Abstract
The Swi5-Mei5 complex and its homologues are involved in specialized recombination pathways in budding and fission yeasts. Although the fission yeast homologue Swi5-Sfr1 is critical for homologous recombination repair, the budding yeast counterpart Sae3-Mei5 is meiosis-specific, interacts with Dmc1, and promotes assembly of Dmc1 on meiotic chromosomes. Here, we identify and characterize the human SWI5-MEI5 (C9orf119-C10orf78) complex. We showed that SWI5 and MEI5 form a stable complex in vitro and in vivo. The C-terminal Swi5 domain of SWI5 and the middle coiled-coil region of MEI5 dictate this conserved interaction. In addition, SWI5-MEI5 directly interacts with RAD51 in vitro. Depletion of SWI5 or MEI5 in human cells causes defects in homologous recombination repair. Finally, SWI5- or MEI5-depleted cells display enhanced sensitivity to ionizing radiation, consistent with the role of this complex in HR repair. Our results suggest that human SWI5-MEI5 has an evolutionarily conserved function in homologous recombination repair.
Keywords: DNA Damage, DNA Recombination, DNA Repair, Protein-Protein Interactions, Tumor Suppressor
Introduction
The human genome is continuously challenged by all kinds of genotoxic stress, such as ultraviolet light, ionizing radiation, and endogeneous processes, including recombination during normal immunological response and at stalled replication forks (1–3). Severe DNA lesions, such as double-strand breaks (DSBs)2 and DNA cross-links, have to be appropriately repaired for cell survival. Inefficient or inaccurate repair of these lesions often lead to genomic instability and ultimately initiate cancer development (4, 5). DSBs are repaired mainly via two parallel repair pathways: the nonhomologous end-joining pathway and homologous recombination (HR) pathway. HR is particularly important for the repair of DSBs due to its ability to restore the genetic information, whereas repair via nonhomologous end-joining may potentially lead to deletions and mutations.
The central component in the HR pathway is RAD51, which is the major recombinase in mitotic cells and also plays a critical role during meiosis. RAD51 is the human homologue of Escherichia coli RecA. It ensures high fidelity DNA repair by facilitating strand exchange between homologous DNA segments (6, 7). BRCA2 is another key protein in HR, as it mediates the loading of RAD51 onto single-stranded DNA and stabilizing RAD51 filaments (8–10). BRCA2 is encoded by a tumor suppressor gene that, when mutated, greatly elevates risks for breast and ovarian cancer. Recently, another tumor suppressor PALB2 (partner and localizer of BRCA2) was also found to associate with BRCA2 and be required for the loading of the BRCA2-RAD51 repair complex onto DNA (11). PALB2 also serves as the molecular scaffold to link BRCA2 with the BRCA1 tumor suppressor (12, 13). In addition to BRCA2/PALB2, other important HR mediators are the five RAD51 paralogues (RAD51B, RAD51C, RAD51D, XRCC2, and XRCC3), which are required for the assembly of DNA damage-induced RAD51 foci, and cell lines with defects in any of these RAD51 paralogues are defective in HR (14–16). Given the importance of HR in the maintenance of genomic stability, it is not surprising that germ line mutations in many of the HR repair components, such as BRCA1, BRCA2, and PALB2, are associated with various human genetic disorders and cancers. Two recent studies identified biallelic mutations in RAD51C, which lead to Fanconi anemia-like disorder, and a monoallelic mutation in RAD51C that is associated with increased risk of breast and ovarian cancer (17, 18).
Studies in budding and fission yeasts have identified another protein complex Swi5-Mei5, which has an important role in HR (19–24). In budding yeast, the complex is named Sae3-Mei5. The Sae3-Mei5 complex is meiosis-specific, interacts with Dmc1, the meiosis-specific RecA homologue, and promotes the assembly of Dmc1 on meiotic chromosomes (20, 21, 24). In fission yeast, the Sae3 homologue is Swi5. There are two Swi5-containing complexes in fission yeast, Swi5-Sfr1 and Swi5-Swi2, with Sfr1 and Swi2 as budding yeast Mei5 homologues. Although Swi5-Sfr1 complex participates in an Rhp51 (the fission yeast RAD51 homologue)-dependent HR pathway (22), the Swi5-Swi2 complex is required for mating type switching (22, 25).
Although these studies suggest that Swi5-Mei5 is a conserved protein complex involved in HR, the human counterparts of these proteins have never been identified and characterized. In this study, we reported the discovery of the human SWI5-MEI5 complex. We showed that human SWI5 and MEI5 form a stable complex. SWI5-MEI5 directly interacts with RAD51 and plays a critical role in HR repair.
