Abstract
A critical hallmark of cancer cell survival is evasion of apoptosis. This is commonly due to overexpression of anti-apoptotic proteins such as Bcl-2, Bcl-XL, and Mcl-1, which bind to the BH3 α-helical domain of pro-apoptotic proteins such as Bax, Bak, Bad, and Bim, and inhibit their function. We designed a BH3 α-helical mimetic BH3-M6 that binds to Bcl-XL and Mcl-1 and prevents their binding to fluorescently labeled Bak- or Bim-BH3 peptides in vitro. Using several approaches, we demonstrate that BH3-M6 is a pan-Bcl-2 antagonist that inhibits the binding of Bcl-XL, Bcl-2, and Mcl-1 to multi-domain Bax or Bak, or BH3-only Bim or Bad in cell-free systems and in intact human cancer cells, freeing up pro-apoptotic proteins to induce apoptosis. BH3-M6 disruption of these protein-protein interactions is associated with cytochrome c release from mitochondria, caspase-3 activation and PARP cleavage. Using caspase inhibitors and Bax and Bak siRNAs, we demonstrate that BH3-M6-induced apoptosis is caspase- and Bax-, but not Bak-dependent. Furthermore, BH3-M6 disrupts Bcl-XL/Bim, Bcl-2/Bim, and Mcl-1/Bim protein-protein interactions and frees up Bim to induce apoptosis in human cancer cells that depend for tumor survival on the neutralization of Bim with Bcl-XL, Bcl-2, or Mcl-1. Finally, BH3-M6 sensitizes cells to apoptosis induced by the proteasome inhibitor CEP-1612.
Keywords: Anticancer Drug, Caspase, Cell Death, Drug Design, Protease Inhibitor
Introduction
Apoptosis, a form of programmed cell death, is a highly conserved process in all multicellular organisms and is essential for embryonic development and adult tissue homeostasis. Deregulation of apoptosis contributes to several diseases including cancer (1). Apoptosis is primarily controlled by two major pathways, namely the death receptor (extrinsic) and the mitochondrial (intrinsic) pathways (2). The former is mediated by members of the tumor necrosis factor (TNF)6 receptor superfamily, while the latter largely depends on multiple Bcl-2 family proteins, which affect the integrity of the mitochondrial outer membrane (MOM) (3). Both pathways converge on common cysteine proteases of the caspase family, which are responsible for the execution of apoptosis (4).
The Bcl-2 family consists of anti-apoptotic and pro-apoptotic proteins. Anti-apoptotic proteins, such as Bcl-2, Bcl-XL, Bcl-w, Mcl-1, and Bfl-1 (Bcl-2A1) contain four Bcl-2 homology (BH) domains, while the pro-apoptotic members are divided into proteins with three BH domains BH1-BH3 (Bax, Bak, and Bok), and proteins with only a BH3 domain (e.g. Bim, Bad, Bik, Bmf, Bid, Noxa, and Puma) (5). Multi-domain pro-apoptotic proteins Bax and Bak are absolutely required for apoptosis (2). In response to cellular stress, they induce the release from mitochondria of apoptogenic factors such as cytochrome c, which then cooperate with APAF-1 to induce caspase-9 activation, followed by caspase-mediated apoptosis (6). BH3-only proteins act upstream of Bax and Bak and are important for the initiation of apoptosis. Importantly, the BH3 domain is essential for the killing function of pro-apoptotic proteins (7).
An important feature of the Bcl-2 proteins is that they can homo- and heterodimerize, giving rise to three competing, but not necessarily exclusive models that could explain how the balance between pro- and anti-apoptotic proteins regulates apoptosis (7). For instance, upon receiving an apoptotic signal, BH3-only proteins directly or indirectly induce Bax and Bak activation and homo-oligomerization in the MOM, which is thought to be responsible for MOM permeabilization, resulting in the release of cytochrome c and the initiation of intrinsic apoptosis. However, activated Bax and Bak still can be kept in check by binding to anti-apoptotic Bcl-2 proteins (8–10). X-ray diffraction and nuclear magnetic resonance (NMR) studies have shown that the amphipathic α-helices of pro-apoptotic proteins such as Bak or Bad BH3 domains fit into a hydrophobic pocket formed by the BH1, BH2, and BH3 domains of Bcl-2, Bcl-XL, and Mcl-1 (11). When BH3-only proteins bind to anti-apoptotic Bcl-2 proteins, multi-domain proteins Bak or Bax become free to induce apoptosis (12). BH3-only proteins Bim, Bid, and Puma can engage all Bcl-2 anti-apoptotic proteins, and are thus the most efficient killers (7). This mechanism is known as the indirect activation model (6, 13). Additionally, certain BH3-only proteins (t-Bid, Bim and potentially Puma) can directly activate Bax, and this is known as the direct activation model (14). Thus, it was recently demonstrated that the Bim-derived BH3 α-helix activates Bax through binding to a site that is distinct from the hydrophobic pocket of the anti-apoptotic proteins (13). A third model suggests that cells can be “poised for death” but survive if their anti-apoptotic proteins sequester sufficient amounts of pro-apoptotic BH3-only proteins (15).
The fact that overexpression of anti-apoptotic Bcl-2 proteins contributes to oncogenesis and drug resistance (5, 16, 17) prompted the search for antagonists of these proteins as novel anti-cancer drugs. One possible approach is to identify compounds that mimic the BH3 domain of pro-apoptotic proteins and use them to disrupt the binding of BH3-containing proteins to anti-apoptotic Bcl-2 proteins, thus enabling the free BH3-containing proteins to initiate intrinsic apoptosis. The first study supporting this concept used a constrained BH3 peptide to induce apoptosis in cancer cells and to retard the growth of transplanted leukemia (18). Since then, several non-peptidic small molecule inhibitors have been identified (11, 19, 20). To date, the most extensively studied and promising small molecule BH3 mimetic is ABT-737, which occupies the BH3 binding groove of Bcl-2, Bcl-XL, and Bcl-w with high affinity, but only binds weakly to Mcl-1 and Bfl-1 (21, 22). Although much progress has been made over the last decade, further investigation is required to generate inhibitors targeting a broad class of anti-apoptotic Bcl-2 proteins (23). This is important as both anti-apoptotic family subclasses, Bcl-2/Bcl-XL/Bcl-w and Mcl-1/Bfl-1, must be neutralized for apoptosis to occur (5, 24, 25). In this manuscript, we report on “pan-Bcl-2” inhibitor BH3-M6, a synthetic terphenyl scaffold with functional groups that mimic the nature and the spatial configuration of the key amino acids in the BH3 α-helix. BH3-M6 disrupts Bcl-2, Bcl-XL, and Mcl-1 binding to Bax, Bak, Bad, or Bim, freeing up pro-apoptotic proteins, which leads to the release of cytochrome c, activation of caspases and induction of apoptosis in a Bax- and Bim-dependent manner in human cancer cells.
EXPERIMENTAL PROCEDURES
Antibodies
Antibodies to human proteins were from the following sources: cytochrome c (BD PharMingen, San Diego, CA); Cox IV and poly(ADP-ribose) polymerase (PARP) (Roche, Indianapolis, IN); GST, Bcl-XL, Bax (N20), Bcl-2, Mcl-1 (Santa Cruz Biotechnology, Santa Cruz, CA); Bax (6A7), HA, FLAG-M2 (Sigma); Bim (Epitomics, Burlingame, CA); Bak (Millipore, Temecula, CA).
