Abstract
T-cell stimulating activity of Staphylococcal enterotoxin B (SEB) is an important factor in the pathogenesis of certain staphylococcal diseases including SEB mediated shock. SEB is one of the most potent superantigens known and treatment of SEB induced shock remains a challenge. We generated and characterized murine monoclonal antibodies (mAbs) to SEB in mice. We tested mAbs neutralize mitogenic effects of SEB in vitro and in vivo with T-cell proliferation assays and 2 murine models for SEB induced lethal shock (SEBILS). Epitope mapping suggests that all these mAbs recognize conformational epitopes that are destroyed by deleting the C terminus of the protein. Further site-directed mutagenesis identified potential residues involved in binding to SEB that differ between Methicillin resistant and sensitive Staphylococcus aureus strains. Only mAb 20B1 was effective as a monotherapy in treating SEBILS in HLA DR3 transgenic mice, which exhibit enhanced sensitivity to SEB. It is noteworthy that mAbs, 14G8 and 6D3 were not protective when given alone in the HLA DR3 mice but their efficacy of protection could be greatly enhanced when mAbs were co-administered simultaneously. Our data suggest combinations of defined mAbs may constitute a better treatment strategy and provide a new insight for the development of passive immunotherapy.
Keywords: Antibodies, Bacterial Toxins, Epitope Mapping, Immunology, Protein Conformation, Protein Purification, T-cell Receptor
Introduction
The Staphylococcal enterotoxins (SEs)2 comprise a family of distinct toxins (A–E) all of which are excreted by various strains of Staphylococcus aureus (S. aureus) (1). Staphylococcal enterotoxin B (SEB) is a well characterized 28 kDa protein that is related to SEC1–3 on the basis of sequence homology (1, 2). SEB is a superantigen that triggers cytokine production and T-cell proliferation by cross-linking MHC class II molecules on antigen presenting cells and T-cell receptors (TCR) (2–5). In humans, SEB can trigger toxic shock, profound hypotension and multi-organ failure. SEB is the major enterotoxin associated with non-menstrual toxic shock syndrome and accounts for the majority of intoxications that are not caused by toxic shock syndrome toxin 1 (TSST-1). In addition, some reports indicate that SEB induces an IgE response and thereby might contribute to the pathogenesis of asthma, chronic rhinitis, and dermatitis (6–9). SEB is considered a select agent. The quantities needed to produce a desired effect are much lower than with synthetic chemicals. Also SEB can be easily produced in large quantities (10).
Currently there are no therapies available for treating enterotoxin-induced shock, but clinical data suggests that immunoglobulins can alleviate disease (11). Moreover, passive administration of pooled human immunoglobulin, as well as murine and chicken antibodies (Abs) can protect against SEB induced lethal shock (SEBILS) in murine and primate animal models as well as against SEB triggered release of cytokines by SEB stimulated T-cells (12, 13). The efficacy of humoral immunity in protection against SEB was established by demonstrating an inverse relationship between susceptibility and antibody (Ab) titer (13–16) and protection in mice and non-human primates. Protection correlated with the titer of Ab to SEB (17–19). The C terminus of the protein has been proposed to be the predominant epitope recognized by human B-cells (20).
We generated and characterized murine mAbs to SEB. We investigated their toxin neutralizing efficacy in two murine models of SEBILS. Site-directed mutagenesis provide new insight into the complexity of the epitope, and neutralization studies in murine models highlight ways to decrease dose and improve efficacy.
EXPERIMENTAL PROCEDURES
S. aureus Toxins
The toxins SEA, SEB, and TSST-1 were purchased from Toxin Technology (Sarasota, FL) in accordance with CDC biosafety regulations. Recombinant full-length SEB and SEB deletion mutants were generated in compliance with 42CFR Parts 72, 73, and health and safety regulations. The commercially available SEB toxin is derived from a methicillin-sensitive S. aureus strain (MSSA).
mAbs
mAbs to SEB were generated from SEB-immunized BALB/c mice in the Hybridoma Facility of Albert Einstein College of Medicine (AECOM) as described. All mice were immunized with full-length SEB (MSSA derived) in complete Freund adjuvans (CFA). The mouse with the highest Ab titer to SEB was selected for spleen harvest and hybridoma generation. Hybridoma supernatants were screened for reactivity to SEB by ELISA, with positive reactivity being defined as absorbance 3-fold higher than background. Four mAbs, 20B1, 14G8, 6D3, and 4C7 were selected and used in this study. Specificity of mAb for SEB was determined by Western blot according to standard methods with purified SEA, SEB, and TSST-1.
T-cell Proliferation and Cytokine Assays
T-cells were isolated from donor blood using RosetteSep CD4+ T-cell enrichment mixture (Stemcell Tech) and T- cell proliferation was measured using the ViaLight HS Cell Proliferation kit (Cambrex BioScience), both according to manufacturer's instructions. Briefly, T-cells (5 × 104/ well) were stimulated in 96 well culture plates with 100 pm of purified SEB (Toxin Technology). SEB-specific mAbs (500 nm) were added concurrently with SEB. Cells were incubated at 37 °C with 10% CO2 for 48 and 96 h. Next, 100 μl per well of nucleotide releasing reagent was added and incubated for 10 min to lyse cells followed by 20 μl of ATP monitoring reagent. The plates were immediately read with 1s integrated read times. For cytokine induction assays, purified T-cells were mixed 1:1 with donor matched PBMCs. Supernatants were removed after 8 h of co-incubation with SEB and mAbs and measured by ELISA for human IL-2 and IFN-γ (21).
Sequence Analysis of Variable (V) Region of mAb
RNA was isolated from hybridoma culture cells with a Qiagen RNeasy kit and cDNA was prepared using Superscript II (Invitrogen). Amplification of variable regions was done by PCR using previously published primers (22). The resulting amplification products were gel purified and sequenced in both directions using M13 primers. Sequence was analyzed using BLAST 2 sequence and amino acid sequence was generated using the program “Translate” from ExPASy proteomic server. The sequences obtained for heavy and light chain V regions were further analyzed for homologous germline variable region genes in the database using IMGT (International ImMunoGeneTics Information System) software program. The AID generated SHM of Immunoglobulin variable (V) regions was analyzed by SHM tool webserver (23).
