Abstract
Factor XIII (FXIII) is a pro-transglutaminase found in the plasma as well as intracellularly in platelets and macrophages. Plasma FXIII is activated by thrombin cleavage (FXIIIa*) and acts in the final stages of blood coagulation cascade. In contrast, the function and activation of cellular FXIII are less characterized. Cellular FXIII relies on a conformational activation of the protein. The nonproteolytic activation of FXIII to FXIIIa° induced by Ca2+ alone is well known, but up until now it has been discussed under which conditions the process can be induced and whether it can be reversed. Here, we study the nature of the Ca2+-induced FXIII activation. Previously used methods to evaluate FXIII activity detect both FXIIIa* and FXIIIa° because they rely on occurrence of enzyme activity or on active site Cys-314 solvent accessibility. Therefore, an analytical HPLC method was developed that separates zymogen recombinant FXIII (rFXIII) from rFXIIIa°. The data demonstrate that nonproteolytic activation and deactivation are highly dependent on Ca2+ concentration, buffer, and salt components. Moreover, it is established that Ca2+ activation of rFXIII is fully reversible, and only 2–5 mm CaCl2 is sufficient to retain full rFXIIIa° activity. However, below 2 mm CaCl2 the rFXIIIa° molecule deactivates. The deactivated molecule can subsequently undergo a new activation round. Furthermore, it is demonstrated that thermal stress of freeze-dried rFXIII can induce a new predisposed form that activates faster than nonstressed rFXIII.
Keywords: Blood Coagulation Factors, Enzyme Catalysis, Enzyme Inactivation, Hemostasis, HPLC, Enzyme Activation, Factor XIII
Introduction
Factor XIII (FXIII)2 (EC 2.3.2.13) is a pro-transglutaminase found in the plasma as well as intracellularly in platelets, monocytes/macrophages, and bone marrow precursors of these cell types. Activated FXIII catalyzes the formation of ϵ-(γ-glutamyl)lysine cross-links between the glutamine γ-carboxymide group and lysine ϵ-amino-group, resulting in intra- and intermolecular isopeptide cross-links.
Plasma FXIII (pFXIII) is a heterotetrameric zymogen consisting of two catalytic A subunits and two carrier B subunits. Activation of pFXIII during the blood coagulation cascade takes place when thrombin cleaves the 37-residue N-terminal activation peptide from the A subunits. In the presence of fibrin and Ca2+, the FXIII-A subunits undergo conformational changes and are restructured, and the activation peptide and B subunits are released. This results in the thrombin-activated FXIII (FXIIIa*). FXIIIa* is essential for stabilizing the hemostatic clot by cross-linking the fibrin monomers (1–4).
In contrast, the function and activation of a cellular FXIII (cFXIII) are less characterized. However, cFXIII, in particular cFXIII from platelets, accounts for approximately half of the FXIII found in the body (5, 6). cFXIII is a homodimeric zymogen consisting solely of catalytic A subunits. cFXIII has been reported to be involved in cytoskeleton remodeling during platelet activation (7, 8), as well as other aspects of tissue remodeling in e.g. wound healing and bone development (for review, see Refs. 9–11).
Furthermore, the activation of cFXIII to active transglutaminase is distinct from the thrombin activation of pFXIII. Being located intracellularly, cFXIII is not accessible to cleavage by thrombin. Several reports have demonstrated that cFXIII is activated without cleavage of the activation peptide, resulting in the FXIIIa° molecule (12–14). Furthermore, this nonproteolytic activation of FXIII has been demonstrated to occur in cells e.g. upon platelet activation (12, 13). In vitro studies have shown that Ca2+ alone is able to induce full activation of cFXIII to FXIIIa°; however, nonphysiologically high concentrations of Ca2+ (50–200 mm) were used (15–18). The Ca2+ dependence of the FXIIIa° production can be modulated by several factors because addition of chaotropic ions such as KSCN, p-toluenesulfonate, iodide, or bromide all lowered the Ca2+ concentration required for activation (18). Only 2 mm CaCl2 was required to induce FXIIIa° in the presence of 1 m NaCl or KCl (14). This is still higher than the net intracellular Ca2+ concentration of 10−4 mm in e.g. keratinocytes and resting platelets (19, 20). Higher concentrations are, however, reached during Ca2+ fluxes induced as a result of various stimuli (19, 21, 22), and FXIIIa° is indeed generated in the cytosols of monocytes or activated platelets (12, 13, 23). FXIII-B subunits appear to prevent the nonproteolytic activation of pFXIII in plasma as indicated by in vitro experiments showing that titration of cFXIII with stoichiometric amounts of B subunit strongly inhibits the activation induced by NaCl/Ca2+ (14).
It is obvious that FXIIIa* generated as a result of proteolytic activation cannot revert to zymogen FXIII. In contrast, reversibility of the nonproteolytic activation of FXIII to FXIIIa° induced by Ca2+ is still an option as has been suggested by previous investigators (18). Irreversibility of the activation was, on the other hand, apparently presumed by other investigators who studied the functional properties of FXIIIa° at submillimolar Ca2+ concentrations (24–26).