Molecular Modeling
Compound docking was carried out using the GLIDE (Grid Based Ligand Docking from Energetics) Program from Schrödinger, L.L.C (26, 27). The Jorgensen OPLS-2001 force field was applied in the GLIDE program. The optimal binding geometry for each model was obtained by utilization of Monte Carlo sampling techniques coupled with energy minimization. GLIDE uses a scoring method based on ChemScore but with additional terms added for greater accuracy. GLIDE 4.5 SP (Standard Precision mode) was used to dock each chemical structure of these compounds followed by GLIDE 4.5 XP (Extra Precision mode) docking to find probable conformational hits. An x-ray crystal structure of mouse Bcl-XL in complex with mouse Bim BH3 at 1.65 Å resolution (1PQ1.pdb) (28) was used for Bcl-XL docking, and an x-ray crystal structure of human Mcl-1 in complex with human Bim BH3 at 1.55 Å resolution (2NL9.pdb) (29) was used for Mcl-1 docking. An NMR solution structure of the human Bcl-2 in complex with an acyl-sulfonamide-based ligand (2O2F.pdb) (30) was used for Bcl-2 docking.
Cell Culture
Human and simian cells were obtained from the American Type Culture Collection. All cell culture media were supplemented with 10% fetal calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin, and maintained at 37 °C and 5% CO2. In addition, H1299 cells were supplemented with 1% sodium pyruvate, 1% HEPES, and 1.1% glucose.
Co-immunoprecipitation
HEK293T cells were co-transfected with 5 μg of pcDNA3, pCMV2, pcDNA3-HA-Bcl-XL or pCMV2-Flag-BimEL expression vectors using TransFectinTM reagent (Bio-Rad Laboratories) and, after 18 h, exposed to DMSO, BH3-M6, or TPC (terphenyl control) for 2 h at 37 °C. Cell lysates were prepared in Nonidet P-40 lysis buffer (10 mm HEPES pH 7.5, 142.5 mm KCl, 5 mm MgCl2, 1 mm EGTA, 0.2% Nonidet P-40, 2 mm Na3VO4, 2 mm phenylmethylsulphonyl fluoride (PMSF), 6.4 mg/ml p-nitrophenylphosphate, and 1X HaltTM EDTA-free protease inhibitor mixture (Pierce). Bim was immunoprecipitated from cell lysates containing 100 μg of protein with 2 μg of FLAG-M2 beads (Sigma) in 500 μl of the same lysis buffer at 4 °C overnight. Beads were washed four times with lysis buffer, boiled for 5 min in Laemmli sample buffer and analyzed by Western blotting.
A549 cells were serum-starved for 20 h and treated with TPC or different concentrations of BH3-M6 for 1 h at 37 °C. Whole cell lysates were prepared as described above and subjected to immunoprecipitation. In case of A549 cells, 230 μg protein was incubated with 1.5 μg of Bcl-XL antibody in 250 μl of Nonidet P-40 lysis buffer overnight at 4 °C. Immunoprecipitates were collected by adding 25 μl of protein A/G-agarose beads (Santa Cruz Biotechnology) for 2 h at 4 °C, followed by centrifugation for 2 min at 3,000 × g. The beads were processed as described above.
MDA-MB-468 cells expressing Bcl-XL-IRES-Bim, Bcl-2-IRES-Bim, Mcl-1-IRES-Bim, and H1299 cells were treated with TPC or different concentrations of BH3-M6 for 24 h at 37 °C. Whole cell lysates were prepared as described above and subjected to immunoprecipitation by incubating 500 μg of protein with 1.5 μg of Bcl-XL, Bcl-2, Mcl-1, or 4 μg of Bak antibodies in 250 μl of Nonidet P-40 lysis buffer overnight at 4 °C. The remainder of the procedure was the same as above.
To detect conformationally changed Bax, 20 h serum-starved A549 cells were treated with TPC or different concentrations of BH3-M6 for 1 h, whole cell lysates were prepared in CHAPS lysis buffer containing protease inhibitors, and 200 μg of protein was incubated with 2 μg Bax 6A7 antibody for 2 h at 4 °C. Then 20 μl of protein G-agarose (Millipore) was added into the reactions and incubated at 4 °C for an additional 2 h, followed by washing with the same lysis buffer, and the immunoprecipitates were immunoblotted with Bax N20 polyclonal antibody.
In Vitro Protein-protein Interactions (GST Pull-down Assay)
Whole cell lysates of HEK293T and A549 cells were prepared using Nonidet P-40 lysis buffer (see above). For in vitro binding assay, 1 μg of GST, GST-Bcl-XL, or GST-Mcl-1 fusion proteins were coupled to 40 μl of prewashed glutathione-SepharoseTM 4B beads (GE Healthcare, Piscataway, NJ) in 250 μl of Nonidet P-40 lysis buffer with continuous rocking for 1 h at room temperature. The beads were centrifuged and washed four times with cold PBS and once with Nonidet P-40 lysis buffer. DMSO, TPC, or different concentrations of BH3-M6 in 100 μl Nonidet P-40 lysis buffer were added and incubated at 4 °C for 1 h with continuous rocking. Whole cell lysates from HEK293T or A549 cells containing 500 μg of protein were added to beads and again incubated at 4 °C for 3 h with continuous rocking. The beads were then processed for Western blotting as described above.
Fluorescence Microscopy Analysis
COS-7 cells were seeded onto glass cover slips in 24-well plates and transfected with 1 μg of pEGFP-Bad expression vector alone or co-transfected with 0.5 μg of pEGFP-Bad and 0.5 μg of pcDNA3-HA-Bcl-XL using TransFectinTM reagent. After 18 h, cells were treated with DMSO, 100 μm BH3-M6, or TPC for 4 h in the presence of a caspase inhibitor (50 μm z-VAD-fmk) to prevent apoptosis. After three washes with 500 μl of cold PBS, cells were fixed with 3.7% paraformaldehyde for 5 min at room temperature. Cells were washed again three times with 500 μl of cold PBS, then permeabilized with 0.5% Triton X-100 in PBS for 3 min at room temperature. Following another wash with cold PBS, the coverslips were mounted with DAPI-containing mount media (Vector Laboratories, Burlingame, CA) and analyzed by fluorescence microscopy.
Previously Published Procedures
BH3-M6, the terphenyl BH3 α-helical mimetic, and the unsubstituted terphenyl compound TPC were prepared as previously described (31).
To determine the ability of BH3-M6 to interfere with the binding of Bak to Bcl-XL or Bim to Mcl-1 by fluorescence polarization we used FITC-labeled peptides derived from the Bak BH3 domain (GQVGRQLAIIGDDINR) (32) or the Bim BH3 domain (Ahx-DMRPEIWIAQELRRIG DEFNAYYAR) (21, 22).
Release of cytochrome c from isolated mitochondria was measured as described by Takahashi et al. (33). Cell-free caspase-3/-7 activity and the extent of PARP cleavage was determined as described previously (34). MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) and TUNEL assays were performed according to Kazi et al. (35). Bax and Bak expression levels were silenced by siRNA using a transfection protocol previously described by us (36).
RESULTS
Docking of BH3-M6 to Bcl-XL, Bcl-2, and Mcl-1
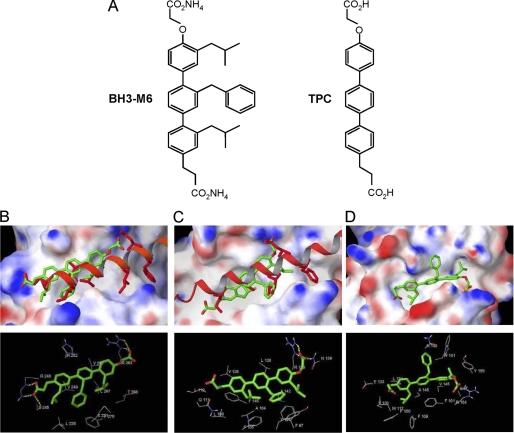
The BH3 α-helical mimic BH3-M6 and the corresponding unsubstituted terphenyl TPC (31) (Fig. 1A) were docked to Mcl-1, Bcl-XL, and Bcl-2 (Fig. 1, B–D). The computational docking studies suggest that the BH3-M6 head carboxylate group is near the Asp-67 position of human Bim BH3 α-helix and forms hydrogen bonds with Arg-263, the same human Mcl-1 residue that binds Bim Asp-67. However, BH3-M6 does not interact with the hydrophobic pockets on Mcl-1 that bind Ile-65 and Phe-69 of the Bim BH3 α-helix. The computational docking studies suggest that one of the isobutyl groups of BH3-M6 is in the position of Leu-62 of Bim and interacts with the hydrophobic pocket formed by Mcl-1 residues Met-231 (X231 in the figure, as modified Met in the crystal structure obtained from the PDB file), Val-253, Leu-267, and Phe-270. Although Bim Leu-62 binds to the same hydrophobic pocket, the Leu-62 side chain reaches into this pocket deeper than the BH3-M6 isobutyl side chain. The benzyl substituent of BH3-M6 mimics the side chain of Ile-58 of Bim interacting with Leu-235, and Val-249 of Mcl-1. The additional isobutyl lies in close proximity to Leu-235 and Val-249. Finally, the tail terminal carboxylate group forms hydrogen bonds with Ser-245 and Arg-248 that further stabilize the complex. The relative binding energy of BH3-M6 to Mcl-1 was −11.3 kcal/mol. In contrast, docking of the control TPC, which lacks hydrophobic side chains, is significantly less favorable (by 4.3 kcal/mol) than that of BH3-M6, consistent with experimental data (see below).