Sequence Analysis and Deletion Mutation Analysis
Sequence analysis of SEB gene in clinical MSSA and methicillin-resistant (MRSA) S. aureus isolates was performed by isolating DNA by Qiagen DNeasy blood and tissue kit (Qiagen) according to manufacturer's instructions. PCR amplification of the SEB gene was done using specific primers (SEB-for 5′-GAGAGTCAACCAGATCCTAA-3′ and SEB-rev 5′-GCAGGTACTCTATAAGTGCCTGC). Purified PCR products were ligated into TOPO-TA cloning vector (Invitrogen) and transformed in Top-10 E. coli competent cells and purified by standard methods for sequencing. Sequences were aligned in ClustalW with SEB gene sequence of S. aureus (M11118).
Purification of SEB
Full-length SEB gene from MRSA and MSSA encoding the residues 1–239 and SEB deletion mutants 1–7 were subcloned into H-MBP-T vector (24) using the primers shown in the supplemental materials. H-MBP-T-SEB plasmid was then transformed into Escherichia coli BL-21(DE3) Codon Plus (Stratagene) cells for protein expression. Cells were grown for ∼18 h at 15 °C in LB media after inducing with 0.5 mm IPTG at 0.6 OD. Cells were harvested and re-suspended in 20 mm Tris, pH 7.5 and lysed with 1× BugBuster. The clear supernatant was incubated with 5 ml of Talon affinity resin (Clontech) for 1 h. The resin was washed with the lysis buffer and the fusion protein was eluted with the lysis buffer supplemented with 200 mm imidazole. The eluted protein was digested with thrombin overnight at 4 °C to cleave the H-MBP fusion tag and the excess imidazole was removed by dialysis into 20 mm Tris, pH 7.5. The fusion tag and other impurities were removed by using a HiTrap Q Sepharose ion-exchange column (GE HealthCare). The fractions, which contained SEB, were pooled and passed through a size exclusion column pre-equilibrated with buffer (20 mm Tris, pH 7.5) to remove high molecular weight soluble aggregates. The protein was found to be >99% pure by SDS-PAGE. Similarly, all other deletion mutants were cloned into H-MBP-T vector and expressed and purified as mentioned above. Full-length SEB, mutant-1 and mutant-2 proteins were successfully expressed as soluble fraction, however mutants 3–7 expressed as insoluble fraction.
Amino Acid Substitutions of SEB by Site-directed Mutagenesis
Selected amino acids residues on SEB were mutated by site-directed mutagenesis using Quickchange XL Site-directed Mutagenesis kit (Stratagene, La Jolla, CA). Based on computer-assisted modeling, we gave precedence to positions where the residues are hydrogen bonded between the backbone C-terminal residues. Fig. 10, A and B shows the expanded view of the β-sheet formed by the three strands. To avoid disrupting the overall folding of SEB, 7 AA positions were mutated to alanine, 135-Arg, 137-Phe, 186-Tyr, 188-Lys, 229-Lys, 231-Glu, 233-Tyr. We also generated mutant-MRSA by adding an extra residue (T) at base position 703. PCR primers were designed using QuickChange® Primer Design Program and PCR was conducted according to manufacturer's instructions. Purified PCR products of mutated clones were ligated into H-MBP-T vector and transformed into Escherichia coli XL-10 gold cells. Substitution of amino acids in all mutant constructs was confirmed by sequencing. Expression and purification of mutant SEBs were done as described above.
FIGURE 10.
Schematic representation of the potential residues recognized by SEB specific mAbs 20B1, 14G8, 6D3, and 4C7. All mAbs recognize non-continuous residues that are likely to contribute to conformational epitopes. A, schematic illustration of the three-dimensional structure of SEB recognizing potential residues of mAbs. B, schematic diagram of expanded view of the β-sheet formed by the three strands, which could disrupt by deleting C-terminal residues. C, surface plot of SEB shows mutated residues (red color) which are distinct from D the MHC surface (rotating 180 degrees around vertical axis) shows in cyan (residues 43, 44, 45, 46, 47, 65, 67, 89, 92, 94, 96, 98, 115, 209, 211, 215) and TCR surface in green (residues 18, 19, 20, 22, 23, 26, 60, 90, 91, 177, 178, and 210).
SDS-PAGE and Western Blotting
The crude induced and uninduced lysates of SEB, mutant 1–3 and single point mutation proteins were dissolved in 30 μl of sample loading buffer and boiled for 10 min. After centrifugation for 30 s, the proteins were resolved on a 10% SDS-polyacrylamide gel under denaturaing condition and stained with Coomassie Brilliant Blue R-250. For immunoblotting, the proteins were separated on a 10% SDS-polyacrylamide gel, and the fractionated proteins were transferred from the gel onto the PVDF membrane (Millipore) in a semi-dry transblot apparatus. The membrane was blocked in blocking buffer (1× PBS, 0.05% Tween 20, 5% milk) for 2 h. The blots were washed and incubated with 1:20,000 dilution of 10 μg/μl concentration mAbs (20B1 or 14G8 or 6D3) for 45 min. Later, the blots were washed twice in PBST and one in PBS and further incubated for 45 min with HRP (horseradish peroxidase)-conjugated anti-mouse IgG (1:10,000). After washing, development was performed by chemiluminescence method according to manufacturer's instructions (Thermo Scientific).
Further binding to mutant proteins and C-terminal decapeptide were investigated under native conditions using dot blot analysis. Briefly 2 μg of synthesized 10-mer peptide (Genscript Corporation), SEB and the mutant-1 and 2 protein were spotted onto the nitrocellulose membrane and dried for 10 min. Membranes were further blocked by soaking in blocking buffer for 2 h. Membranes were washed with PBST twice and incubated with 1:10,000 dilution of 10 μg/μl concentration mAbs (20B1 or 14G8 or 6D3) for 45 min. Blots were further washed with PBST twice and incubated with HRP-conjugated anti-mouse IgG1 (1:10,000) and developed as before.
ELISA
Standard ELISA to measure SEB concentration was performed as described (21). To establish relative affinity of mAbs decreasing levels of mAb (0.1–0.001 μg) as well as decreasing levels of SEB toxin (0.1 and 0.001 μg) were used in ELISA assay. ELISA was performed with WT-SEB and purified SEB mutants protein (1 and 2) and point mutation proteins by coating the plate with purified protein, followed by unlabeled mAbs 20B1 or 14G8 or 6D3 or 4C7, which further binds to AP-conjugated anti-mouse IgG1 and was developed by PNPP tablets.