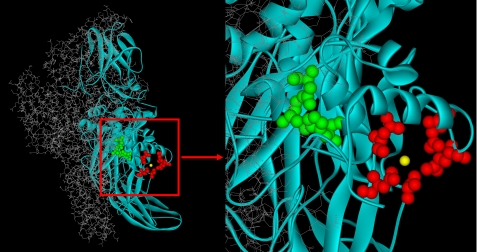
The zymogen structure of recombinant cellular FXIII (rFXIII) has been solved several times by x-ray crystallography (27–30), and the active site and the primary Ca2+ binding site have been identified (27) (Fig. 1) and shown to bind Ca2+ with a KD of 0.1 mm (31). Furthermore, 43Ca NMR studies of rFXIII, rFXIIIa*, and rFXIIIa° suggested additional low affinity Ca2+ binding sites. These additional Ca2+ binding sites were proposed to play a part in the restructuring occurring upon enzyme activation (32).
FIGURE 1.
Crystal structure of rFXIII. One FXIII-A is presented in gray sticks and the other FXIII-A in turquoise ribbons. Residues Asn-436, Asp-438, Ala-457, Glu-485, and Glu-490 form the Ca2+ binding site (red ball and stick, Ca2+ in yellow). Residues Cys-314, His-373, and Asp-396 form the catalytic site (green ball and stick). The structure is from Protein Data Bank entry 1GGU (27).
Here, we study the nature of the nonproteolytic Ca2+-induced activation of rFXIII to rFXIIIa°. Because previously used methods rely on the occurrence of enzyme activity or on active site Cys-314 titration (e.g. (12, 14, 15), these methods do not distinguish between FXIIIa* and FXIIIa°. Therefore, an analytical HPLC method was developed that separates zymogen rFXIII from rFXIIIa°. The data demonstrate that activation and deactivation of rFXIIIa° are highly dependent on Ca2+ concentration, buffer, and salt components, as well as thermal stress of the rFXIII molecule. Moreover, it is established that Ca2+ activation of rFXIII is fully reversible and that the molecule subsequently can undergo a new activation round.
EXPERIMENTAL PROCEDURES
rFXIII Samples
rFXIII, which is equivalent to cFXIII, was obtained from Novo Nordisk A/S (33). rFXIII was buffer exchanged to 30 mm Tris/HCl, pH 8.0, final concentration 7.5 mg/ml. This provided the rFXIII starting material for Ca2+ activation experiments. The rFXIII was diluted to 5 mg/ml in a test solution, divided in two, and freeze-dried. One of these samples was stored at −20 °C (unstressed sample). The other sample was stored for 1 month at 37 °C (heat-stressed sample). These two freeze-dried samples were used in the activation rate studies. rFXIII concentrations were determined using A280 (extinction coefficient at 1% = 14.8 liters g−1 cm−1).
Ca2+ Activation of rFXIII
Several activation conditions with different concentrations of rFXIII, Ca2+, and NaCl have previously been published (14, 15, 18, 25, 26). These conditions were optimized to minimize precipitation of rFXIII both during and after the activation process. The rFXIII starting material was diluted to 1 mg/ml in either Tris/NaCl/CaCl2; Tris/NaCl; Tris/CaCl2; Tris/ArgHCl/CaCl2, or borate/NaCl/CaCl2. Concentrations used were always 30 mm Tris/HCl, pH 8.0, 150 mm NaCl, 50 mm CaCl2, 200 mm ArgHCl, pH 8.0, and 20 mm borate, pH 8.3. See also Table 1. All samples were then incubated at 37 °C for 1 h to allow activation to occur.
TABLE 1.
AIEC and enzymatic assay results for various activation and deactivation conditions of rFXIII
| Sample | Incubated ina | Exchanged tob | AIECc |
Enzymatic assay | |
|---|---|---|---|---|---|
| Recoveryd | rFXIIIa°, | ||||
| % | % | % rFXIIIa | |||
| 1 | Tris/NaCl/CaCl2 | 91 | 100 | 112 | |
| 2 | Tris/NaCl | 100 | 0 | 0.5 | |
| 3 | Tris/CaCl2 | 65 | 54 | NAe | |
| 4 | Tris/NaCl/CaCl2 | Tris/NaCl/50 mm CaCl2 | 65 | 100 | NA |
| 5 | Tris/NaCl/CaCl2 | Tris/50 mm CaCl2 | 34 | 99.6 | NA |
| 6 | Tris/NaCl/CaCl2 | Tris/NaCl/0.33 mMCaCl2 | 65 | 14 | 29 |
| 7 | Borate/NaCl/CaCl2 | 88 | 100 | 107 | |
| 8 | Borate/CaCl2 | 56 | 52 | NA | |
| 9 | Borate/NaCl/CaCl2 | Borate/NaCl/0.33 mMCaCl2 | 61 | 0.7 | NA |
| 10 | Borate/NaCl/CaCl2 | Borate/0.33 mm CaCl2 | 33 | 0.7 | NA |
| 11 | Tris/ArgHCl/CaCl2 | 58 | 100 | 113 | |
| 12 | Tris/ArgHCl | 92 | 0 | NA | |
a The concentrations of the individual components used were: 30 mm Tris, pH 8.0; 20 mm borate, pH 8.3; 150 mm NaCl; 200 mm ArgHCl, pH 8.0; 50 mm CaCl2. Note that sample 2 does not contain CaCl2. 1 mg/ml rFXIII is prepared and incubated in this buffer for 1 h at 37 °C prior to analysis.
b After incubation, the sample was buffer exchanged to this buffer and thereafter analyzed. The CaCl2 concentrations were either 50 mm or 0.33 mm as indicated.
c All data presented here were obtained in the same analytical run to facilitate a direct comparison of results and to eliminate the analytical variation between runs.
d The recovery is calculated relative to the UV signal of rFXIII (sample 2).
e NA, not analyzed.