FIGURE 1.
Molecular docking studies of the interactions between BH3-M6 and Mcl-1, Bcl-XL, and Bcl-2. A, chemical structures of BH3-M6 and TPC. B–D, BH3-M6 docked to Mcl-1, Bcl-XL, and Bcl-2. B, crystal structure of human Bim BH3 helix bound to human Mcl-1 with the side chains of Bim Glu-55, Ile-58, Leu-62, Ile-65, Asp-67, and Phe-69, relative to the BH3-M6 position. The protein Mcl-1 is shown by its molecular surface. Area with positive electrostatic potential is colored blue and that with negative electrostatic potential colored red (the same presentation is used for proteins Bcl-XL and Bcl2). B, lower panel, residues of Mcl-1 interacting with BH3-M6. Yellow dotted lines indicate hydrogen bonds. C, crystal structure of mouse Bim BH3 helix bound to mouse Bcl-XL with the side chains of Bim Ile-90, Leu-94, Ile-97, and Asp-99, relative to the BH3-M6 position. C, lower panel, residues of Bcl-XL interacting with BH3-M6. Yellow dotted lines indicate hydrogen bonds. D, BH3-M6 docked to the original NMR structure of Bcl-2. D, lower panel, residues of Bcl-2 that interact with BH3-M6. The alignment between human and mouse Bim BH3 α-helix is as follows: Human (58-I A Q E L R R I G D E F N A Y-72) Mouse (90-I A Q E L R R I G D E F N E T-104).
The BH3-M6 head carboxylate group is in the same position as Asp-99 of the mouse Bim helix and forms hydrogen bonds with Asn-136 and Arg-139 of mouse Bcl-XL (Fig. 1C). However, the docking results suggest that BH3-M6 does not interact significantly with the hydrophobic pocket on Bcl-XL that binds Phe-101 of the Bim BH3 α-helix. The first BH3-M6 isobutyl group is near Bcl-XL residues Phe-97 and Tyr-101. The BH3-M6 benzyl group of BH3-M6 mimics the Ile-97 of Bim and interacts with the hydrophobic pocket of Bcl-XL that is formed by Phe-97, Tyr-101, and Ala-104, the same residues that bind Bim Ile-97. The other isobutyl group is in the position of Leu-94 of Bim and interacts with the hydrophobic pocket of Bcl-XL that is formed by Phe-105, Val-126, and Phe-146. The hydrophobic part of the propionic acid substituent is in the position of Ile-90 of Bim and rests near hydrophobic residues Leu-108 and Leu-112. The relative binding energy of BH3-M6 to Bcl-XL was −10.4 kcal/mol. As in the previous example, docking of the control compound TPC is significantly less favorable (by 4.1 kcal/mol) than that of BH3-M6, consistent with experimental data (see below).
The BH3-M6 head carboxylate group lies near human Bcl-2 residue Arg-104 but too far to interact through hydrogen bonding (Fig. 1D). The first BH3-M6 isobutyl side chain is inserted into the binding pocket formed by Bcl-2 residues Ala-97, Trp-141, and Tyr-199. The BH3-M6 benzyl side group is located near residues Asn-140 and Trp-141. The last BH3-M6 isobutyl side chain is in the hydrophobic pocket of Phe-109, Val-130, Leu-134, and Phe-150. Finally, the BH3-M6 carboxylate tail is projected near residues Met-112 and Glu-133. The relative binding energy of BH3-M6 to Bcl-2 was −6.3 kcal/mol. As in the previous examples, TPC docking is significantly less favorable (by 3.6 kcal/mol) than that of BH3-M6, consistent with experimental data (see below).
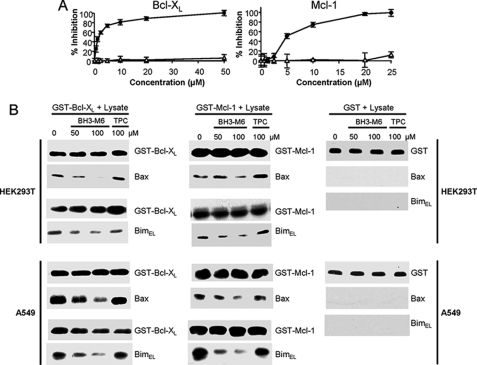
BH3-M6 Inhibits the Binding of FITC-Bak-BH3 Peptide to Bcl-XL and FITC-Bim-BH3 Peptide to Mcl-1 in Vitro and the Binding of Bcl-XL and Mcl-1 to Bax and Bim in Cell-free GST Pull-down Assays
The docking studies described above suggest that BH3-M6 binds to anti-apoptotic proteins in a similar manner to that of the α-helix of the Bim BH3 domain. We reasoned that if BH3-M6 binds anti-apoptotic proteins in this manner, then it should disrupt their binding to pro-apoptotic proteins. To evaluate this possibility, we first determined whether BH3-M6 disrupts the binding of Bcl-XL and Mcl-1 to Bak or Bim in vitro. Using fluorescence polarization assays as described by us (32), we showed that BH3-M6 inhibited the interaction between a FITC-labeled Bak-BH3 peptide and GST-Bcl-XL or a FITC-labeled Bim-BH3 peptide and GST-Mcl-1 in a dose-dependent manner with IC50 values of 1.5 or 4.9 μm, respectively, whereas control TPC showed lack of displacement (Fig. 2A). We next determined whether in lysates from HEK293T or A549 cells, BH3-M6 was able to disrupt the interaction between full-length Bax and Bim to full-length Bcl-XL and Mcl-1, respectively, in GST pull-down assays. Fig. 2B (left panels) shows that pre-incubation of the GST-Bcl-XL beads with BH3-M6 resulted in a dose-dependent inhibition of the binding of Bcl-XL to Bax and Bim in both HEK293T and A549 cell lysates. In contrast, pre-incubation of the beads with the unsubstituted control TPC did not inhibit binding of Bcl-XL to Bax or Bim (Fig. 2B, left panels). Similar results were observed with Mcl-1, where BH3-M6, but not TPC, disrupted the binding of Mcl-1 to Bax and Bim from HEK293T and A549 cells (Fig. 2B, middle panels). As expected the GST-only beads did not bind Bax or Bim from HEK293T or A549 cell lysates (Fig. 2B, right panel).
FIGURE 2.
BH3-M6, but not TPC, inhibits the binding of anti-apoptotic proteins to pro-apoptotic proteins in vitro. A, BH3-M6 blocks the binding of Bak BH3 α-helix to Bcl-XL (left) or Bim BH3 α-helix to Mcl-1 (right) as measured by fluorescence polarization assay, black circles: BH3-M6, open triangles: TPC. B, BH3-M6 inhibits the interaction of Bcl-XL or Mcl-1 with Bax and Bim as measured by GST pull-down assay. Purified GST, GST-Bcl-XL, and GST-Mcl-1 fusion proteins immobilized on glutathione-Sepharose beads were incubated for 1 h with 0, 50, and 100 μm BH3-M6 or 100 μm TPC, followed by addition of cell extracts from HEK293T or A549 cells for 1 h. Proteins associated with beads were eluted and analyzed by Western blotting with Bax and Bim antibodies. We used GST antibodies either for negative controls (in lysates incubated with GST beads) or to demonstrate equal input (in lysates incubated with GST-Bcl-XL or GST-Mcl-1 beads).