A modified competition ELISA was done to determine if two mAbs could bind to SEB simultaneously. This assay involved coating the plate with anti-IgG1 Ab, followed by unlabeled SEB specific mAb (mAbs 20B1 or 6D3 or 14G8 or 4C7) and SEB Ag. After washing another mAb (mAbs 14G8 or 6D3 or 20B1 or 4C7) was added and incubated for 1 h and further captured with a labeled anti-mouse IgG1. Alternatively, this ELISA was also performed with directly labeled mAbs.
Animal Experiments
All animal experiments were carried out with the approval of the Animal Institute Committee (AIC), in accordance with the rules and regulations set forth by the AECOM AIC. Protective efficacy of mAbs was tested in 2 murine models for SEBILS. BALB/c mice, injected intraperitoneal with 25 mg of d-galactosamine in PBS, followed by 20 μg of purified SEB (Toxin technology) die with 48 h. Transgenic mice expressing HLA-DR3 in the absence of endogenous MHC class II (a generous gift of Dr. David Chella, Mayo Clinic) were injected intraperitoneal with two doses of 50 μg of SEB 48 h apart and die within 4–5 days. To test protective efficacy, mice were injected intraperitoneal once with different doses of mAbs 20B1, 14G8, 6D3, and 4C7, or in combinations 10 min prior to administration of SEB. Control mice were treated with PBS, isotype-specific mAb 18B7 or NSO ascites, which was made by injecting mice with the myeloma cell partner NSO and thus provides an ascites control without specific antibodies. Murine blood was obtained from retro-orbital bleeding at 2, 8, and 24 h post-toxin injection according to animal institute guidelines as outlined by AIC. Serum was separated by centrifugation from clotted blood at 3000 rpm × 10 min and frozen prior to measurement by ELISA.
RESULTS
Generation of mAbs to SEB
All mice immunized with full-length SEB (MSSA-derived) in CFA responded to immunization. Eleven hybridomas were successfully stabilized after two soft agar cloning steps that allowed selection for efficient Ab producers with strong binding to SEB. To identify good candidates that could be further developed as potential therapeutic reagents, hybridomas were characterized for isotype and protective efficacy in vivo in BALB/c mice co-injected with SEB and d-galactosamine (Table 1). d-Galactosamine potentiates the SEB effect in mice, which by nature are resistant to SEB. These experiments identified 3 mAbs that conveyed protection, 5 mAbs that conveyed partial protection and 3 mAbs that exhibited no protection against SEBILS. We focused on four IgG1 mAbs (20B1, 6D3, 14G8, and 4C7), which showed different degrees of protection. Their respective hybridomas had good in vitro growth parameters. Furthermore, IgGs have a long serum half-life time, which makes them suitable candidates for in vivo application.
TABLE 1.
List of SEB-specific mAbs and their efficacy to protect against SEBILS in vivo
| mAb | Isotype | Protection in vivo (BALB/c) |
|---|---|---|
| 20B1 | IgG1 | 100% |
| 6D3 | IgG1 | 40–60% |
| 3B4 | IgM | 100% |
| 10F1 | IgA | 100% |
| 14G8 | IgG1 | 0% |
| 14B9 | IgG2a | 60% |
| 11B4 | IgG2a | 60% |
| 17C12 | IgG2a | 60% |
| 4D4 | IgG1 | 20% |
| 12A1 | IgG1 | 20% |
| 4C7 | IgG1 | 0% |
Characterization of mAbs to SEB: Specificity
Specificity of mAbs for SEB was evaluated by their binding to SEA, SEB, and TSST-1. Western blot analysis showed that mAbs 20B1 14G8, 4C7, and 6D3 bound to SEB but not to SEA or TSST-1 (Fig. 1).
FIGURE 1.

Western blot analysis of mAbs 20B1, 14G8, 6D3, and 4C7 shows specificity of mAbs for SEB and not for SEA and TSST.
SEB Sequence from Clinical Isolates
Sequence analysis of SEB genes derived from 9 MRSA and 3 MSSA clinical isolates was performed and compared with the SEB sequence of MSSA strain M11118. An additional nucleotide was found at position 703 in all MRSA but not in any MSSA strain. This addition results in three amino acid changes at positions 235, 236, and 238 (tyrosine-threonine, asparagine-threonine, and glutamine-lysine) (Fig. 2). Multilocus sequence typing (MLST) and spa typing assigned all 9 MRSA isolates to CC8 spa7 type whereas the MSSA strains were assigned to CC5 spa2, CC8 spa 139, and CC8 spa7 type.
FIGURE 2.
Schematic of SEB sequence in MRSA and MSSA strains demonstrate the additional nucleotide thymidine found in all MRSA strains at position 703 which results in 3-aa residues change in the C-terminal part of the protein.
Ig Gene Utilization
The germ line genes encoding 3 of the 4 mAbs are shown in Table 2. These data demonstrate that each of the 3 mAbs studied were different. The heavy chain V region sequence (VH) of 14G8 and 6D3 is identical to the IGHV5–6-3*1 family member whereas the mAb 20B1 VH was identical to the sequence of IGHV5–9-3*1 family member. The light chain V sequence (VL) was different for each of the 3 mAbs and contains mutations of 5 (20B1), 4 (14G8), and 6 residues (6D3) when compared with IGKV9–124*01, IGKV5–39*01 and IGKV8–19*01 germline kappa sequences, respectively. All 3 mAbs used the same J segment genes for the VH and different J segments in the VL. To test if the activation-induced deaminase (AID) and Pol-η mediated error prone repair contributed to somatic hypermutation (SHM) observed in the VL sequence, further analysis was done using SHM tool. The results showed that 67, 40, and 90% of the mutations are in AID and Pol-η associated hotspots in the VL regions of mAbs 20B1, 14G8, and 6D3, respectively (Table 3).
TABLE 2.
Identification of germline variable region genes for the SEB-specific mAbs
TABLE 3.