Buffer Exchange/Removal of CaCl2
After activation by CaCl2, the rFXIIIa° could be desalted on a NAP-5 column (GE Healthcare). The procedure recommended by the vendor was modified essentially to eliminate any buffer carryover. Briefly, the rFXIII solution (500 μl) was added to a preequilibrated NAP-5 column. 200 μl of the elution buffer was added to the column and the effluent discarded. Hereafter, the protein was eluted using 600 μl of elution buffer.
Ca2+-dependent Activation Rate
The two freeze-dried samples (unstressed and stressed samples) were reconstituted at 5 mg/ml and diluted to 1 mg/ml rFXIII in 30 mm Tris/HCl, pH 8, 150 mm NaCl containing either 2 mm or 11 mm CaCl2. The samples were placed in the HPLC autosampler at 30 °C immediately after preparation, and the first injection was performed within 10 min after adding the CaCl2-containing buffer.
Analytical Anion Exchange HPLC (AIEC)
rFXIII was separated from rFXIIIa° using a TSKgel DEAE-NPR 4.6 × 35-mm + guard 4.6 × 5-mm column (Tosoh Bioscience Ltd., UK), held at 25 °C. The flow rate was 1 ml/min with buffer A: 50 mm Tris, pH 7.5, + 3 mm CaCl2; and buffer B: 50 mm Tris, pH 7.5, + 500 mm NaCl + 5 mm CaCl2. A gradient of 1–27% B at 2–17.2 min was used. The autosampler was either kept at ambient temperature (∼21 °C) or at 30 °C in the activation rate studies; 20 μl containing 10–20 μg of protein was injected, and the absorbance at 215 nm was followed.
Enzymatic Assay
rFXIIIa transglutaminase activity was measured using the fluorescent FXIIIa substrate, A101 (Zedira), which is internally quenched and becomes fluorescent upon action of the transglutaminase activity of FXIIIa (34). For each sample, the transglutaminase activity was measured in the presence of 2 mm Ca2+ in both the absence and presence of 0.24 NIH units/ml thrombin. The relative amount of activity that was independent of thrombin was calculated.
RESULTS
Specific Detection of rFXIIIa° by Analytical AIEC
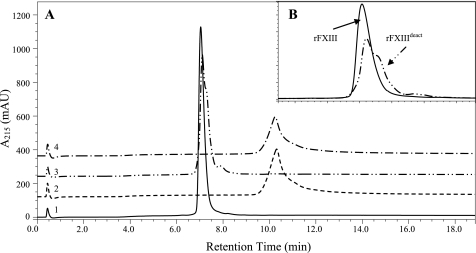
FXIIIa, regardless of activation mechanism, can be detected by measuring transglutaminase activity in numerous different activity assays. Furthermore, the active site cysteine, Cys-314, becomes solvent-accessible upon activation. This has previously been utilized for measuring FXIIIa by cysteine reactive reagents (12, 18). However, none of these methods distinguished between activation pathways nor identified the activation on the molecular level. Biophysical characterization has revealed that conformational changes in the secondary structure of rFXIIIa° alter its surface charge and hydrophobicity relative to native rFXIII.3 These changes were utilized in the development of an analytical AIEC that separated rFXIII (retention time approximately 7.1 min; Fig. 2A, trace 1) and rFXIIIa° (retention time approximately 10.5 min; Fig. 2A, trace 2). rFXIIIa° was eluted from the column with the activity intact as measured in enzymatic assay (data not shown). Given the reversibility of this activation process (see section below) and the absence of any contaminating proteolytic activation, it is apparent that we are observing a true nonproteolytic activation.
FIGURE 2.
AIEC traces from four rFXIII samples. A, rFXIII (trace 1), rFXIIIa° (trace 2), rFXIIIdeact (trace 3), and rFXIIIa° (trace 4). rFXIIIa° (trace 2) was produced by activation in 50 mm Ca2+, 37 °C, 1 h. rFXIIIdeact (trace 3) was produced by removal of Ca2+ from rFXIIIa°. rFXIIIa° from trace 4 was produced by reactivating rFXIIIdeact in Ca2+ similarly to rFXIIIa° in trace 2. B, enlargement of the area around the retention times of rFXIII and rFXIIIdeact (i.e. traces 1 and 3). rFXIII elutes as a one smooth peak whereas rFXIIIdeact seems to contain at least two components.
rFXIIIa° Activation and Deactivation Are Reversible
rFXIIIa° (Fig. 2A, trace 2) was produced by incubating rFXIII (Fig. 2A, trace 1) for 1 h at 37 °C in 50 mm CaCl2 as described under “Experimental Procedures. ” Upon removal of CaCl2 from rFXIIIa°, the resulting sample was recovered at a retention time similar to rFXIII with only a minor peak, 0.6%, at the position of rFXIIIa° (Fig. 2A, trace 3). Importantly, the nonproteolytic Ca2+-induced activation of rFXIII therefore seemed fully reversible. This Ca2+-activated and subsequently deactivated molecule will herein be termed rFXIIIdeact. By reincubation in 50 mm CaCl2, rFXIIIdeact could be reactivated to rFXIIIa° (Fig. 2A, trace 4). Thus, both the activation and the deactivation process were shown to be fully reversible. The results were confirmed by enzymatic activity assay showing <0.4% activity for zymogen rFXIII, full activity for rFXIIIa° samples, and 2.7% activity for rFXIIIdeact (data not shown).