EXPERIMENTAL PROCEDURES
Antibodies
The antibody against RAD51 was described previously (12, 26). Anti-RPA2 antibody was obtained from Abcam. The anti-Myc antibody was obtained from Santa Cruz Biotechnology. Anti-FLAG (M2) were obtained from Sigma. Anti- maltose binding protein (MBP) antibody was raised by immunizing rabbits with purified full-length MBP protein. Antisera were affinity-purified using AminoLink plus Immobilization and purification kit (Pierce).
Cell Culture, Transfection, and siRNAs
U2OS and 293T cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin. Plasmid transfection was performed using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. The sequence for RAD51 siRNA was described previously (26, 27). ON-TARGETplus siRNA Sets for SWI5 (C9orf119) and MEI5 (C10orf78) were purchased from Dharmacon. The sequences of human SWI5 siRNAs were as follows: 18, CUGAAAUGUCGCAGUGAUAUU; 19, GAACCAAGACUUACCCGAAUU; 20, AGAGUUGUAUCCAGAGUUUUU; and 21, GUUCGUAUCUGAAGGCUACUU. The sequences of human MEI5 siRNAs were: 9, AUACAAAUAGUUCCCGAAAUU; 10, AAACAAAGAUUAAACGCUGUU; 11, ACUAUGGGUUAGAUGAUAAUU; and 12, CUGAUAGUCUAGCAGGUAAUU. The siRNA transfection was performed using Oligofectamine (Invitrogen) following the manufacturer's instructions. Transfection was repeated once with an interval of 24 h to achieve maximal RNA interference effect.
RT-PCR
RT-PCR was performed by using ProtoScript M-MuLV TaqRT-PCR Kit (New England Biolabs) following the manufacturer's instructions. The primers for SWI5 were 5′-TCTCTAGGACTGAACCAAGAC-3′ (forward) and 5′-CTCGCTGGAAACTCTGTAGGTG-3′ (reverse), yielding a 439-bp PCR product. The primers for MEI5 were 5′-CAAATTGGTGAAGCAGGTTCAG-3′ (forward) and 5′-CATTGGGATACCTTCCTCTGAG-3′ (reverse), yielding a 623-bp PCR product.
Constructs
Human SWI5 (C9orf119) cDNA was obtained from OriGene Technologies (catalogue no. RG211457; RefSeq, NM_001040011). Human MEI5 (C10orf78) cDNA was obtained from human ORFeome collection (hORFeome version 5.1, GenBankTM accession no. BC020892). All cDNAs were subcloned into pDONR201 (Invitrogen) as entry clones and were subsequently transferred to gateway compatible destination vectors for the expression of N-terminal-tagged fusion protein. SFB (triple-epitope of S-protein, FLAG, and streptavidin binding peptide), Myc, MBP, and GST-tagged proteins were used in this study as described in the text. All point or deletion mutants were generated using the QuikChange site-directed mutagenesis kit (Stratagene) and verified by sequencing.
Binding Assays
For co-immunoprecipitation assays, constructs encoding SFB-tagged and Myc-tagged proteins were transiently co-transfected into 293T cells. Cells were lysed in NETN buffer (20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm EDTA, 0.5% Nonidet P-40) on ice for 30 min, cleared by centrifugation, and incubated with S protein beads for 2 h at 4 °C. Beads were washed, boiled in 2× Laemmli buffer, and separated on SDS-PAGE. Membranes were blocked in 5% milk in Tris-buffered saline/Tween buffer and then probed with antibodies as indicated. For direct binding assays, bacterially purified MBP-tagged and GST-tagged proteins were incubated together in NETN buffer containing glutatione-agarose beads for 2 h at 4 °C. Beads were washed, boiled in 2× Laemmli buffer, and separated on SDS-PAGE.
Immunostaining
Cells cultured on coverslips were treated with ionizing radiation (IR) and then allowed to recover. Cells were then washed with PBS, pre-extracted with solution containing 0.5% Triton X-100 for 3 min and fixed with 3% paraformaldehyde for 12 min. Coverslips were washed with PBS and then immunostained with primary antibodies in 5% goat serum for 30 min at room temperature. Coverslips were washed and incubated with secondary antibodies conjugated with rhodemine or FITC for 30 min. Cells were subsequently stained with DAPI for the visualization of nuclear DNA. The coverslips were mounted onto glass slides with anti-fade solution and visualized under a Nikon ECLIPSE E800 fluorescence microscope with a Nikon Plan Fluor 40× oil objective lens (numerical aperture, 1.30) at room temperature. Cells were photographed and analyzed using a SPOT camera (Diagnostic Instruments, Inc.) and Adobe Photoshop software.