Percentage of mutations located in AID and Pol η associated hotspots
WRC (W = A/T, r = A/G, Y = C/T, S = G/C and the underlined C is the mutated base.
| 20B1-VL region (6 mutations) | 14G8-VL region (10 mutations) | 6D3-VL region (10 mutations) | |||
|---|---|---|---|---|---|
| AID | Hotspot | WRC | 0 | 0 | 3 (30%) |
| GYW | 3 (50%) | 1 (10%) | 2 (20%) | ||
| Coldspot | SYC | 0 | 0 | 0 | |
| GRS | 0 | 2 (20%) | 0 | ||
| Pol η Hotspot | WA | 0 | 0 | 4 (40%) | |
| TW | 1 (16%) | 3 (30%) | 0 |
Inhibition of T-cell Proliferation and Cytokine Induction with SEB-specific mAbs
SEB acts as a potent T-cell mitogen that binds to the Vβ chain of the TCR and induces T-cell proliferation and cytokine production. Because the human MHC-II complex has the highest affinity for SEB, humans are more sensitive than mice. Therefore, neutralizing efficacy was also tested in vitro in human T-cells. The effect of SEB-specific mAbs alone or in combination on SEB induced T-cell proliferation and cytokine production in human T-cells from a normal donor was measured. MAbs 20B1, 14G8, and 6D3 each demonstrated comparable levels of inhibition of SEB induced T-cell proliferation after 48 and 96 h (Fig. 3, A and B) whereas the effect of 4C7 treatment was only half that of the positive controls. Inhibition of cytokine induction was also measured after 8 h and as expected T-cells produced less IFN-γ (Fig. 3C) and IL-2 (Fig. 3D) if treated with SEB specific mAb when compared with untreated T-cells. These assays also demonstrated comparable inhibition of IFN-γ by mAbs including 4C7. Inhibition of IL-2 excretion was less complete and not observed in mAb 4C7-treated T-cells. Enhanced inhibition of T-cell proliferation and IL-2 production could not be shown for when mAbs were used in combination, however mAb 4C7 used in combination with mAb 20B1 lessened the potent neutralizing effect of mAb 20B1.
FIGURE 3.
Inhibition of T-cell proliferation and cytokine production by treatment with SEB specific mAb 20B1, 14G8, 6D3, and 4C7 individually or in different combinations. A, SEB-induced T-cell proliferation was measured by ViaLight HS Cell Proliferation kit after 48 h (A) and 96 h (B) and inhibited in the presence of all three mAb except 4C7. IFNγ (C) and IL-2 (D) were measured by ELISA in the supernatant of SEB stimulated T-cells (n = 3 wells per condition). Cytokines were significantly (p < 0.05 by t test) lower in the presence of mAbs relative to conditions with no specific antibody. The bars represent the S.D. derived from triplicate wells from same experiment.
SEB-specific mAbs Protect Mice against SEBILS
Next, the protective efficacy of mAbs 20B1, 14G8, 6D3 and 4C7 mAbs was explored in vivo in two different models of SEBILS, one in BALB/c and the other in HLA-DR3 transgenic mice. In contrast to in vitro assays, these animal experiments demonstrated significant differences in toxin neutralization for the different mAbs as well as for combinations of mAbs. Protection also differed between the two models. Two of the four mAbs (6D3 and 20B1) demonstrated consistent levels of protection in the d-galactosamine potentiated BALB/c model (Fig. 4A). Treatment with doses of mAb 20B1 as low as 100 μg per mouse conveyed protection (Fig. 4B). Enhanced protection was observed when mAb 20B1 was given in combination with mAbs 6D3 or 14G8 in doses as low as 50 μg, which were not protective when used as monotherapy. In the BALB/c model 20B1 demonstrated superior efficacy compared with 6D3, which was less protective when used alone in HLA-DR3 (Fig. 4C). MAbs 14G8 and 4C7 treatment did not protect mice from SEBILS in either mouse model. However, mAb14G8 enhanced protection when used in combination with mAb 20B1 or 6D3 in HLA-DR3 as well as in BALB/c mice whereas 4C7 lowered the efficacy of mAb 20B1 in a manner analogous to that observed for in vitro neutralization assays. In the HLA-DR3 model combination of two non-protective mAbs resulted in 60–100% protection whereas treatment with either one of the mAb could not protect mice from SEBILS (Fig. 4D). Lastly, we also investigated the protective efficacy of mAbs in mice that were injected with MRSA-derived SEB protein. These mice died in the same time frame as those injected with MSSA derived SEB. Although these mice were protected by treatment with mAbs 20B1, efficacy was decreased as low doses of 100 μg could not convey protection whereas they did when mice were injected with MSSA-derived SEB (Fig. 5).
FIGURE 4.
Protection against SEBILS was tested in BALB/c and HLA-DR3 mice (n = 10 per group) that were injected intraperitoneal with 20 μg of SEB for BALB/c (0 h) (A and B), or 50 μg of SEB for HLA-DR3 mice (0 and 48 h) (C and D). Analysis of survival data were performed using Mantel-Cox Test. In the BALB/c model mAb 20B1 was protective at doses of 500 μg (p = <0.0001) as well as 100 μg (p = 0.0003). HLA DR-3 mice that were treated intraperitoneal with 500 μg 20B1 at the same time were 100% protected whereas all SEB-injected mice treated with PBS or up to 1000 μg of mAbs 14G8 or 6D3 (HLA/DR3) died within 6 days (p = <0.0001). In contrast, mice treated with combination of mAbs 6D3 and 14G8 survived although monotherapy with the individual mAb was not protective. Similar enhanced protection was observed in the BALB/c mouse model when 20B1 was combined either with 6D3 or 14G8. No enhanced protection was found when 4C7 was administered.
FIGURE 5.
Protection against MRSA-derived SEB protein induced lethal shock was also determined in BALB/c mice by treatment with mAb 20B1 (p = 0. 0109). n = 10 each group. Analysis of survival data were performed using Mantel-Cox Test.
SEB serum levels measured by ELISA were consistently higher in mice (both murine models), treated with mAbs compared with non-treated control mice (Fig. 6, A and B). Of note, SEB serum levels in mice correlated with protection. Treatment with one mAb did not interfere with the accurate quantification of SEB in serum but quantification could not be accurately carried out in the setting of combination therapy.
FIGURE 6.
SEB level in the serum of (A) BALB/c and (B) HLA-DR3 mice (n = 10 per group) was measured by ELISA. Note that mice injected with SEB and mAb 20B1 exhibited the highest SEB serum levels both in BALB/c and HLA/DR3 mice. Bars are averages of SEB measurements in the serum of five mice in each group and brackets denote intra-assay S.D. The experiment was repeated and yielded similar differences. Gala, galactosamine.