Although rFXIII and rFXIIIdeact samples were generally indistinguishable in enzymatic assay, the AIEC revealed that peak shapes and thus surface charge of the samples were not completely identical (Fig. 2B). These data implied that upon deactivation, rFXIIIdeact did not fully regain its native structure and/or surface charge compared with zymogen rFXIII.
Salts and Buffers Impact Both the Activation and the Solubility of rFXIIIa°
The trends in activation and deactivation due to salt and buffer compositions are summarized in Table 1. All data presented here were obtained in the same analytical run to facilitate a direct comparison of the results and to eliminate the analytical variation between runs. rFXIII was fully activated to rFXIIIa° in 50 mm CaCl2 at 150 mm NaCl (Table 1, sample 1). Furthermore, the recovery of rFXIIIa° was close to 90% compared with native rFXIII incubated without CaCl2 (Table 1, sample 2). In the absence of NaCl, only 65% protein was recovered with only 54% as the rFXIIIa° form. The remaining 46% was unchanged (Table 1, sample 3). Thus, NaCl affects both the activation process and the solubility of rFXIII/rFXIIIa°.
rFXIIIa° samples were then passed over an NAP-5 column to examine the effect of various buffer exchanges (Table 1, samples 4–10). The general protein recovery was lower in these samples due to the modified NAP-5 buffer exchange procedure designed to eliminate buffer carryover. First, and as expected, the molecule was maintained on the rFXIIIa° form when the buffer composition was unchanged (Table 1, samples 4 and 5), showing that passing of the NAP5 column per se did not result in deactivation. Very low concentrations of CaCl2 did, however, result in lowered rFXIIIa° levels (Table 1, sample 6). Previous rFXIII activation and characterization studies were performed with borate buffers (24–26). We therefore also tested the effect of borate in absence and presence of NaCl. Activation of rFXIII in borate buffer showed results similar to those found with Tris buffer (Table 1, samples 7 and 8).
In contrast, Tris and borate have differential effects on the deactivation process. Upon desalting of the rFXIIIa° sample to 0.33 mm CaCl2, complete deactivation to rFXIIIdeact occurred in borate buffer whereas 14% rFXIIIa° remained when the experiment was performed in Tris buffer (Table 1, samples 6, 9, and 10). This suggests that previously reported characterizations using 0.33 mm CaCl2 might have been performed on rFXIIIdeact and not rFXIIIa° as expected (24–26). Other salts have been reported to have an effect similar to that of NaCl on FXIII activation (18). Arginine is often used in formulation and stabilization of proteins, and replacing NaCl with ArgHCl in the activation procedure still resulted in 100% rFXIIIa° in the presence of CaCl2 and no rFXIIIa° in the absence of CaCl2 (Table 1, samples 11 and 12). However, the recovery of rFXIIIa° was poor (58%; Table 1, sample 11), suggesting lower stability or solubility of rFXIIIa° in the presence of ArgHCl.
Effects of Ca2+ Concentration on Deactivation of rFXIIIa°
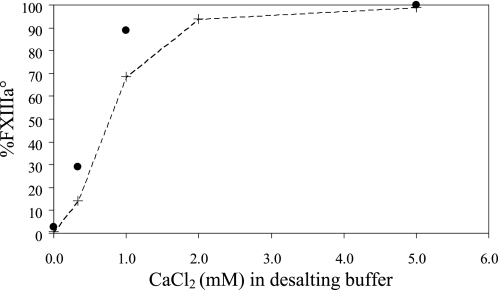
The Ca2+-induced activation of rFXIIIa° seemed to revert upon complete removal of CaCl2 (Fig. 2 and Table 1). We therefore examined the effect of intermediate concentrations of Ca2+ on rFXIIIa°. rFXIIIa° was desalted to various concentrations of CaCl2 and analyzed for remaining percentage FXIIIa° by both AIEC and enzymatic activity assay (Fig. 3). Percentage FXIIIa° is defined as (rFXIIIa°/(rFXIII + rFXIIIa°)) × 100%. Upon desalting to 2–5 mm CaCl2, nearly all rFXIIIa° was retained. However, a reduction in rFXIIIa° content was observed in samples containing <2 mm CaCl2, indicating that the level of rFXIIIa° in the sample was effectively controlled by the CaCl2 concentration. Enzymatic activity assay confirmed the trend observed by AIEC; however, it consistently measured higher values than the AIEC. The discrepancy between the two analytical methods could arise from differences in analysis time, approximately 11 min for AIEC compared with approximately 40 min for the enzymatic activity assay.
FIGURE 3.
Content of rFXIIIa° upon desalting to various CaCl2 concentrations. Fully active rFXIIIa° was buffer-exchanged to a lower concentration of CaCl2. The buffer-exchanged samples were analyzed for relative content of rFXIIIa° by both AIEC (+) and enzymatic activity assay (●).
