzamarias and Struhl, 1994). This is fully consistent with our model that Tup1p works by targeting Srb7p. Tup1p might bind Srb7p and block Med6p binding to Srb7p directly, but a change in conformation of Srb7p or the holoenzyme upon Tup1p binding is plausible too.
A comparison of the available Srb7p sequences from Drosophila melanogaster (DmSrb7p), Homo sapiens (HsSrb7p), S.cerevisiae (ScSrb7p) and Schizosaccharo myces pombe (SpSrb7p) revealed that the N-terminal 10 amino acids are extremely well conserved among all species (Figure 8). In this region, human Srb7p is 100% identical to fly Srb7p and 80% identical to the two yeasts. Also, the first 30 amino acids represent a region with a high degree of homology (50% identity between man and S.cerevisiae), whereas the next 30 amino acids display only a low degree of homology (13% identity). The central 30 amino acids are again better conserved (37% identity). We tried to affect the interaction between Tup1p and Srb7p by mutating single amino acids in the N-terminus of Srb7p, but all point mutants tested failed to disrupt the protein interaction and displayed no obvious phenotype. The most likely explanation is that because two different regions of Tup1p interact independently with the N-terminus of Srb7p, single point mutations affecting one interaction are compensated for by the other interaction. Future experiments will involve testing the Srb7p mutants with the two different Tup1p regions separately. Mutations affecting one interaction will be combined with mutations affecting the other interaction and double mutants will be tested for phenotypes. Also, we have tried to see whether there is an interaction between HsSrb7p and HirAp, a human Tup1p homologue. In the split-ubiquitin assay, Tup1p interacted very strongly with itself, probably reflecting the higher order chromatin complexes described previously (Ducker and Simpson, 2000), and HirAp interacted strongly with Tup1p. However, HirAp was not able to complement a TUP1 deletion, and no interaction between HirAp and HsSrb7p was observed (E.Rojo-Niersbach and N.Lehming, unpublished). This is not surprising, since the homology between Tup1p and HirAp is found in the C-terminal WD repeats and not in the N-terminal halves of the proteins.
Fig. 8. Sequence alignment of Srb7 proteins from D.melanogaster (DmSrb7p), H.sapiens (HsSrb7p) and S.pombe (SpSrb7p) to Srb7p of S.cerevisiae (ScSrb7p). Residues identical to the consensus sequence shown on top are shaded black. The sequences were derived from the DDBJ/EMBL/GenBank and the alignment was made with the program MegAlign.
Tup1p causes a decreased occupancy of the TATA-binding protein (TBP) at the promoters of Tup1p-repressed genes like SUC2 and MFA1 (Kuras and Struhl, 1999). How does the Tup1p–Srb7p interaction cause a repressive condition at Tup1p-repressed promoters if TBP (and presumably the rest of the holoenzyme) is not present? Tup1p causes a complex chromatin structure (Ducker and Simpson, 2000), and we suggest that this structure hinders TBP binding. However, even in the presence of Tup1p, binding of TBP to the Tup1p-repressed promoters can still be detected, and the differences in the occupancies of these promoters by TBP are much smaller than the differences in transcription (Kuras and Struhl, 1999). Apparently, the holoenzyme of transcription still binds to these promoters to some extent despite the complex structure, and transcription is repressed because Tup1p blocks the holoenzyme of transcription. If the block is removed by the use of Srb7p derivatives that Tup1p cannot bind, transcription can take place, but the level of transcription is reduced to ∼50%, reflecting the influence of the decreased TBP occupancy. Alternatively, the interaction between Tup1p and Srb7p might be important to stop transcription at any given moment, but it might not prevent recruitment of subsequent holoenzymes to the promoter. Perhaps the interaction halts transcription initiation long enough to facilitate chromatin remodelling, which would then provide a stable repressed state. That would also explain why neither histone mutations nor SRB mutations give full derepression of Tup1p-regulated genes.
Tup1p causes a complex chromatin structure along the entire repressed gene (Ducker and Simpson, 2000), and this structure cannot easily be reconstructed in vitro. This is presumably the reason why repression by Tup1p-dependent repressors in vitro is ∼2% of that observed in vivo (Herschbach et al., 1994). A factor of four has been measured for repression by α2 in vitro, and since the effect we have observed for disrupting the Tup1p–Srb7p interaction is less than that observed for deleting the entire TUP1 gene, we cannot expect more than a factor of two for currently available in vitro systems. Therefore, it is currently not possible to confirm by in vitro transcription the effects of Srb7p on transcriptional repression that we have observed in vivo.