Mapping of SEB-specific Ab Binding Sites
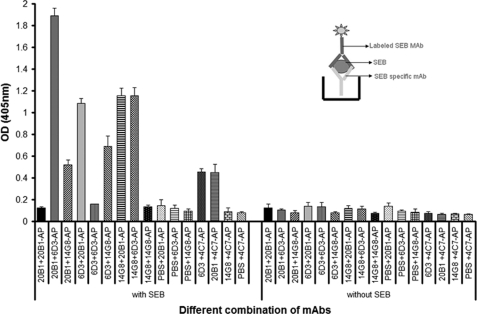
First, the capture ELISA was modified to determine if mAbs recognized distinct epitopes. The results demonstrated that mAbs 20B1, 14G8, and 6D3 each recognized different epitopes and thus can bind in any combination of two of the three mAbs simultaneously (Fig. 7) whereas mAbs 4C7 and 14G8 cannot bind simultaneously. Also apparent from these experiments was that there is only one epitope present per toxin molecule as binding inhibited additional binding of the same mAb. Competition ELISA where one mAb was kept constant while the other was varied in concentration indicated some concentration-dependent inhibition of binding in the setting of two mAbs (data not shown), which was most significant for mAbs 4C7 and 20B1.
FIGURE 7.
Capture ELISA with mAbs shows that two different SEB-specific mAbs can bind to SEB at the same time. Bars represent the average of three absorbance units at wavelength 405 nm and brackets denote intra-assay S.D. Inset, schematic diagram of ELISA, which applies to this experiment.
Deletion Mutational Analysis of SEB-specific mAbs Binding
To investigate the domains recognized by the various mAbs to SEB, mutant proteins were cloned in accordance with select agent regulations (42CFR73). Full-length SEB, SEB-MRSA, three C-terminal deletions of 5, 11, 15, residues (mutants 1–3) and mutants of aa 1–209, 1–189, 1–149, 46–149 (mutant 4–7) (Fig. 8A) were successfully expressed. All mAbs recognized the full-length SEB protein, deletion mutant-1 (5 terminal residues deleted) and the MRSA-derived SEB protein (addition of thymidine at 703). Further deletion of the C terminus (11 and 15 residues) eliminated binding by Western blot (Fig. 8C) and ELISA (Fig. 8E). Dot blot analysis comparing binding of mAbs to the decapeptide (227–236), SEB and mutant-1 demonstrated binding of the mAbs to the decapeptide (Fig. 8D) but not mutant-2, however binding efficiency was variable. Given that the C terminus distal 10 residue epitope would be too small to accommodate distinct binding of 4 mAbs we hypothesized that the actual mAb binding domain was more complex and included conformational epitopes to which distantly located residues contribute. Consequently the C terminus would be either directly part of several conformational epitopes each binding one of the mAbs or contribute indirectly to their stability.
FIGURE 8.
A, schematic diagram of SEB deletion mutants. B, SDS-PAGE shows the expression of SEB and deletion mutants (M, marker, 1, uninduced cells, 2, induced SEB, 3, induced mutant-1 (5del SEB), 4, induced mutant-2 (11 del SEB), 5, induced mutant-3 (15 del SEB). C, Western blot with mAbs and SEB deletion mutants shows that all three mAbs fail to bind to mutant 2 (11 residue deletion) and 3 (15 residue deletion). Not shown is that these mAbs also do not bind to the shorter SEB fragments. D, dot blot analysis shows binding of 10-mer peptide with all three mAbs with SEB and mutant-1 and no binding with mutant-2. The binding affinity for the 10-mer peptide was low. E, ELISA with purified SEB mutants protein (1 and 2) confirmed no binding of mutant 2 by mAbs 20B1, 14G8, and 6D3. FL, full-length.
Site-directed Mutagenesis
To identify individual amino acids that could be potentially involved in epitope structure, we focused on 7 residues based on computer-assisted three-dimensional modeling derived from crystal structure of SEB (Fig. 10) (2, 25) (Brookhaven Protein Data Bank (accession code 3SEB) that were hydrogen-bonded to the residues of the C terminus and make up a centrally located β-stranded sheet. The Tyr, Phe, and Lys side chains of these Aa are solvent exposed and therefore could interact with V region of mAbs. By site-directed mutagenesis the residues (135-Arg, 137-Phe, 186-Tyr, 188-Lys, 229-Lys, 231-Glu, 233-Tyr) were replaced by Ala and the binding of mAbs to the mutated proteins, WT SEB and MRSA-derived SEB protein was compared by ELISA (Fig. 9). These assays demonstrated that the binding of the mAbs was differentially affected by site-directed mutagenesis of these residues, with the most common outcome being decreased binding relative to WT SEB. Based on decreases in binding, residues 135-R, 137-F, 186-Y, 235- and 236-T interacted with mAb 20B1 (Fig. 9A), whereas mAb 14G8 interacted with residues 135-R, 137-F, 186-Y, 188-K, 231-E, 233-Y, and 235, 236-T (Fig. 9B). The residues 135-R, 186-Y were required for the interaction with mAb 6D3 (Fig. 9C), and 135-R, 137-F, 186-Y, 188-K, and 235, 236-T were involved in the binding of mAb 4C7 (Fig. 9D). An interesting finding was that the binding of mAb 4C7 was enhanced by certain mutations. Overall, these data also support previous dot blot data that suggested enhanced binding of mAb 14G8 to the decapeptide when compared with mAb 20B1. The latter mAb uses only 235 and 236 residues in the C-terminal whereas mAb 14G8 binds also to residues 231 and 233. Consistent with a difference in neutralizing efficacy evident in animal models of SEBILS, these assays also underscored the differences of MRSA- and MSSA-derived SEB. Our findings demonstrate the complexity of SEB epitopes recognized by neutralizing and non-neutralizing antibodies.
FIGURE 9.
ELISA shows the effect of binding using different site directed mutagenesis proteins. Mutant proteins were coated in polystyrene plates at a concentration of 0.5 μg/ml. Further mAb 20B1 or 14G8 or 6D3 or 4C7 was added, detected by alkaline phosphatase (AP)-conjugated goat anti-mouse IgG1 and developed by PNPP tablets. The x-axis represents absorbance at 405 nm and y-axis represents the log of antibody concentration (in μg). Results identify different critical residues, which could interact with the individual SEB specific mAbs. For mAb 20B1 mutation of residue 135-R, 137-F, 186-Y, 235 & 236-T affected binding. The residues 135-R, 186-Y were required for the interaction with mAb 6D3. mAb 14G8 bind to residues 135-R, 137-F, 186-Y, 188-K, 231-E, 233-Y, and 235, 236-T, whereas mAb 4C7 interact with 135-R, 137-F, 186-Y, 188-K, and 235, 236-T.