Effects of Ca2+ Concentration on Activation Rates
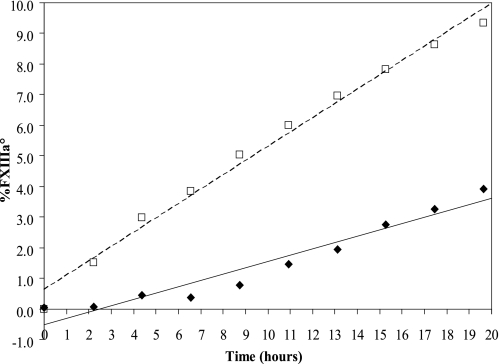
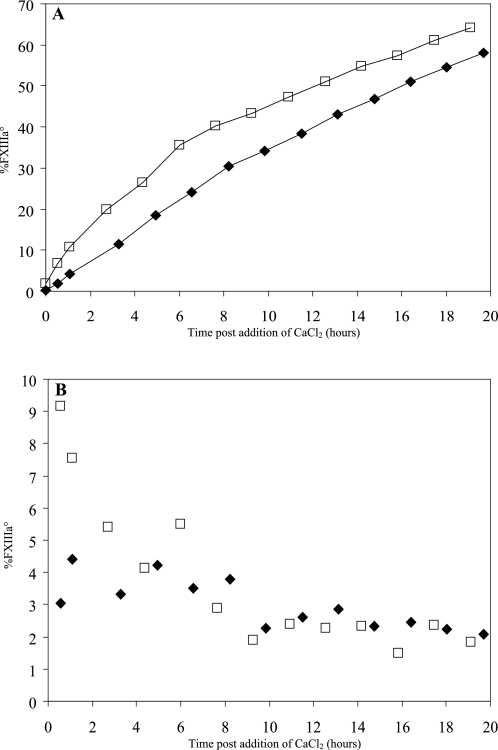
All rFXIIIa° activations described thus far were performed in the presence of 50 mm CaCl2. Because the deactivation process was affected by Ca2+ (Fig. 3) we also studied the activation of rFXIII to rFXIIIa° over time at 2 mm or 11 mm CaCl2 (Figs. 4 and 5A). A slowly advancing activation of zymogen rFXIII to rFXIIIa° occurred even at the low 2 mm concentration of CaCl2 (Fig. 4). The sample contained 3–4% rFXIIIa° after a 20-h incubation. Correspondingly, >55% rFXIIIa° was present in the sample after a 20-h incubation in the presence of 11 mm CaCl2 (Fig. 5A). These data demonstrated that rFXIIIa was activated to rFXIIIa° at a relatively low Ca2+ concentration and that the activation rate increased with the Ca2+ concentration. Finally, we noted that rFXIIIdeact obtained by Ca2+ activation and subsequent deactivation retained its propensity to be activated to rFXIIIa° when presented with Ca2+ (data not shown). This was despite the fact that rFXIIIdeact did not fully regain the zymogen shape (Fig. 2B).
FIGURE 4.
rFXIII activation to rFXIIIa° in 2 mm CaCl2. Two freeze-dried rFXIII samples were reconstituted and incubated at 30 °C in the presence of 2 mm CaCl2. The content of rFXIIIa° was followed by AIEC. The heat-stressed rFXIII sample (□) activated faster than the unstressed sample (♦).
FIGURE 5.
rFXIII activation to rFXIIIa° in 11 mm CaCl2. Two freeze-dried rFXIII samples, heat-stressed (□) and unstressed (♦), were reconstituted and incubated at 30 °C in the presence of 11 mm CaCl2. A, the content of rFXIIIa° was followed by AIEC. Note the difference in y axis scale compared with Fig. 4. B, the activation rates expressed as percentage rFXIIIa°/h were calculated after every injection and plotted. A subpopulation of the molecules in the heat-stressed rFXIII is more prone to activate upon addition of CaCl2.
Predisposal for Ca2+-induced Activation Due to Thermal Stress
In addition to Ca2+, we found that thermal stress of rFXIII also affected the activation rate. rFXIII samples freeze-dried in an exploratory formulation were stored at either 20 °C (unstressed sample) or incubated at 37 °C for 1 month (stressed sample). Upon reconstitution and dilution to 1 mg/ml in a buffer containing 2 mm CaCl2, the content of rFXIIIa° increased faster in the stressed sample with an activation rate of 0.4–0.5% rFXIIIa°/h (Fig. 4). In comparison, the unstressed sample activated considerably slower, i.e. approximately 0.2% rFXIIIa°/h (Fig. 4). Both samples were subjected to a similar analysis at 11 mm CaCl2, and again a higher overall activation rate was observed for the heat-stressed sample (Fig. 5A). Furthermore, the activation rate of the heat-stressed sample in 11 mm CaCl2 seemed to change over time (Fig. 5B). Within the first 6 h the activation rates were higher for the heat-stressed sample compared with the unstressed sample. However, after 6 h the activation rates were comparable in the two samples. Thus, the sample seemed to contain a subpopulation of rFXIII molecules that, as a result of the thermal stress, displayed faster activation rate when exposed to high concentration of CaCl2.
DISCUSSION
Ca2+ and the Activation of Transglutaminases
FXIII gains transglutaminase activity following activation by either a proteolytic thrombin-dependent pathway or a nonproteolytic pathway. Both pathways are found in vivo (3, 12, 13). Ca2+ is crucial in the conformational change of both the thrombin-cleaved FXIIIa′ as well as the zymogen FXIII molecule into the activated forms, FXIIIa* and FXIIIa°, respectively. Activation following proteolytic cleavage is common for both the allosteric and catalytic proteins of the blood coagulation cascade including pFXIII (3). Conversely, cFXIII and several other human transglutaminases rely solely on a conformational change for activation (e.g. Ref. 11), thus resembling the nonproteolytic activation of rFXIII. Importantly, Ca2+ is crucial for the activation and enzymatic activity of all human transglutaminases. In line with this observation, our data demonstrate an unambiguous effect of the Ca2+ concentration on both the rFXIII activation rate (Figs. 4 and 5) and the deactivation process (Fig. 3). The data also clearly reveal that complete deactivation of rFXIIIa° is possible while still maintaining the potential for subsequent reactivation (Fig. 2). The Ca2+ concentrations needed for both effective activation of rFXIII to rFXIIIa° as well as maintenance of rFXIIIa° determined here in vitro (i.e. 2 mm Ca2+) are several orders of magnitude higher than the intracellular net Ca2+ concentration of 10−4 mm in cells, e.g. keratinocytes and resting platelets (19, 20). However, several parameters such as chaotropic ions, NaCl and general ion strength can affect and considerably decrease the apparent concentration of Ca2+ necessary for activation of FXIII or other transglutaminases (14, 18, 35). Moreover, Ca2+ fluxes reaching higher concentrations occur in cells as a result of several stimuli (e.g. (19, 21, 22).