Other members of the holoenzyme of transcription are necessary for transcriptional repression by Tup1p in vivo too. For example, the deletion of SRB10 leads to the derepression of genes repressed by Tup1p (Wahi and Johnson, 1995). Srb10p is a kinase, and its function is needed for Tup1p to repress its target genes (Kuchin and Carlson, 1998). However, we have seen no interaction between Srb10p and Tup1p, either in vivo or in vitro. And Med3p, a holoenzyme component that interacts with Tup1p in vivo and in vitro (Papamichos-Chronakis et al., 2000), is not necessary for transcriptional repression by Tup1p. The deletion of MED3 does not lead to the derepression of any of the genes repressed by Tup1p (Papamichos-Chronakis et al., 2000). MED3 is not essential and the effects of deleting MED3 on transcription are not dramatic (Piruat et al., 1997). Therefore, it is not possible that no derepression was observed because transcription was completely abolished in the MED3 deletion strain. Hence, deleting MED3 reduces transcriptional activation of a GAL1-LacZ reporter 5-fold (Piruat et al., 1997), whereas activation of GAL1 is eliminated in an Srb7ts strain at the restrictive temperature (Gromöller and Lehming, 2000). Thus, if Med3p were a physiological target of Tup1p, the deletion of MED3 should lead to the derepression of Tup1p-repressed genes, as observed for the Nub–Srb7Δ7p strain. Interestingly, overexpression of Med3p causes flocculence and derepression of FLO11 (Papamichos-Chronakis et al., 2000). Therefore, even though Med3p does not mediate repression by Tup1p, the interaction between Tup1p and Med3p is biologically relevant, and Med3p might be involved in counteracting Tup1p, similar to the role we are suggesting for Med6p here. Sfl1p, a Tup1p-dependent repressor that binds and represses the SUC2 promoter, co-immunoprecipitates with holoenzyme components like Srb9p (Song and Carlson, 1998). However, neither an involvement of Tup1p in the co-immunoprecipitation nor a direct protein interaction between Sfl1p, Tup1p and Srb9p has been demonstrated, and we have not seen an interaction between Tup1p and Srb9p in our assays. We conclude that Srb7p is the first holoenzyme component necessary for Tup1p-dependent repression for which a physical interaction with Tup1p has been demonstrated.
Materials and methods
Parental S.cerevisiae strains used were JD52 and JD53 (Dohmen et al., 1995). The split-ubiquitin system has been described previously (Wittke et al., 1999). The Cub-RGFP reporter was constructed by replacing the URA3 open reading frame of the Cub-RUra3p reporter by GFP with the help of the oligonucleotides GCCAGGCCTCATGAGTAAAGGAGAAGAACT and GCCATGCATTATTTGTATAGTTCATCCA. The respective genes were cloned as PCR fragments into the Nub and Cub fusion vectors and the constructs were sequenced by the ADIS service unit of the MPIZ. GST-pulldown experiments were performed as described previously (Wellhausen and Lehming, 1999). Expression vectors used were GEX5X1 (Pharmacia) and pET11a (Invitrogen). The α-HA antibody used for the detection of bound proteins in western blots was obtained from Babco. SRB7 was deleted with the help of the LYS2 marker from JD53 in the presence of the URA3-marked vector YCplac33 (Gietz and Sugino, 1988) containing the entire SRB7 gene, and TUP1 and SRB10 were deleted from JD53 with the help of the previously deleted ADE2 marker. SRB7 was also disrupted in the unrelated strain NLY2 (Saha et al., 1993), with the same results as presented here. Northern blots were performed following standard procedures (Ausubel et al., 1998). For the mating assay, the HIS3 gene of the JD52 strain was repaired and diploids were selected on plates lacking histidine and lysine (for the ΔSRB7 strains) or on plates lacking histidine and adenine (for the ΔTUP1 and wild-type strains). β-galactosidase assays were performed as described previously (Ausubel et al., 1998). The strains containing the GAL1-LacZ fusion were assayed after the cells were grown on YPDA plates instead of in liquid glucose medium, because under these conditions derepression by the deletion of TUP1 was more pronounced.