DISCUSSION
We present data on four murine mAbs to SEB, which bind to conformational epitopes that are destroyed by deletion of the distal C-terminal 11 amino acids. Three of four mAbs inhibited SEB induced T-cell proliferation as well as IL-2 and IFN-γ production by human T-cells in vitro. However, when tested in murine models these mAbs differed in their protective efficacy against SEBILS. In addition, our data are the first to show that MRSA-derived SEB contains an addition in the C-terminal, which affects binding of certain protective Abs. We also demonstrated enhanced protection against SEBILS when two non-protective mAbs were combined in vivo even if they were not protective in monotherapy. Our findings support the concept that mAb combination treatment should be further investigated, even if the individual Abs are not effective as they may be useful in toxin clearance and neutralization when combined.
Sequence analysis of the V region of mAb 20B1, 14G8, and 6D3 revealed that mAbs 20B1 and 14G8 use VH genes from the same germline gene (7183 family) but differ in VL usage and the number of mutations that resulted from somatic hypermutation. Because most of them are located in WRC hotspots, these differences reflect mutation and selection. Taken together, documented differences in antigen binding sites are consistent with the observed differences in protective efficacy. Furthermore sequence analysis of the V region will facilitate the generation of single chain variable fragments (ScFv) in the future.
Several studies have shown that Abs can protect against SEBILS in diverse animal models and species (14, 15, 26–29). Although vaccination would be a very effective method to protect humans from toxins, it carries a risk, is costly, and not necessary for all people, as natural immunity could be present and effective (30, 31). Therefore, in recent years major efforts have been undertaken to develop passive immunization therapies against a variety of toxins including potential biological weapons (32). The major advantage of mAbs is that they are biochemically defined reagents that can be readily manufactured in unlimited supply. Although some mAbs have been generated for SEB, most of these studies demonstrate only efficacy or binding in vitro (33–35). In other studies mAbs were generated by vaccination with SEB fragments that recognize the MHC II or Vβ TCR binding site on SEB (13). In our study, we vaccinated mice with MSSA-derived full-length SEB.
In this study mAb protection induced by SEBILS was investigated in two animal models; BALB/c (5, 36) and HLA-DR3. There are valid concerns with using the first model to determine the efficacy of reagents for SEBILS (37), because the contribution of potentiating reagents like d-galactosamine or LPS to the pathogenesis of SEBILS is not understood and could conceivably cause death or morbidity directly or indirectly affect clearance of the toxin. In addition, the murine MHC complex binds SEB with a significantly lower affinity than human HLA and consequently mice are less susceptible to SEBILS, which makes them not an ideal model to test therapeutic reagents. Accordingly, a number of studies have proposed that the transgenic HLA-DR3 mouse model is the superior animal model for SEBILS (38–40). In this study, 100% protection was achieved in both murine models against SEBILS only with mAb 20B1. In contrast, mAb 14G8 was not protective and mAb 6D3 was partially protective only in BALB/c mice. No protection was achieved in HLA-DR3 mice administered either mAb 14G8 or 6D3, even when using high doses. In contrast, protection was achieved in both murine models when combinations of one protective and one non-protective mAb (20B1 + 14G8 or 20B1 + 6D3) or two non-protective mAbs only (14G8 and 6D3) were administered simultaneously even when lower doses were used. This is the first demonstration of enhanced protection against SEBILS in the BALB/c as well as HLA-DR3 model when two non-protective mAbs (14G8 and 6D3) are combined. Additionally our experiments with MRSA-derived SEB protein suggest that mAb 20B1 can be used for protection of both MSSA- and MRSA-derived SEB intoxication although higher doses are required for neutralization of MRSA-derived SEB.
Previous studies have proposed that the C-terminal residues constitutes the predominant epitope recognized by human polyclonal serum (20). Our studies have only partially validated these conclusions. Instead, we demonstrate that the C terminus constitutes a complex region involved in correct folding of the SEB. Binding studies with the decapeptide indicate that the C-terminal region of SEB may include some linear epitopes (particularly residues 235 and 236 for mAbs 20B1, 14G8, and 4C7), but mostly these residues are critical for maintaining the conformational structure of this region of SEB that is part of a larger conformational epitope. It is evident from the crystal structures that the C-terminal region is well folded and forms an anti-parallel β-sheet as shown in Fig. 10B (41). Previous mutational studies have demonstrated that the C-terminal region of SEB does not bind to MHC class II or TCR (3) but is critical for the conformation of the SEB molecule (42). Our studies give further support to this conclusion as the loss of the last 11 residues result in loss of mAb binding, whereas deleting the last 5 residues did not cause any loss of binding or toxicity. Presumably the conformation of epitopes is disrupted as the deletion of the last 11 residues removes a central strand from the β-sheet, which destabilizes the overall fold of SEB. This needs to be confirmed by NMR analysis.
Modified capture ELISA in this study demonstrated that 2 mAbs can bind simultaneously to SEB, which would not be expected if the epitope was solely 11 residue long linear sequence. Hence we generated point mutation SEB clones using site-directed mutagenesis and confirmed that binding of these mAbs is also affected by residues that are not in the linear part of the C-terminal region, but rather interact with the correctly folded C-terminal, thus contributing to more complex conformational epitopes of SEB. Site-directed mutagenesis identified several residues that affect binding of the individual mAbs differentially. We propose that two mAbs can bind simultaneously because they bind to secondary and tertiary conformational eptiopes in this region. This finding is relevant because mAbs administered simultaneously confer enhanced protection. Furthermore these assays confirm that each epitope is present only once on a SEB toxin molecule. The detection of an additional nucleotide at position 703 in the SEB of all clinical MRSA strains tested, and not in MSSA strains, may affect folding and Ab neutralization resulting in biological advantages that promoted its selection. Detection of toxin sequence variation is relevant because it highlights potential mechanisms of evasion of the immune response that have to be taken into consideration when passive immunotherapy and vaccination is designed.