Ca2+ is involved in activation of all human transglutaminases. Early studies demonstrated transglutaminase activity in human erythrocytes, upon Ca2+ induction (36, 37). Furthermore, dependent on exposure, time latent transglutaminase could be activated with as little as 0.01 mm Ca2+ (38, 39).
However, human transglutaminases can also be activated, regulated, and inhibited by compounds besides Ca2+. TG2, TG3, and TG5 are inhibited by the presence of GTP/GDP nucleotides (40–43), and TG1 and TG3 are moreover dependent on proteolytic cleavage for activity (44, 45). The additional regulation of TG2 by GTP/GDP acts opposite Ca2+, and low concentrations of Ca2+ and physiological concentrations of GTP represent a double protection against inapt TG2 activation. Similar to this situation, Mg2+ ions compete for the Ca2+ binding sites in TG3 and thereby counteract activation and increase the apparent Ca2+ concentration requirements of TG3 (46).
Therefore, our data suggest that intracellular Ca2+ concentration is essential for regulating the catalytic activity and activation of intracellular FXIII. However, as demonstrated in the analogous transglutaminase systems, other, yet unidentified, parameters may influence the activation process of FXIII.
Structural Changes upon Activation of rFXIII
The conformation of FXIIIa° is evidently distinct from that of zymogen FXIII. This is demonstrated here with the differential binding to the AIEC column as well as previous reports on solvent accessibility of the active site Cys-314 in FXIIIa° (12, 18). In addition, conformational alterations in rFXIIIa° compared with zymogen rFXIII have also been observed by hydrogen-deuterium exchange (47).
Although rFXIII has been crystallized both in the presence and absence of Ca2+ (27), little information could be gained concerning the conformational changes taking place upon activation. The active site Cys-314 was inaccessible to solvent in all structures (27, 28, 30). Even under conditions that would normally yield rFXIIIa*, i.e. thrombin cleavage in the presence of Ca2+, the crystals contained rFXIIIa′ (29), thus suggesting that the zymogen structure is preferentially crystallized.
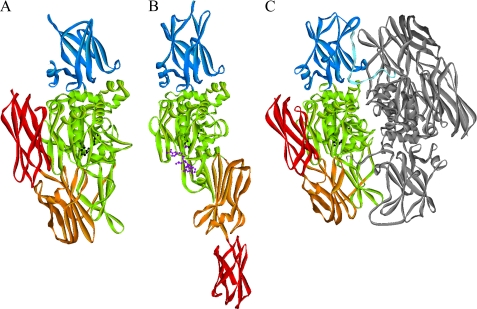
Zymogen human TG2 and TG3 have tertiary structures similar to zymogen rFXIII (40, 48) (Fig. 6, A and C). TG3 has also been crystallized under several different Ca2+ and Mg2+ conditions as well as cleaved for activation (46, 48). Ca2+ is an activator of TG3, and a minor adjustment of the structure around the active site was observed. However, the active site was not exposed, and the overall domain structure was similar to the zymogen form of TG3 (46, 48). Importantly, TG2 has also been crystallized in the absence of inhibitory nucleotide and with a peptide bound in the active site (49). This activated TG2 structure is remarkably different from zymogen TG2 (Fig. 6A), and recent studies have demonstrated that FXIII presumably is able to undergo the same conformational changes (47, 50). The secondary structure and individual domains are largely intact in activated TG2 (49). However, the four domains are oriented differently relative to each other, and the two C-terminal β-barrel domains are now positioned in an elongated structure not blocking the active site entrance (Fig. 6, A and B).
FIGURE 6.
Crystal structures of zymogen TG2 (A), active TG2 (B), and zymogen FXIII (C). The structures are from Protein Data Bank entries 1KV3, 2Q3Z, and 1GGU, respectively (27, 40, 49). The domains from the N terminus are activation peptide (FXIII only, light blue), β-sandwich (blue), catalytic domain (green), β-barrel 1 (orange), and β-barrel 2 (red). The active site residues are shown in black ball and stick, and the active site peptide inhibitor is in purple ball and stick (active TG2 (B)). The second monomer of FXIII dimer is shown in gray. TG2 is monomeric.
Overall, the conformational changes of rFXIII upon activation as observed here by a major shift in retention time in AIEC of rFXIIIa° are not likely to be explained by the small structural adjustments observed in the TG3 structures. However, a major conformational reorganization as seen in TG2 could offer a plausible explanation
Intermediate Activation States: rFXIIIdeact and the Predisposed Form of rFXIII
The Ca2+-activated and then deactivated rFXIII molecule was termed rFXIIIdeact. Although the enzymatic properties of rFXIIIdeact were similar to zymogen rFXIII, rFXIIIdeact displayed a slightly altered AIEC mobility (Fig. 2B). This might suggest that a minor population of the sample was characterized by a different surface charge and that perhaps a conformational change had occurred, possibly leaving the molecules in neither the zymogen nor the activated state. Provided rFXIII follows the same scheme as the conformational changes in TG2, this suggests that the domain reorganization occurring as a result of Ca2+ desalting is not complete such that the domains do not dock completely back into the zymogen structure. Similarly, a minor population of rFXIII performs differently upon thermal stress in an early exploratory freeze-dried formulation. This population activates faster to rFXIIIa° upon Ca2+ addition compared with zymogen rFXIII. Correspondingly, this minor population could have a structure that is intermediate between zymogen and activated states, and thus it may be more prone to domain reorganization upon Ca2+ exposure.