BH3-M6 Disrupts Complex Formation between Pro- and Anti-apoptotic Proteins in Intact Cells
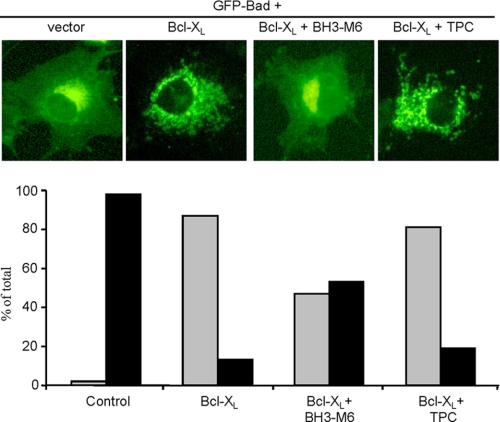
To determine whether BH3-M6 is active in intact cells, we used two methods: fluorescence microscopy for cells exogenously expressing Bcl-XL and GFP-Bad, as well as co-immunoprecipitation of endogenous proteins. First, as shown in Fig. 3, exogenous expression of GFP-Bad alone in COS-7 cells resulted in a diffuse pattern of fluorescence. In contrast, co-expression of GFP-Bad with HA-Bcl-XL resulted, as expected (37), in a punctate pattern of Bad suggesting that Bcl-XL bound Bad and localized it to the mitochondria. BH3-M6 treatment of COS-7 cells co-expressing GFP-Bad and Bcl-XL induced a diffuse pattern suggesting that BH3-M6 was able to penetrate intact cells and inhibit Bcl-XL/Bad interaction, preventing Bad from localizing into the mitochondria (Fig. 3, upper panel). Quantification of cells with diffuse versus punctate patterns indicated that BH3-M6, but not TPC, inhibited Bcl-XL-induced Bad mitochondrial localization by 50% (Fig. 3, lower panel).
FIGURE 3.
BH3-M6 inhibits the Bcl-XL interaction with Bad in intact cells. BH3-M6 inhibits Bcl-XL-induced GFP-Bad mitochondrial localization. COS-7 cells were grown in DMEM supplemented with 10% FBS and antibiotics, transfected with the indicated constructs, treated and analyzed as described under “Experimental Procedures.” The upper panel shows cells expressing pEGFP-Bad and pcDNA3-HA-Bcl-XL. The lower panel represents the quantification of the fluorescence data (black: diffuse, gray: punctate cells). At least 85 transfected cells per sample were counted in three different fields of view.
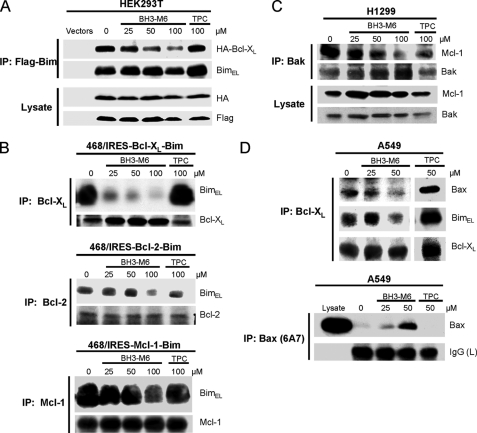
The second approach involved co-immunoprecipitation of exogenously as well as endogenously expressed proteins. First, in HEK293T cells co-transfected with HA-Bcl-XL and Flag-Bim, BH3-M6, but not TPC, inhibited the association of Flag-Bim with HA-Bcl-XL in a dose-dependent manner (Fig. 4A). Similarly, in human breast cancer MDA-MB-468 cells stably co-expressing either Bcl-XL and Bim, Bcl-2 and Bim or Mcl-1 and Bim, BH3-M6, but not TPC, disrupted the interaction of Bim with Bcl-XL, Bcl-2, and Mcl-1 in a dose-dependent manner (Fig. 4B). Taken together, the results obtained by two different approaches (Figs. 3 and 4) suggest that BH3-M6 disrupts the interactions of exogenously expressed pro- with anti-apoptotic proteins in intact cells. Next we determined whether BH3-M6 could disrupt these interactions of the corresponding endogenous proteins. Treatment of H1299 cells with BH3-M6, but not TPC, resulted in a dose-dependent inhibition of the interaction of endogenous Bak with endogenous Mcl-1 (Fig. 4C). Likewise, in A549 cells BH3-M6 was able to disrupt the interaction of Bcl-XL/Bax and Bcl-XL/Bim (Fig. 4D, upper panel). Finally, a mechanism by which freed Bax is believed to induce apoptosis is through a conformational change, which can be detected by a specific antibody. Treatment of A549 cells with BH3-M6, but not TPC, increased the levels of Bax in its pro-apoptotic conformation (Fig. 4D, lower panel).
FIGURE 4.
BH3-M6 inhibits the interaction between Bcl-XL, Bcl-2, and Mcl-1 with Bim, Bak, or Bax in intact cells. A, co-immunoprecipitation from HEK293T cells. HEK293T cells were co-transfected with HA-Bcl-XL and Flag-BimEL for 18 h. Cells were exposed to 0, 25, 50, and 100 μm of BH3-M6 or 100 μm of TPC for 2 h at 37 °C, lysed, and subjected to immunoprecipitation with anti-FLAG-M2 beads. The resulting immune complexes, as well as total lysates, were analyzed by Western blotting with the indicated antibodies. B, co-immunoprecipitation from MDA-MB-468 cells expressing Bcl-XL-IRES-BimEL, Bcl-2-IRES-BimEL, and Mcl-1-IRES-BimEL. Cells were grown in 100-mm plates and treated with 0, 25, 50, and 100 μm BH3-M6 or 100 μm TPC for 24 h at 37 °C, lysed, and subjected to immunoprecipitation with Bcl-XL, Bcl-2, and Mcl-1 antibodies. The resulting immune complexes were analyzed by Western blotting with Bim, Bcl-XL, Bcl-2, and Mcl-1 antibodies. C, co-immunoprecipitation from H1299 cells. H1299 non-small lung cancer cells were grown in RPMI 1640 medium plus 10% FBS, antibiotics and 1% sodium pyruvate, 1% HEPES, and 1.1% glucose. They were seeded in 100-mm plates and treated with 0, 25, 50, and 100 μm BH3-M6 or 100 μm TPC for 24 h at 37 °C, lysed and subjected to immunoprecipitation with Bak antibody. The resulting immune complexes were analyzed by Western blotting with the indicated antibodies. D, co-immunoprecipitation from A549 cells. A549 cells were grown in F-12K medium plus 10% FBS and antibiotics and then serum-starved for 20 h, followed by treatment with 0, 25, and 50 μm of BH3-M6 or 50 μm of TPC for 1 h at 37 °C. Cells were then lysed and subjected to immunoprecipitation with Bcl-XL antibody (upper panel) and Bax 6A7 antibody (lower panel). The resulting immune complexes were analyzed by Western blotting with the antibodies indicated on the right.