Several Abs that recognize conformational epitopes have been described, such as the mAbs that are employed in diagnosing misfolded prion proteins (43). Conformational epitopes are inherently difficult to study. These studies highlight the need for sophisticated structural biology studies to further characterize the interaction of mAb with conformational epitopes in solution. Future studies employing NMR analysis of SEB binding to ScFv of the individual mAbs are in process to better define the epitopes of this immunodominant part of SEB. They will determine if mAb binding to SEB can promote conformational changes of SEB and destabilize the MHC-TCR-SEB trimer formation, which is critical to confer toxicity.
Clearance of toxin is an important aspect for successful toxin neutralization assay. Although earlier studies have shown that SEB is excreted renally (44), it is not known if mAb treatment can affect renal clearance. Our study indicates that in experimental SEBILS the SEB serum levels in are consistently higher in mice treated with SEB-specific mAb than in control mice. SEB serum levels differed for the individual mAbs but correlated with protective efficacy. Experiments done 50 years ago with SEB specific polyclonal sera also demonstrated prolonged clearance of SEB in blood of injected monkeys (45). At first, it is counterintuitive to think that prolonged serum life correlates with protection, but binding to SEB by mAbs may induce conformational changes and prevent further interaction with cellular receptors and or renal clearance. This mechanism could be operative even though the MHC class II and TCR binding sites on SEB are distant from the epitope that presumably binds the mAbs. mAbs 14G8 and 6D3 achieved protection to SEBILS in HLA-DR3 mice only when administered in combination and never alone, even at higher doses. Unfortunately SEB levels in mice treated with 2 mAbs cannot be accurately determined as combination of mAbs interfered with the ELISA. Cooperative binding of mAbs may induce conformational changes in the toxin thereby altering affinities (allosteric effect) or promote FCR mediated uptake of the immunocomplex, which we could not investigate with FCR knock-out mice because they exhibit inconsistent sensitivity to SEBILS. In pneumococcal pneumonia, treatment with combination of two protective mAbs also enhanced protection against the devastating effects of pneumolysin (46). Furthermore, investigators have shown that in the treatment of viral diseases including rabies and SARS, combination of mAbs against wild-type epitope and variant epitope can prevent the emergence of escape variants (47, 48). Moreover several studies have shown that targeting more than one adhesion protein with mAb in S. aureus infection can be beneficial (49, 50).
The finding that mice were better protected against SEBILS by the combination of protective and non (or less)-protective mAbs may have important implications for current FDA regulations which state that “non- or low protective mAb when used individually, fail to show efficacy would not be further considered even though they may be highly effective when used in combination against a potentially lethal disease. In the setting of intoxications, toxin clearance could be of pivotal importance and further improved by mutating the Fc portion of mAbs, which would affect FcγR binding and FcγR-mediated uptake. Future studies are warranted that will dissect these aspects of Ab mediated protection against toxins. These findings could be highly relevant for fine-tuning an old successful treatment modality and thus be applied to many diseases that are primarily caused by toxins and lack successful treatment regimens.
Supplementary Material
Acknowledgments
We thank the hybridoma facility especially Susan Buhl at AECOM for technical support, and Sergio Roa and Richard Chahwan for help with the SHM tool. We thank Dr. David Chella at the Mayo Clinic for supplying us with a breeding pair of HLA-DR3 mice.
This work was supported, in whole or in part, by National Institutes of Health Grants U54-AI057158-Lipkin and 5T32AI007506-Casadevall, the U.S. Dept. of Defense Grant W911NF0710053; and the New York Structural Biology Center (NYSBC). NYSBC is a STAR center supported by the New York State Office of Science, Technology, and Academic Research.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig./Table.
- SE
- Staphylococcal enterotoxin
- SEB
- Staphylococcal enterotoxin B
- TSST-1
- toxic shock syndrome toxin
- SEBILS
- SEB-induced lethal shock
- Ab
- antibody
- mAb
- monoclonal antibody
- FcγR
- Fc gamma receptor.
REFERENCES
- 1. Iandolo J. J., Shafer W. M. (1977) Infect. Immun. 16, 610–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Swaminathan S., Furey W., Pletcher J., Sax M. (1992) Nature 359, 801–806 [DOI] [PubMed] [Google Scholar]
- 3. Kappler J. W., Herman A., Clements J., Marrack P. (1992) J. Exp. Med. 175, 387–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Faulkner L., Cooper A., Fantino C., Altmann D. M., Sriskandan S. (2005) J. Immunol. 175, 6870–6877 [DOI] [PubMed] [Google Scholar]
- 5. Miethke T., Wahl C., Heeg K., Echtenacher B., Krammer P. H., Wagner H. (1992) J. Exp. Med. 175, 91–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bachert C., Zhang N., Patou J., van Zele T., Gevaert P. (2008) Curr. Opin. Allergy Clin. Immunol. 8, 34–38 [DOI] [PubMed] [Google Scholar]
- 7. Liu T., Kong W., Yang P., Wang B. (2006) J. Huazhong. Univ. Sci. Technolog. Med. Sci. 26, 63–67 [DOI] [PubMed] [Google Scholar]
- 8. Rossi R. E., Monasterolo G. (2004) Int. Arch. Allergy Immunol. 133, 261–266 [DOI] [PubMed] [Google Scholar]
- 9. Rossi R. J., Muralimohan G., Maxwell J. R., Vella A. T. (2004) Int. Immunol. 16, 1751–1760 [DOI] [PubMed] [Google Scholar]