The following oligonucleotides were used for cloning of fusions containing Srb7: 5′-GCCGAATTCCATATGACAGATAGATTAACACAAT-3′, 5′-GCCGTCGACTTATGTGCTCTTTTTGAGTTT-3′, 5′-GGCGGCGTCGACCCATGGTTCTTATCTATGTA-3′ and 5′-GCCGTCGACGTGCTCTTTTTGAGTTTGCAA-3′; Med6: 5′-GCCAGATCTCCATGAACGTGACACCGTTGG-3′, 5′-GCGGCGGCCGCTCATATGTAGTTTGGGGTG-3′, 5′-GCCGAATTCATGAACGTGACACCGTTGG-3′ and 5′-GCCGTCGACACTATGTAGTTTGGGGTGGATC-3′; Tup1: 5′-GGCGTCGACTTAATTTGGCGCTATTTTTTT-3′, 5′-GCCCCCGGGCAATTGATGACTGCCAGCGTTTCGAA-3′, 5′-GCCCTCGAGTCGACGCATTTGGCGCTATTTTTTTATAC-3′ and 5′-GCCGCCTGATCATGACTGCCAGCGTTTCGA-3′; MFA1: 5′-GCCCTCGAGAAACTACGCAC-3′ and 5′-GCCTGGCCAGTTGCATTTCTATTCGATGGC-3′; STE2: 5′-GCCCTCGAGATCCAATATCACCTGACCTT-3′ and 5′-GCTTGGCCAAAGCCGCATCAGACATTTTTG-3′; SUC2: 5′-GCCCTCGAGTAAAAAAAAAACTAAGTTTTC-3′ and 5′-GCCTGGCCAAAGCTTGCAAAAGCATCATA-3′; GAL1: 5′-GCCCTCGAGGAACTTTCAGTAATACG-3′ and 5′-GCCTGGCCAAGCTTGGGCACTTTTCGGCCA-3′.
Acknowledgments
Acknowledgements
We thank Stephanie Heck for excellent technical help, Nils Johnsson for providing helpful suggestions and reagents, and Benno Müller-Hill for critical comments on the manuscript. This work was supported by grants from the Bundesministerium für Forschung und Bildung and the Max-Planck-Gesellschaft to N.L.
References
- Ausubel F., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1998) Current Protocols in Molecular Biology. John Wiley & Sons, Inc., New York, NY. [Google Scholar]
- DeRisi J.L., Iyer,V.R. and Brown,P.O. (1997) Exploring the metabolic and genetic control of gene expression on a genomic scale. Science, 278, 680–686. [DOI] [PubMed] [Google Scholar]
- Dohmen R.J., Stappen,R., McGrath,J.P., Forrova,H., Kolarov,J., Goffeau,A. and Varshavsky,A. (1995) An essential yeast gene encoding a homolog of ubiquitin-activating enzyme. J. Biol. Chem., 270, 18099–18109. [DOI] [PubMed] [Google Scholar]
- Ducker C.E. and Simpson,R.T. (2000) The organized chromatin domain of the repressed yeast a cell-specific gene STE6 contains two molecules of the corepressor Tup1p per nucleosome. EMBO J., 19, 400–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson D.G., Smith,M.M. and Roth,S.Y. (1996) Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev., 10, 1247–1259. [DOI] [PubMed] [Google Scholar]
- Gietz R.D. and Sugino,A. (1988) New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene, 74, 527–534. [DOI] [PubMed] [Google Scholar]
- Gromöller A. and Lehming,N. (2000) Srb7p is essential for the activation of a subset of genes. FEBS Lett., 484, 48–54. [DOI] [PubMed] [Google Scholar]
- Han S.J., Lee,Y.C., Gim,B.S., Ryu,G.H., Park,S.J., Lane,W.S. and Kim,Y.J. (1999) Activator-specific requirement of yeast mediator proteins for RNA polymerase II transcriptional activation. Mol. Cell. Biol., 19, 979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschbach B.M., Arnaud,M.B. and Johnson,A.D. (1994) Transcriptional repression directed by the yeast α2 protein in vitro. Nature, 370, 309–311. [DOI] [PubMed] [Google Scholar]
- Huang L., Zhang,W. and Roth,S.Y. (1997) Amino termini of histones H3 and H4 are required for a1-α2 repression in yeast. Mol. Cell. Biol., 17, 6555–6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson N. and Varshavsky,A. (1994) Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl Acad. Sci. USA, 91, 10340–10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keleher C.A., Redd,M.J., Schultz,J., Carlson,M. and Johnson,A.D. (1992) Ssn6–Tup1 is a general repressor of transcription in yeast. Cell, 68, 709–719. [DOI] [PubMed] [Google Scholar]
- Koh S.S., Ansari,A.Z., Ptashne,M. and Young,R.A. (1998) An activator target in the RNA polymerase II holoenzyme. Mol. Cell, 1, 895–904. [DOI] [PubMed] [Google Scholar]
- Koleske A.J. and Young,R.A. (1995) The RNA polymerase II holoenzyme and its implications for gene regulation. Trends Biochem. Sci., 20, 113–116. [DOI] [PubMed] [Google Scholar]
- Kuchin S. and Carlson,M. (1998) Functional relationships of Srb10-Srb11 kinase, carboxy-terminal domain kinase CTDK-I, and transcriptional corepressor Ssn6–Tup1. Mol. Cell. Biol., 18, 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras L. and Struhl,K. (1999) Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature, 399, 609–613. [DOI] [PubMed] [Google Scholar]