In summary, we found that the nonproteolytic activation of rFXIII and the rate thereof are very dependent on the Ca2+ concentration, buffer, salt, and thermal stress of the rFXIII molecule. Moreover, it is proven that Ca2+ activation is fully reversible and that the molecule subsequently can undergo a new activation round. The activated, and presumably also predisposed, forms of the rFXIII molecule seem to have a different conformation compared with zymogen rFXIII.
Acknowledgments
We thank Helle Holton, Lene Villadsen, and Ulla Wahlers for excellent technical assistance. We also thank Drs. Lene Hørlyck and Hans Holmegård Sørensen for incentive interest and valuable discussions and Drs. Lene Hørlyck, Lars Christian Petersen, and Christian Rischel for critical reading of the manuscript.
G. K. Kristiansen, J. T. Bukrinsky, and M. Persson, unpublished data.
- FXIII
- Factor XIII
- AIEC
- anion exchange HPLC
- cFXIII
- cellular FXIII (i.e. consisting of the A2 subunits only)
- FXIIIa
- activated FXIII (regardless of the mode of activation)
- FXIIIa*
- thrombin-activated FXIII
- FXIIIa°
- FXIII activated without proteolysis
- FXIIIa′
- thrombin cleaved but not active
- FXIIIdeact
- FXIIIa° that has been deactivated e.g. by removal of Ca2+
- pFXIII
- plasma FXIII (i.e. consisting of the A2B2 subunits)
- rFXIII
- recombinant cellular FXIII (i.e. consisting of the A2 subunits only)
- TG
- transglutaminase. FXIII nomenclature is in accordance with Ref. 51.
REFERENCES
- 1. Ariëns R. A., Lai T. S., Weisel J. W., Greenberg C. S., Grant P. J. (2002) Blood 100, 743–754 [DOI] [PubMed] [Google Scholar]
- 2. Lorand L. (2005) J. Thromb. Haemost. 3, 1337–1348 [DOI] [PubMed] [Google Scholar]
- 3. Muszbek L., Yee V. C., Hevessy Z. (1999) Thromb. Res. 94, 271–305 [DOI] [PubMed] [Google Scholar]
- 4. Muszbek L., Bagoly Z., Bereczky Z., Katona E. (2008) Cardiovasc. Hematol. Agents Med. Chem. 6, 190–205 [DOI] [PubMed] [Google Scholar]
- 5. Lopaciuk S., Lovette K. M., McDonagh J., Chuang H. Y., McDonagh R. P. (1976) Thromb. Res. 8, 453–465 [DOI] [PubMed] [Google Scholar]
- 6. Sixma J. J., van den Berg. A., Schiphorst M., Geuze H. J., McDonagh J. (1984) Thromb. Haemost. 51, 388–391 [PubMed] [Google Scholar]
- 7. Serrano K., Devine D. V. (2002) Thromb. Haemost. 88, 315–320 [PubMed] [Google Scholar]
- 8. Asijee G. M., Muszbek L., Kappelmayer J., Polgár J., Horváth A., Sturk A. (1988) Biochim. Biophys. Acta 954, 303–308 [DOI] [PubMed] [Google Scholar]
- 9. Adány R., Bárdos H. (2003) Cell. Mol. Life Sci. 60, 1049–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Iismaa S. E., Mearns B. M., Lorand L., Graham R. M. (2009) Physiol. Rev. 89, 991–1023 [DOI] [PubMed] [Google Scholar]
- 11. Lorand L., Graham R. M. (2003) Nat. Rev. Mol. Cell Biol. 4, 140–156 [DOI] [PubMed] [Google Scholar]
- 12. Muszbek L., Haramura G., Polgár J. (1995) Thromb. Haemost. 73, 702–705 [PubMed] [Google Scholar]
- 13. Muszbek L., Polgár J., Boda Z. (1993) Thromb. Haemost. 69, 282–285 [PubMed] [Google Scholar]
- 14. Polgár J., Hidasi V., Muszbek L. (1990) Biochem. J. 267, 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Credo R. B., Curtis C. G., Lorand L. (1978) Proc. Natl. Acad. Sci. U.S.A. 75, 4234–4237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. De Backer-Royer C., Traoré F., Meunier J. C. (1992) Int. J. Biochem. 24, 91–97 [DOI] [PubMed] [Google Scholar]
- 17. Hornyak T. J., Shafer J. A. (1992) Biochemistry 31, 423–429 [DOI] [PubMed] [Google Scholar]
- 18. Lorand L., Credo R. B., Janus T. J. (1981) Methods Enzymol. 80, 333–341 [DOI] [PubMed] [Google Scholar]
- 19. Li L., Tucker R. W., Hennings H., Yuspa S. H. (1995) Cell Growth Differ. 6, 1171–1184 [PubMed] [Google Scholar]
- 20. Poch E., Fernández-Llama P., Botey A., Gaya J., Darnell A., Rivera F., Revert L. (1995) Nephrol. Dial. Transplant. 10, 366–371 [PubMed] [Google Scholar]
- 21. Lee C. H., Poburko D., Kuo K. H., Seow C. Y., van Breemen C. (2002) Am. J. Physiol. Heart Circ. Physiol. 282, H1571–1583 [DOI] [PubMed] [Google Scholar]
- 22. Rink T. J., Sage S. O. (1990) Annu. Rev. Physiol. 52, 431–449 [DOI] [PubMed] [Google Scholar]