- 10. Ulrich R. G., Sidell S., Taylor T. J., Wilhelmsen C. L., Franz D. R. (2001) Textbook of Military Medicine: Medical Aspects of Chemical and Biological Warfare, Borden Institute, Washington D. C [Google Scholar]
- 11. Darenberg J., Söderquist B., Normark B. H., Norrby-Teglund A. (2004) Clin Infect. Dis. 38, 836–842 [DOI] [PubMed] [Google Scholar]
- 12. Bavari S., Ulrich R. G., LeClaire R. D. (1999) J. Infect. Dis. 180, 1365–1369 [DOI] [PubMed] [Google Scholar]
- 13. Hamad A. R., Herman A., Marrack P., Kappler J. W. (1994) J. Exp. Med. 180, 615–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. LeClaire R. D., Bavari S. (2001) Antimicrob. Agents Chemother. 45, 460–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. LeClaire R. D., Hunt R. E., Bavari S. (2002) Infect. Immun. 70, 2278–2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Boles J. W., Pitt M. L., LeClaire R. D., Gibbs P. H., Ulrich R. G., Bavari S. (2003) Vaccine 21, 2791–2796 [DOI] [PubMed] [Google Scholar]
- 17. Bavari S., Dyas B., Ulrich R. G. (1996) J. Infect. Dis. 174, 338–345 [DOI] [PubMed] [Google Scholar]
- 18. Ulrich R. G. (1998) Trends Microbiol. 6, 134. [DOI] [PubMed] [Google Scholar]
- 19. Ulrich R. G., Olson M. A., Bavari S. (1998) Vaccine 16, 1857–1864 [DOI] [PubMed] [Google Scholar]
- 20. Nishi J. I., Kanekura S., Takei S., Kitajima I., Nakajima T., Wahid M. R., Masuda K., Yoshinaga M., Maruyama I., Miyata K. (1997) J. Immunol. 158, 247–254 [PubMed] [Google Scholar]
- 21. Cook E., Wang X., Robiou N., Fries B. C. (2007) Clin Vaccine Immunol. 14, 1094–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dattamajumdar A. K., Jacobson D. P., Hood L. E., Osman G. E. (1996) Immunogenetics 43, 141–151 [DOI] [PubMed] [Google Scholar]
- 23. MacCarthy T., Roa S., Scharff M. D., Bergman A. (2009) DNA Repair 8, 137–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Alexandrov A., Dutta K., Pascal S. M. (2001) BioTechniques 30, 1194–1198 [DOI] [PubMed] [Google Scholar]
- 25. Papageorgiou A. C., Tranter H. S., Acharya K. R. (1998) J. Mol. Biol. 277, 61–79 [DOI] [PubMed] [Google Scholar]
- 26. Stiles B. G., Garza A. R., Ulrich R. G., Boles J. W. (2001) Infect. Immun. 69, 2031–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Arad G., Hillman D., Levy R., Kaempfer R. (2004) Immunol. Lett. 91, 141–145 [DOI] [PubMed] [Google Scholar]
- 28. Woody M. A., Krakauer T., Stiles B. G. (1997) Vaccine 15, 133–139 [DOI] [PubMed] [Google Scholar]
- 29. DaSilva L., Welcher B. C., Ulrich R. G., Aman M. J., David C. S., Bavari S. (2002) J. Infect. Dis. 185, 1754–1760 [DOI] [PubMed] [Google Scholar]
- 30. Holtfreter S., Broker B. M. (2005) Arch. Immunol. Ther. Exp. 53, 13–27 [PubMed] [Google Scholar]
- 31. Holtfreter S., Roschack K., Eichler P., Eske K., Holtfreter B., Kohler C., Engelmann S., Hecker M., Greinacher A., Broker B. M. (2006) J. Infect. Dis. 193, 1275–1278 [DOI] [PubMed] [Google Scholar]
- 32. Casadevall A. (2002) Nat. Biotechnol. 20, 114. [DOI] [PubMed] [Google Scholar]
- 33. Alakhov V. Y., Kabanov A. V., Batrakova E. V., Koromyslova I. A., Levashov A. V., Severin E. S. (1990) Biotechnol. Appl. Biochem. 12, 94–98 [PubMed] [Google Scholar]
- 34. Alakhov V., Klinsky E., Kolosov M. I., Maurer-Fogy I., Moskaleva E., Sveshnikov P. G., Pozdnyakova L. P., Shemchukova O. B., Severin E. S. (1992) Eur. J. Biochem. 209, 823–828 [DOI] [PubMed] [Google Scholar]
- 35. Li H. Y., Yao Y. M., Shi Z. G., Dong N., Yu Y., Lu L. R., Sheng Z. Y. (2003) Shock 20, 257–263 [DOI] [PubMed] [Google Scholar]
- 36. Galanos C., Freudenberg M. A., Reutter W. (1979) Proc. Natl. Acad. Sci. U.S.A. 76, 5939–5943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rajagopalan G., Sen M. M., David C. S. (2004) Infect. Immun. 72, 6733–6737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rajagopalan G., Sen M. M., Singh M., Murali N. S., Nath K. A., Iijima K., Kita H., Leontovich A. A., Gopinathan U., Patel R., David C. S. (2006) Shock 25, 647–656 [DOI] [PubMed] [Google Scholar]
- 39. Rajagopalan G., Smart M. K., Cheng S., Krco C. J., Johnson K. L., David C. S. (2003) Tissue Antigens 62, 149–161 [DOI] [PubMed] [Google Scholar]
- 40. Cheng S., Smart M., Hanson J., David C. S. (2003) J. Autoimmun. 21, 195–199 [DOI] [PubMed] [Google Scholar]
- 41. Li H., Llera A., Tsuchiya D., Leder L., Ysern X., Schlievert P. M., Karjalainen K., Mariuzza R. A. (1998) Immunity 9, 807–816 [DOI] [PubMed] [Google Scholar]
- 42. Metzroth B., Marx T., Linnig M., Fleischer B. (1993) Infect. Immun. 61, 2445–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Caughey B. (2003) Nat. Med. 9, 819–820 [DOI] [PubMed] [Google Scholar]
- 44. Vabulas R., Bittlingmaier R., Heeg K., Wagner H., Miethke T. (1996) Infect. Immun. 64, 4567–4573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Normann S. J. (1970) J. Immunol. 104, 673–678 [PubMed] [Google Scholar]
- 46. García-Suárez Mdel M., Cima-Cabal M. D., Flórez N., García P., Cernuda-Cernuda R., Astudillo A., Vázquez F., De los Toyos J. R., Méndez F. J. (2004) Infect. Immun. 72, 4534–4540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bakker A. B., Marissen W. E., Kramer R. A., Rice A. B., Weldon W. C., Niezgoda M., Hanlon C. A., Thijsse S., Backus H. H., de Kruif J., Dietzschold B., Rupprecht C. E., Goudsmit J. (2005) J. Virol. 79, 9062–9068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Goudsmit J., Marissen W. E., Weldon W. C., Niezgoda M., Hanlon C. A., Rice A. B., Kruif J., Dietzschold B., Bakker A. B., Rupprecht C. E. (2006) J. Infect. Dis. 193, 796–801 [DOI] [PubMed] [Google Scholar]
- 49. Arrecubieta C., Matsunaga I., Asai T., Naka Y., Deng M. C., Lowy F. D. (2008) J. Infect. Dis. 198, 571–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Welsch J. A., Ram S., Koeberling O., Granoff D. M. (2008) J. Infect. Dis. 197, 1053–1061 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.