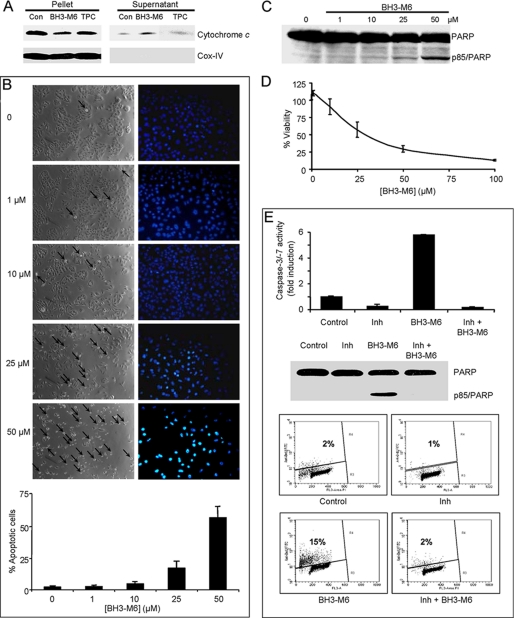
BH3-M6 Induces Cytochrome c Release, Caspase-3/-7 Activation, Cell Death, and Apoptosis
Figs. 1–4 strongly suggest that BH3-M6 disrupts the binding of pro-apoptotic to anti-apoptotic proteins in cell-free systems and in intact cells. We next determined if this disruption results in triggering apoptosis. Because cytochrome c release from mitochondria to the cytosol and subsequent activation of caspases represent key steps during intrinsic apoptosis, we first determined whether BH3-M6 affected this process in A549 cells. Fig. 5A shows that BH3-M6, but not TPC, triggered the release of cytochrome c from isolated mitochondria. In addition, BH3-M6 induced apoptosis as measured by DAPI staining (Fig. 5B) and the appearance of an 85-kDa PARP fragment in a concentration-dependent manner (Fig. 5C). Furthermore, treatment of A549 cells with BH3-M6 induced activation of caspase-3 and -7 by 2.7 ± 0.1-fold (n = 3) compared with controls (data not shown) and it inhibited cell viability as measured by MTT assay in a dose-dependent manner (Fig. 5D). Taken together, these data suggest that BH3-M6-mediated disruption of complexes between anti- and pro-apoptotic proteins releases pro-apoptotic factors to induce intrinsic apoptosis. Next, we determined whether BH3-M6 requires caspase activity to induce apoptosis. In DoHH2 cells, which express high levels of Bcl-2, the BH3-M6-induced caspase-3/-7 activity (∼5-fold), PARP cleavage and apoptosis (∼7.5-fold) were abrogated by a pan-caspase inhibitor (Fig. 5E). These results suggest that BH3-M6-mediated induction of apoptosis requires caspase activation.
FIGURE 5.
BH3-M6 induces apoptosis in a caspase-dependent manner. A, BH3-M6 releases cytochrome c from mitochondria. Mitochondria from A549 cells were incubated at 37 °C with 0, 50 μm BH3-M6, or TPC for 60 min. Cytochrome c release was determined as described under “Experimental Procedures.” B–D, A549 cells were treated with the indicated BH3-M6 concentrations for up to 72 h and then processed for various assays as described under “Experimental Procedures.” B, BH3-M6 induces apoptosis after 48 h of exposure to BH3-M6, as shown by phase images of cells (left panel) or DAPI nuclear staining (right panel). The arrows point toward cells in the process of rounding. The images from phase and DAPI are from different experiments. The graph below the images shows the percent of apoptotic cells and is the quantification of DAPI nuclear staining. Error bars indicate standard deviations of triplicates. C, BH3-M6 induces PARP cleavage after 48 h of drug treatment. D, BH3-M6 decreases the viability of A549 cells in a dose-dependent manner as measured by MTT assay after 72 h of treatment. Error bars indicate standard deviations of triplicates. E, upper panel, activation of caspase-3/-7 by BH3-M6 is blocked by a pan-caspase inhibitor. DoHH2 cells were pretreated with 50 μm Boc-d-FMK, a pan-caspase inhibitor, followed by a co-treatment with 50 μm BH3-M6 for 24 h. Caspase-3/-7 activity was determined as described under “Experimental Procedures.” Results shown represent one of two independent experiments in triplicate. Error bars indicate standard deviations of triplicates. E, middle panel, PARP cleavage was measured by Western blotting, lower panel, extent of apoptosis was measured by TUNEL assay.
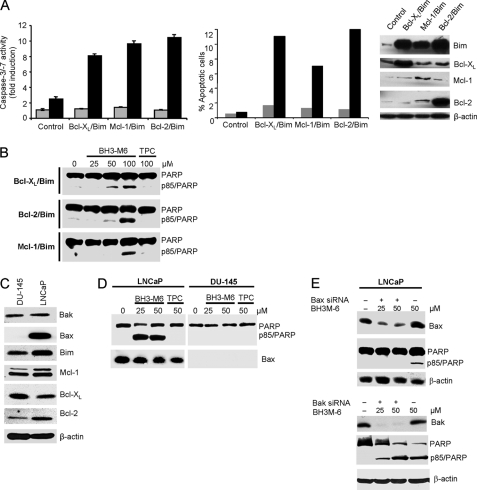
BH3-M6 Induces Apoptosis in a Bcl-XL/Bim-, Bcl-2/Bim-, and Mcl-1/Bim-dependent Manner
To investigate whether BH3-M6-induced apoptosis depends on disrupting the interaction between pro- and anti-apoptotic proteins, we used three MDA-MB-468 cell lines that depend for survival on the interaction between Bcl-XL and Bim, Bcl-2 and Bim or Mcl-1 and Bim (in these cells, Bim induces apoptosis when it is freed from Bcl-XL, Bcl-2, or Mcl-1, respectively (38)). In MDA-MB-468 control cells, BH3-M6 induced little apoptosis as measured by caspase-3/-7 activation and TUNEL assays (Fig. 6A). However, in MDA-MB-468 cells expressing Bcl-XL/Bim, Bcl-2/Bim, Mcl-1/Bim proteins, BH3-M6 induced apoptosis by 8–10-fold by caspase-3/-7 activation assay (Fig. 6A, left panel), and 7–12-fold by TUNEL assay (Fig. 6A, middle panel). Furthermore, BH3-M6, but not TPC, induced PARP cleavage in these three cell lines (Fig. 6B). Coupled with the fact that BH3-M6 disrupted the interaction of Bcl-XL/Bim, Bcl-2/Bim, Mcl-1/Bim in these three cell lines (see Fig. 4B), the results suggest that BH3-M6 disrupts the binding of anti-apoptotic proteins to Bim, which once free can induce apoptosis.
FIGURE 6.
BH3-M6 induces apoptosis in a Bim-, Bcl-2-, Bcl-XL-, and Mcl-1-dependent manner. BH3-M6 induces caspase-3/-7 activities and apoptosis in Bcl-XL/Bim-, Bcl-2/Bim-, and Mcl-1/Bim-overexpressing cells. MDA-MB-468 cells transfected with Bcl-XL-IRES-BimEL, Bcl-2-IRES-BimEL, and Mcl-1-IRES-BimEL were grown in 100-mm plates and treated with 0, 25, 50, or 100 μm BH3-M6 or 100 μm of TPC for 24 h at 37 °C. Cells were then harvested and lysed using 0.2% Nonidet P-40 lysis buffer (without protease and phosphatase inhibitors for cell-free caspase assay and with protease and phosphatase inhibitors for Western blot analysis). A, caspase-3/-7 activity was determined by incubating whole-cell extracts with caspase-3/-7 substrate and measuring free AMCs (left panel), apoptosis was quantified by TUNEL assay (middle panel), gray: TPC, black: BH3-M6. BimEL, Bcl-XL, Bcl-2, and Mcl-1 expression levels were determined by Western blotting (right panel). B, PARP cleavage by Western blotting. C, expression levels of different anti- and pro-apoptotic proteins in DU-145 and LNCaP cells as determined by Western blotting. D, BH3-M6 induces apoptosis in LNCaP human prostate cancer cells expressing Bax, but not in DU-145 human prostate cancer cells lacking detectable Bax expression. LNCaP and DU-145 cells were treated with 0, 25, and 50 μm of BH3-M6 or 50 μm of TPC for 24 h. Cells were lysed and subjected to Western blot analysis. E, depletion of Bax, but not Bak by siRNA renders LNCaP cells resistant to apoptosis. LNCaP cells were transfected with 10 nm Bax or Bak siRNA or control siRNA for 48 h, followed by 24 h of treatment with or without 25 or 50 μm BH3-M6. Cells were lysed and subjected to Western blot analysis with the indicated antibodies.