- Laser H., Bongards,C., Schüller,J., Heck,S., Johnsson,N. and Lehming,N. (2000) A new screen for protein interactions reveals that the S. cerevisiae HMG proteins Nhp6A/B are involved in the regulation of the GAL1 promoter. Proc. Natl Acad. Sci. USA, 97, 13732–13737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M., Chatterjee,S. and Struhl,K. (2000) Genetic analysis of the role of pol II holoenzyme components in repression by the Cyc8–Tup1 corepressor in yeast. Genetics, 155, 1535–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.C. and Kim,Y.J. (1998) Requirement for a functional interaction between mediator components Med6 and Srb4 in RNA polymerase II transcription. Mol. Cell. Biol., 18, 5364–5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.C., Park,J.M., Min,S., Han,S.J. and Kim,Y.J. (1999) An activator binding module of yeast RNA polymerase II holoenzyme. Mol. Cell. Biol., 19, 2967–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehming N., Le Saux,A., Schüller,J. and Ptashne,M. (1998) Chromatin components as part of a putative transcriptional repressing complex. Proc. Natl Acad. Sci. USA, 95, 7322–7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai Y., Harashima,S. and Oshima,Y. (1991) AAR1/TUP1 protein, with a structure similar to that of the β subunit of G proteins, is required for a1-α2 and α2 repression in cell type control of Saccharomyces cerevisiae. Mol. Cell. Biol., 11, 3773–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng H.H. and Bird,A. (2000) Histone deacetylases: silencers for hire. Trends Biochem. Sci., 25, 121–126. [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis M., Conlan,R.S., Gounalaki,N., Copf,T. and Tzamarias,D. (2000) Hrs1/Med3 is a Cyc8–Tup1 corepressor target in the RNA polymerase II holoenzyme. J. Biol. Chem., 275, 8397–8403. [DOI] [PubMed] [Google Scholar]
- Peterson C.L. and Workman,J.L. (2000) Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev., 10, 187–192. [DOI] [PubMed] [Google Scholar]
- Piruat J.I., Chavez,S. and Aguilera,A. (1997) The yeast HRS1 gene is involved in positive and negative regulation of transcription and shows genetic characteristics similar to SIN4 and GAL11. Genetics, 147, 1585–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo-Niersbach E., Morley,D., Heck,S. and Lehming,N. (2000) A new method for the selection of protein interactions in mammalian cells. Biochem. J., 348, 585–590. [PMC free article] [PubMed] [Google Scholar]
- Saha S., Brickman,J.M., Lehming,N. and Ptashne,M. (1993) New eukaryotic transcriptional repressors. Nature, 363, 648–652. [DOI] [PubMed] [Google Scholar]
- Song W. and Carlson,M. (1998) Srb/mediator proteins interact functionally and physically with transcriptional repressor Sfl1. EMBO J., 17, 5757–5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. (1999) Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell, 98, 1–4. [DOI] [PubMed] [Google Scholar]
- Trumbly R.J. (1986) Isolation of Saccharomyces cerevisiae mutants constitutive for invertase synthesis. J. Bacteriol., 166, 1123–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzamarias D. and Struhl,K. (1994) Functional dissection of the yeast Cyc8–Tup1 transcriptional co-repressor complex. Nature, 369, 758–761. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. (1996) The N-end rule: functions, mysteries, uses. Proc. Natl Acad. Sci. USA, 93, 12142–12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahi M. and Johnson,A.D. (1995) Identification of genes required for α2 repression in Saccharomyces cerevisiae. Genetics, 140, 79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellhausen A. and Lehming,N. (1999) Analysis of the in vivo interaction between a basic repressor and an acidic activator. FEBS Lett., 453, 299–304. [DOI] [PubMed] [Google Scholar]
- Wittke S., Lewke,N., Müller,S. and Johnsson,N. (1999) Probing the molecular environment of membrane proteins in vivo. Mol. Biol. Cell, 10, 2519–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Rosenblum-Vos,L.S., Lowry,C.V., Boakye,K.A. and Zitomer,R.S. (1991) A yeast protein with homology to the β-subunit of G proteins is involved in control of heme-regulated and catabolite-repressed genes. Gene, 97, 153–161. [DOI] [PubMed] [Google Scholar]