- 23. AbdAlla S., Lother H., Langer A., el Faramawy Y., Quitterer U. (2004) Cell 119, 343–354 [DOI] [PubMed] [Google Scholar]
- 24. Sabo T. M., Brasher P. B., Maurer M. C. (2007) Biochemistry 46, 10089–10101 [DOI] [PubMed] [Google Scholar]
- 25. Turner B. T., Jr., Maurer M. C. (2002) Biochemistry 41, 7947–7954 [DOI] [PubMed] [Google Scholar]
- 26. Turner B. T., Jr., Sabo T. M., Wilding D., Maurer M. C. (2004) Biochemistry 43, 9755–9765 [DOI] [PubMed] [Google Scholar]
- 27. Fox B. A., Yee V. C., Pedersen L. C., Le Trong I., Bishop P. D., Stenkamp R. E., Teller D. C. (1999) J. Biol. Chem. 274, 4917–4923 [DOI] [PubMed] [Google Scholar]
- 28. Weiss M. S., Metzner H. J., Hilgenfeld R. (1998) FEBS Lett. 423, 291–296 [DOI] [PubMed] [Google Scholar]
- 29. Yee V. C., Pedersen L. C., Bishop P. D., Stenkamp R. E., Teller D. C. (1995) Thromb. Res. 78, 389–397 [DOI] [PubMed] [Google Scholar]
- 30. Yee V. C., Pedersen L. C., Le Trong I., Bishop P. D., Stenkamp R. E., Teller D. C. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 7296–7300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lewis B. A., Freyssinet J. M., Holbrook J. J. (1978) Biochem. J. 169, 397–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ambrus A., Bányai I., Weiss M. S., Hilgenfeld R., Keresztessy Z., Muszbek L., Fésüs L. (2001) J. Biomol. Struct. Dyn. 19, 59–74 [DOI] [PubMed] [Google Scholar]
- 33. Bishop P. D., Teller D. C., Smith R. A., Lasser G. W., Gilbert T., Seale R. L. (1990) Biochemistry 29, 1861–1869 [DOI] [PubMed] [Google Scholar]
- 34. Oertel K., Hunfeld A., Specker E., Reiff C., Seitz R., Pasternack R., Dodt J. (2007) Anal. Biochem. 367, 152–158 [DOI] [PubMed] [Google Scholar]
- 35. Nemes Z., Marekov L. N., Steinert P. M. (1999) J. Biol. Chem. 274, 11013–11021 [DOI] [PubMed] [Google Scholar]
- 36. Lorand L., Weissmann L. B., Epel D. L., Bruner-Lorand J. (1976) Proc. Natl. Acad. Sci. U.S.A. 73, 4479–4481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Siefring G. E., Jr., Apostol A. B., Velasco P. T., Lorand L. (1978) Biochemistry 17, 2598–2604 [DOI] [PubMed] [Google Scholar]
- 38. Lorand L. (2007) FASEB J. 21, 1627–1632 [DOI] [PubMed] [Google Scholar]
- 39. Lorand L., Siefring G. E., Jr., Lowe-Krentz L. (1980) in Membrane Transport in Erythrocytes (Lassen U. V., Ussing H. H., Wieth J. O. eds) pp. 285–299, Munksgaard, Copenhagen [Google Scholar]
- 40. Liu S., Cerione R. A., Clardy J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2743–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Begg G. E., Carrington L., Stokes P. H., Matthews J. M., Wouters M. A., Husain A., Lorand L., Iismaa S. E., Graham R. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 19683–19688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Candi E., Paradisi A., Terrinoni A., Pietroni V., Oddi S., Cadot B., Jogini V., Meiyappan M., Clardy J., Finazzi-Agro A., Melino G. (2004) Biochem. J. 381, 313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Smethurst P. A., Griffin M. (1996) Biochem. J. 313, 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim S. Y., Chung S. I., Steinert P. M. (1995) J. Biol. Chem. 270, 18026–18035 [DOI] [PubMed] [Google Scholar]
- 45. Kim I. G., Gorman J. J., Park S. C., Chung S. I., Steinert P. M. (1993) J. Biol. Chem. 268, 12682–12690 [PubMed] [Google Scholar]
- 46. Ahvazi B., Boeshans K. M., Idler W., Baxa U., Steinert P. M. (2003) J. Biol. Chem. 278, 23834–23841 [DOI] [PubMed] [Google Scholar]
- 47. Andersen M. D., Faber J. H. (2011) Int. J. Mass Spectrom., in press [Google Scholar]
- 48. Ahvazi B., Kim H. C., Kee S. H., Nemes Z., Steinert P. M. (2002) EMBO J. 21, 2055–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pinkas D. M., Strop P., Brunger A. T., Khosla C. (2007) PLoS Biol. 5, e327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Komáromi I., Bagoly Z., Muszbek L. (2011) J. Thromb. Haemost. 9, 9–20 [DOI] [PubMed] [Google Scholar]
- 51. Muszbek L., Ariëns R. A., Ichinose A. (2007) J. Thromb. Haemost. 5, 181–183 [DOI] [PubMed] [Google Scholar]