BH3-M6-induced Apoptosis Requires Bax but Not Bak
To determine whether BH3-M6 requires Bax or Bak to induce apoptosis, we treated LNCaP cells containing high levels of Bax and DU-145 cells with undetectable Bax levels with BH3-M6; both cell lines express similar amounts of Bak (Fig. 6C). BH3-M6, but not TPC, induced PARP cleavage in LNCaP cells, but not in DU-145 cells (Fig. 6D). In addition, siRNA-mediated silencing of Bax or Bak expression in LNCaP cells showed that the absence of Bax, but not Bak, prevented BH3-M6-induced PARP cleavage (Fig. 6E). These data suggest that BH3-M6-induced apoptosis requires Bax, but not Bak, in LNCaP cells.
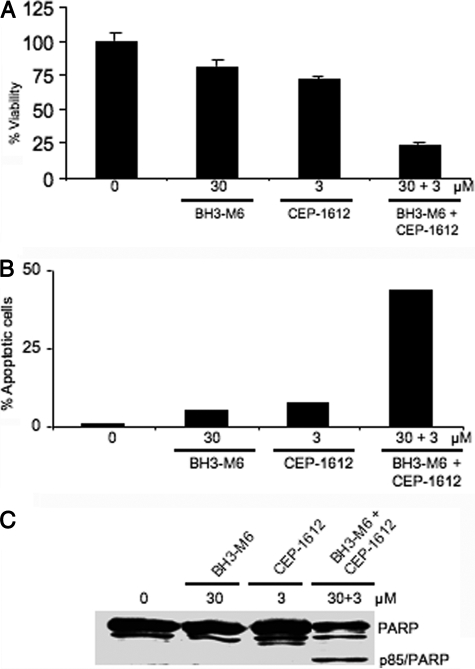
BH3-M6 Sensitizes A549 Cells to the Proteasome Inhibitor CEP-1612
To determine whether BH3-M6 sensitizes cells to apoptosis induced by other stimuli, we treated A549 cells with BH3-M6 and CEP-1612 (39), either alone or in combination. BH3-M6 and CEP-1612 alone had little effect on cell viability (20 and 28%, respectively) (Fig. 7A) and apoptosis (5 and 8%, respectively) (Fig. 7B). However, the combination treatment showed dramatic inhibition of cell viability (77%) and induction of apoptosis (44%) (Fig. 7, A and B). In addition, immunoblot analysis indicated that combination treatment, but not either drug alone induced PARP cleavage (Fig. 7C). The results from Fig. 7C were obtained after 24 h of drug treatment, whereas those from Fig. 5C were obtained after 48 h of drug treatment.
FIGURE 7.
BH3-M6 sensitizes A549 cells to the proteasome inhibitor CEP-1612 to induce apoptosis. A549 cells were treated with the indicated concentrations of either drug alone or in combination and then processed for various assays as described under “Experimental Procedures.” A, cell viability as measured by MTT assay after 72 h of treatment, error bars indicate standard deviations of triplicates. B, apoptosis as determined by TUNEL assay after 48 h of treatment. C, PARP cleavage after 24 h of drug treatment.
DISCUSSION
One of the major determinants of cell survival is the balance between the anti-apoptotic and pro-apoptotic members of the Bcl-2 family, and overexpression of anti-apoptotic proteins such as Bcl-2, Bcl-XL, and Mcl-1 contributes to cancer progression and confers resistance to chemotherapy and radiation therapy (5, 16, 17). For example, Bcl-2 overexpression is a key molecular feature of drug resistance of non-Hodgkin's lymphoma patients to chemotherapy (19, 40, 41), and high Mcl-1 levels in chronic lymphocytic leukemia patients correlate with a decreased complete response to chemotherapy (42, 43). Therefore, an important goal of cancer drug discovery is to target the anti-apoptotic pathways regulated by Bcl-2 family proteins.
The ability of anti-apoptotic proteins to preserve cancer cell survival depends on protein-protein interactions involving the binding of the amphipathic α-helical BH3 domain of pro-apoptotic proteins such as Bax, Bak, Bad, and Bim to a hydrophobic pocket formed by the BH1, BH2, and BH3 domains at the surface of anti-apoptotic proteins (44, 45). This prompted a search for BH3 domain mimics as potential novel anti-cancer drugs (23, 46). Determination of the structure of several anti-apoptotic proteins and the development of high-throughput screening assays led to the identification of many antagonists of anti-apoptotic proteins. However, except for a few such as ABT-737 and ABT-263, a thorough characterization of their specificity has not been carried out, and with some of these antagonists, off-target effects may contribute to their activity (47). Furthermore, some Bcl-2 antagonists such as ABT-737 are highly specific for one subclass of anti-apoptotic proteins (i.e. Bcl-2, Bcl-XL, and Bcl-w) but not the other subclass (i.e. Mcl-1 and Bfl-1) (47). However, apoptosis induction requires antagonizing both subclasses of anti-apoptotic proteins (5, 24, 25). In this report, we describe the development of α-helix mimetic BH3-M6 that disrupts the interactions between Bcl-2, Bcl-XL, and Mcl-1 with Bax, Bak, Bim, or Bad and leads to cytochrome c release, caspase activation, PARP cleavage, and apoptosis in human cancer cells. It induced Bax conformational change and required Bax expression and caspase activation to induce apoptosis.
While several antagonists of anti-apoptotic proteins such as Gossypol, Apogossypolone, TW-37, Obatoclax, ABT-737, ABT-263, HA1–41, Chelerythrine, Antimycin, BHI-1, and others have been identified, only a few have been thoroughly characterized for their specificity to disrupt protein-protein interactions between various class of pro- and anti-apoptotic proteins (23, 46). Furthermore, unlike other antagonists of anti-apoptotic proteins, BH3-M6 was designed based on our α-helix mimicry strategy that uses a terphenyl scaffold to spatially project functionality in a manner similar to that of two turns of the BH3 domain α-helix (32). Our computational docking studies suggest that BH3-M6 binds at the BH3 binding cleft of anti-apoptotic proteins and engages amino acid residues that are involved in binding to the BH3 α-helices of pro-apoptotic proteins. The unsubstituted control compound TPC makes few favorable contacts, confirming that the side chains attached to the terphenyl scaffold are required for interaction with Bcl-2, Bcl-XL, and Mcl-1. Consistent with our docking results, fluorescence polarization assays showed that BH3-M6 displaced the Bcl-XL- and Mcl-1-bound Bak or Bim peptides, suggesting that BH3-M6 disrupts Bcl-XL/Bak and Mcl-1/Bim interactions by binding the BH3 binding site on Bcl-XL and Mcl-1. Furthermore, using several approaches, we show that BH3-M6 disrupted Bcl-XL, Bcl-2, and Mcl-1 interactions with Bax, Bak, Bad, or Bim, suggesting that BH3-M6 is a broad pan-Bcl-2antagonist capable of antagonizing the two distinct subclasses of anti-apoptotic proteins, which is critical for full induction of apoptosis (5, 24, 25). Other published antagonists including ABT-737 and ABT-263 are not pan-Bcl-2 inhibitors and on their own may not be able to induce apoptosis in tumors where both Bcl-XL and Mcl-1 are overexpressed (23, 46). This is consistent with studies that showed that Mcl-1 confers resistance to Bcl-2-, Bcl-XL-, and Bcl-w-selective antagonists such as ABT-737, and that Obatoclax, which antagonizes both subclasses, overcomes this resistance (48). Because BH3-M6 disrupted the interaction of anti-apoptotic proteins with both multi-domain and BH3-only pro-apoptotic proteins, this compound represents an important advantage, since several mechanisms have been proposed for Bcl-2 family-mediated cancer cell survival including direct and indirect ones that involve neutralization by anti-apoptotic proteins of either multi-domain or BH3-only pro-apoptotic proteins (7, 49).
It is also important to note that some antagonists of anti-apoptotic proteins have other targets and may kill cells by mechanisms that are independent of activation of Bax and Bak. Indeed, Gossypol, Antimycin, BHI-1, HA1–41, and Chelerythrine kill wild-type and Bax/Bak-deficient fibroblasts with similar efficacy (47). In contrast, the same study showed that ABT-737 kills only wild-type, but not Bax/Bak-deficient cells, suggesting that this compound is a true BH3 mimic that induces apoptosis by specifically binding anti-apoptotic proteins and releasing pro-apoptotic proteins. The data presented in Fig. 6, A and B suggest that in cells depending for survival on anti-apoptotic proteins neutralizing Bim, BH3-M6-induced apoptosis depends on its ability to disrupt the interaction of Bim with Bcl-XL, Bcl-2, or Mcl-1. This frees up Bim allowing it to induce apoptosis, possibly by directly activating Bax and/or Bak as has been suggested previously (14). Furthermore, the data presented in Fig. 6, C–E suggest that BH3-M6-induced apoptosis depends on Bax. These results suggest that BH3-M6 induces apoptosis by freeing up Bax from anti-apoptotic proteins and/or directly inducing Bax to assume its pro-apoptotic conformation.
The ability of BH3-M6 to inhibit the binding of Bcl-2, Bcl-XL, and Mcl-1 to pro-apoptotic proteins is predicted to free up Bax, Bak, Bim, and Bad. However, these pro-apoptotic proteins can be degraded by the proteasome. Therefore, a combination of BH3-M6 and a proteasome inhibitor would be predicted to be more effective than single agent treatment. Indeed, our findings support this prediction by demonstrating that that the combination of BH3-M6 with the proteasome inhibitor CEP-1612 synergistically kills A549 cells. Our results are consistent with others that show that ABT-737 synergizes with the proteasome inhibitors bortezomib (50) or MG-132 (51).
In summary, a strategy of α-helix mimicry based on a substituted terphenyl scaffold was successfully applied to develop a pan Bcl-2 family antagonist. The disruption of protein-protein interactions between pro- and anti-apoptotic Bcl-2 family members with non-peptidic small molecules that mimic large areas of protein surfaces such as α-helices is a major milestone in the challenging field of protein-protein disruptor discovery. This is an important milestone as protein-protein interactions involving α-helices are implicated in many pathological conditions and are hence promising targets for drug discovery. Furthermore, the ability of BH3-M6 to disrupt interactions of anti-apoptotic proteins with both multi-domain and BH3-only pro-apoptotic proteins, its induction of Bax conformational change and its reliance on caspase and Bax for apoptosis induction suggest that the essential multi-domain pro-apoptotic proteins can be activated to induce apoptosis. These results, along with the fact that the ability of BH3-M6 to induce apoptosis is dependent on disrupting Bcl-XL, Bcl-2, and Mcl-1 interactions with Bim, suggest that BH3-M6 induces apoptosis by the intended mechanism.
Acknowledgments
We thank the Molecular Biology, Microscopy, and Flow Cytometry Core Facilities at the Moffitt Cancer Center for technical contributions. We are also grateful to Emily Cheng (Molecular Oncology, Department of Medicine, Washington University School of Medicine, St. Louis, MO) for the Bcl-XL-IRES-BimEL, Bcl-2-IRES-BimEL, and Mcl-1-IRES-BimEL constructs.
This work was funded, in whole or in part, by National Institutes of Health P01 Grants CA118210 and GM69850.
- TNF
- tumor necrosis factor
- MOM
- mitochondrial outer membrane
- PARP
- poly(ADP-ribose) polymerase
- BH
- Bcl-2 homology.
REFERENCES
- 1. Green D. R., Evan G. I. (2002) Cancer Cell 1, 19–30 [DOI] [PubMed] [Google Scholar]
- 2. Wei M. C., Zong W. X., Cheng E. H., Lindsten T., Panoutsakopoulou V., Ross A. J., Roth K. A., MacGregor G. R., Thompson C. B., Korsmeyer S. J. (2001) Science 292, 727–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Korsmeyer S. J. (1999) Cancer Res. 59, 1693s–1700s [PubMed] [Google Scholar]
- 4. Hengartner M. O. (2000) Nature 407, 770–776 [DOI] [PubMed] [Google Scholar]
- 5. Adams J. M., Cory S. (2007) Oncogene 26, 1324–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chipuk J. E., Green D. R. (2008) Trends Cell Biol. 18, 157–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Willis S. N., Adams J. M. (2005) Curr. Opin. Cell Biol. 17, 617–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yin X. M., Oltvai Z. N., Korsmeyer S. J. (1994) Nature 369, 321–323 [DOI] [PubMed] [Google Scholar]
- 9. Chittenden T., Harrington E. A., O'Connor R., Flemington C., Lutz R. J., Evan G. I., Guild B. C. (1995) Nature 374, 733–736 [DOI] [PubMed] [Google Scholar]
- 10. Lovell J. F., Billen L. P., Bindner S., Shamas-Din A., Fradin C., Leber B., Andrews D. W. (2008) Cell 135, 1074–1084 [DOI] [PubMed] [Google Scholar]
- 11. Pellecchia M., Reed J. C. (2004) Curr. Pharm. Des. 10, 1387–1398 [DOI] [PubMed] [Google Scholar]
- 12. Yang E., Zha J., Jockel J., Boise L. H., Thompson C. B., Korsmeyer S. J. (1995) Cell 80, 285–291 [DOI] [PubMed] [Google Scholar]
- 13. Gavathiotis E., Suzuki M., Davis M. L., Pitter K., Bird G. H., Katz S. G., Tu H. C., Kim H., Cheng E. H., Tjandra N., Walensky L. D. (2008) Nature 455, 1076–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yamaguchi H., Wang H. G. (2002) J. Biol. Chem. 277, 41604–41612 [DOI] [PubMed] [Google Scholar]
- 15. Certo M., Del Gaizo Moore V., Nishino M., Wei G., Korsmeyer S., Armstrong S. A., Letai A. (2006) Cancer Cell 9, 351–365 [DOI] [PubMed] [Google Scholar]
- 16. Reed J. C., Miyashita T., Takayama S., Wang H. G., Sato T., Krajewski S., Aimé-Sempé C., Bodrug S., Kitada S., Hanada M. (1996) J. Cell Biochem. 60, 23–32 [DOI] [PubMed] [Google Scholar]
- 17. Amundson S. A., Myers T. G., Scudiero D., Kitada S., Reed J. C., Fornace A. J., Jr. (2000) Cancer Res. 60, 6101–6110 [PubMed] [Google Scholar]
- 18. Wang J. L., Zhang Z. J., Choksi S., Shan S., Lu Z., Croce C. M., Alnemri E. S., Korngold R., Huang Z. (2000) Cancer Res. 60, 1498–1502 [PubMed] [Google Scholar]
- 19. Mohammad R. M., Goustin A. S., Aboukameel A., Chen B., Banerjee S., Wang G., Nikolovska-Coleska Z., Wang S., Al-Katib A. (2007) Clin. Cancer Res. 13, 2226–2235 [DOI] [PubMed] [Google Scholar]
- 20. Paoluzzi L., Gonen M., Gardner J. R., Mastrella J., Yang D., Holmlund J., Sorensen M., Leopold L., Manova K., Marcucci G., Heaney M. L., O'Connor O. A. (2008) Blood 111, 5350–5358 [DOI] [PubMed] [Google Scholar]
- 21. Oltersdorf T., Elmore S. W., Shoemaker A. R., Armstrong R. C., Augeri D. J., Belli B. A., Bruncko M., Deckwerth T. L., Dinges J., Hajduk P. J., Joseph M. K., Kitada S., Korsmeyer S. J., Kunzer A. R., Letai A., Li C., Mitten M. J., Nettesheim D. G., Ng S., Nimmer P. M., O'Connor J. M., Oleksijew A., Petros A. M., Reed J. C., Shen W., Tahir S. K., Thompson C. B., Tomaselli K. J., Wang B., Wendt M. D., Zhang H., Fesik S. W., Rosenberg S. H. (2005) Nature 435, 677–681 [DOI] [PubMed] [Google Scholar]
- 22. Konopleva M., Contractor R., Tsao T., Samudio I., Ruvolo P. P., Kitada S., Deng X., Zhai D., Shi Y. X., Sneed T., Verhaegen M., Soengas M., Ruvolo V. R., McQueen T., Schober W. D., Watt J. C., Jiffar T., Ling X., Marini F. C., Harris D., Dietrich M., Estrov Z., McCubrey J., May W. S., Reed J. C., Andreeff M. (2006) Cancer Cell 10, 375–388 [DOI] [PubMed] [Google Scholar]
- 23. Azmi A. S., Mohammad R. M. (2009) J. Cell Physiol. 218, 13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen L., Willis S. N., Wei A., Smith B. J., Fletcher J. I., Hinds M. G., Colman P. M., Day C. L., Adams J. M., Huang D. C. (2005) Mol. Cell 17, 393–403 [DOI] [PubMed] [Google Scholar]
- 25. Willis S. N., Chen L., Dewson G., Wei A., Naik E., Fletcher J. I., Adams J. M., Huang D. C. (2005) Genes Dev. 19, 1294–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Friesner R. A., Banks J. L., Murphy R. B., Halgren T. A., Klicic J. J., Mainz D. T., Repasky M. P., Knoll E. H., Shelley M., Perry J. K., Shaw D. E., Francis P., Shenkin P. S. (2004) J. Med. Chem. 47, 1739–1749 [DOI] [PubMed] [Google Scholar]
- 27. Halgren T. A., Murphy R. B., Friesner R. A., Beard H. S., Frye L. L., Pollard W. T., Banks J. L. (2004) J. Med. Chem. 47, 1750–1759 [DOI] [PubMed] [Google Scholar]
- 28. Liu X., Dai S., Zhu Y., Marrack P., Kappler J. W. (2003) Immunity 19, 341–352 [DOI] [PubMed] [Google Scholar]
- 29. Czabotar P. E., Lee E. F., van Delft M. F., Day C. L., Smith B. J., Huang D. C., Fairlie W. D., Hinds M. G., Colman P. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 6217–6222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bruncko M., Oost T. K., Belli B. A., Ding H., Joseph M. K., Kunzer A., Martineau D., McClellan W. J., Mitten M., Ng S. C., Nimmer P. M., Oltersdorf T., Park C. M., Petros A. M., Shoemaker A. R., Song X., Wang X., Wendt M. D., Zhang H., Fesik S. W., Rosenberg S. H., Elmore S. W. (2007) J. Med. Chem. 50, 641–662 [DOI] [PubMed] [Google Scholar]
- 31. Kutzki O., Park H. S., Ernst J. T., Orner B. P., Yin H., Hamilton A. D. (2002) J. Am. Chem. Soc. 124, 11838–11839 [DOI] [PubMed] [Google Scholar]
- 32. Yin H., Lee G. I., Sedey K. A., Kutzki O., Park H. S., Orner B. P., Ernst J. T., Wang H. G., Sebti S. M., Hamilton A. D. (2005) J. Am. Chem. Soc. 127, 10191–10196 [DOI] [PubMed] [Google Scholar]
- 33. Takahashi Y., Karbowski M., Yamaguchi H., Kazi A., Wu J., Sebti S. M., Youle R. J., Wang H. G. (2005) Mol. Cell Biol. 25, 9369–9382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kazi A., Smith D. M., Zhong Q., Dou Q. P. (2002) Mol. Pharmacol. 62, 765–771 [DOI] [PubMed] [Google Scholar]
- 35. Kazi A., Lawrence H., Guida W. C., McLaughlin M. L., Springett G. M., Berndt N., Yip R. M., Sebti S. M. (2009) Cell Cycle 8, 1940–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kazi A., Carie A., Blaskovich M. A., Bucher C., Thai V., Moulder S., Peng H., Carrico D., Pusateri E., Pledger W. J., Berndt N., Hamilton A., Sebti S. M. (2009) Mol. Cell Biol. 29, 2254–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Degterev A., Lugovskoy A., Cardone M., Mulley B., Wagner G., Mitchison T., Yuan J. (2001) Nat. Cell Biol. 3, 173–182 [DOI] [PubMed] [Google Scholar]
- 38. Kim H., Rafiuddin-Shah M., Tu H. C., Jeffers J. R., Zambetti G. P., Hsieh J. J., Cheng E. H. (2006) Nat. Cell Biol. 8, 1348–1358 [DOI] [PubMed] [Google Scholar]
- 39. Sun J., Nam S., Lee C. S., Li B., Coppola D., Hamilton A. D., Dou Q. P., Sebti S. M. (2001) Cancer Res. 61, 1280–1284 [PubMed] [Google Scholar]
- 40. Kramer M. H., Hermans J., Wijburg E., Philippo K., Geelen E., van Krieken J. H., de Jong D., Maartense E., Schuuring E., Kluin P. M. (1998) Blood 92, 3152–3162 [PubMed] [Google Scholar]
- 41. Iqbal J., Neppalli V. T., Wright G., Dave B. J., Horsman D. E., Rosenwald A., Lynch J., Hans C. P., Weisenburger D. D., Greiner T. C., Gascoyne R. D., Campo E., Ott G., Müller-Hermelink H. K., Delabie J., Jaffe E. S., Grogan T. M., Connors J. M., Vose J. M., Armitage J. O., Staudt L. M., Chan W. C. (2006) J. Clin. Oncol. 24, 961–968 [DOI] [PubMed] [Google Scholar]
- 42. Kitada S., Andersen J., Akar S., Zapata J. M., Takayama S., Krajewski S., Wang H. G., Zhang X., Bullrich F., Croce C. M., Rai K., Hines J., Reed J. C. (1998) Blood 91, 3379–3389 [PubMed] [Google Scholar]
- 43. Saxena A., Viswanathan S., Moshynska O., Tandon P., Sankaran K., Sheridan D. P. (2004) Am. J. Hematol. 75, 22–33 [DOI] [PubMed] [Google Scholar]
- 44. Youle R. J. (2007) Science 315, 776–777 [DOI] [PubMed] [Google Scholar]
- 45. Jiang X., Wang X. (2004) Annu. Rev. Biochem. 73, 87–106 [DOI] [PubMed] [Google Scholar]
- 46. Vogler M., Dinsdale D., Dyer M. J., Cohen G. M. (2009) Cell Death Differ 16, 360–367 [DOI] [PubMed] [Google Scholar]
- 47. van Delft M. F., Wei A. H., Mason K. D., Vandenberg C. J., Chen L., Czabotar P. E., Willis S. N., Scott C. L., Day C. L., Cory S., Adams J. M., Roberts A. W., Huang D. C. (2006) Cancer Cell 10, 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nguyen M., Marcellus R. C., Roulston A., Watson M., Serfass L., Murthy Madiraju S. R., Goulet D., Viallet J., Bélec L., Billot X., Acoca S., Purisima E., Wiegmans A., Cluse L., Johnstone R. W., Beauparlant P., Shore G. C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19512–19517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ewings K. E., Wiggins C. M., Cook S. J. (2007) Cell Cycle 6, 2236–2240 [DOI] [PubMed] [Google Scholar]
- 50. Paoluzzi L., Gonen M., Bhagat G., Furman R. R., Gardner J. R., Scotto L., Gueorguiev V. D., Heaney M. L., Manova K., O'Connor O. A. (2008) Blood 112, 2906–2916 [DOI] [PubMed] [Google Scholar]
- 51. Miller L. A., Goldstein N. B., Johannes W. U., Walton C. H., Fujita M., Norris D. A., Shellman Y. G. (2009) J. Invest. Dermatol. 129, 964–971 [DOI] [PubMed] [Google Scholar]