Homologous Recombination Assay
A U2OS cell clone stably expressing HR reporter direct repeat GFP was described previously (28). This reporter consists of two differentially mutated GFP genes oriented as direct repeats. Expression of I-SceI endonuclease will generate a site-specific DSB between the mutated GFP genes, which when repaired by gene conversion, results in a functional GFP gene. Briefly, 2 days after transfection with indicated siRNAs, 1 × 106 U2OS direct repeat GFP cells were electroporated with 20 μg of pCBASce, an I-SceI expression vector described previously (29). Cells were harvested 2 days after electroporation and subjected to flow cytometry analysis to determine percentages of GFP-positive cells, resulted from HR repair induced by I-SceI-induced DSBs.
Cell Survival Assay
A total of 1 × 103 cells were seeded onto a 60-mm dish in triplicate. Twenty-four hours after seeding, cells were irradiated by using a JL Shepherd Mark I-68A 137Cs-irradiator at indicated doses and incubated for 14 days. Resulting colonies were fixed and stained with Coomassie Blue. Numbers of colonies were counted using a GelDoc with Quantity One software (Bio-Rad).
RESULTS
SWI5-MEI5 Is an Evolutionarily Conserved Protein Complex
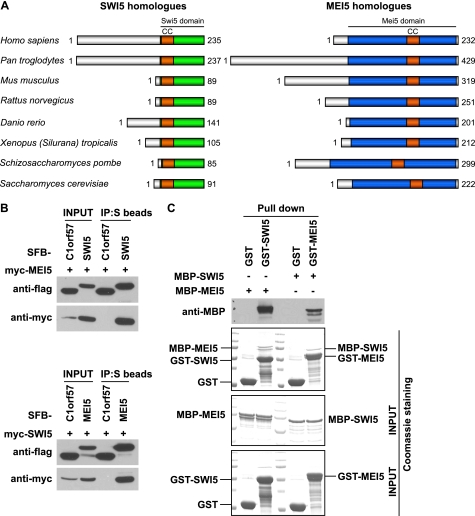
To identify human homologues of yeast Swi5 and Mei5, we used BLAST (Basic Local Alignment Search Tool) to search for human proteins with Swi5 or Mei5 domains, similar to budding yeast Sae3 or Mei5. Two uncharacterized proteins, C9orf119 and C10orf78, are found to contain Swi5 and Mei5 domains, respectively (Fig. 1A). The C9orf119 gene is located at chromosome 9q34.11. This gene encodes a protein of 235 amino acids. The C10orf78 gene is located at chromosome 10q25.1, which encodes a protein of 232 amino acids. Although the Swi5 domain sits at the C terminus of C9orf119, the Mei5 domain occupies a major portion of C10orf78. Here, we designated C9orf119 as human SWI5 and C10orf78 as human MEI5.
FIGURE 1.
SWI5 and MEI5 form an evolutionarily conserved protein complex. A, all SWI5 or MEI5 orthologues contain a Swi5 or Mei5 domain. Conserved coiled-coil motifs (CC) are indicated within both Swi5 and Mei5 domains. The accession numbers of SWI5 orthologues are: NP_001035100.1 for Homo sapiens, XP_001144446.1 for Pan troglodytes, NP_780399.1 for Mus musculus, XP_575112.2 for Rattus norvegicus, XP_002663552.1 for Danio rerio, NP_001107337.1 for Xenopus (Silurana) tropicalis, NP_595453.1 for Schizosaccharomyces pombe, and NP_011947.2 for Saccharomyces cerevisiae. The accession numbers of MEI5 orthologues are: NP_660290.3 for H. sapiens, XP_001135759.1 for P. troglodytes, NP_080653.2 for M. musculus, NP_001041321.1 for R. norvegicus, NP_001076329.1 for D. rerio, XP_002936845.1 for X. (Silurana) tropicalis, NP_595668.1 for S. pombe, and NP_015204.1 for S. cerevisiae. B, the interaction between SWI5 and MEI5 was confirmed by co-immunoprecipitation (IP) experiments. 293T cells were transfected with plasmids encoding Myc-tagged SWI5 or MEI5 together with plasmids encoding SFB-tagged MEI5 or SWI5 as indicated. SFB-tagged C1orf57 was used here as a control. C, SWI5 and MEI5 directly interact with each other. MBP-tagged and GST-tagged proteins were expressed and purified from E. coli. Pulldown assays were performed by incubating purified proteins together with glutathione-agarose beads in NETN buffer. Beads were washed, associated proteins were eluted and detected by Coomassie staining and immunoblotting using antibodies as indicated.
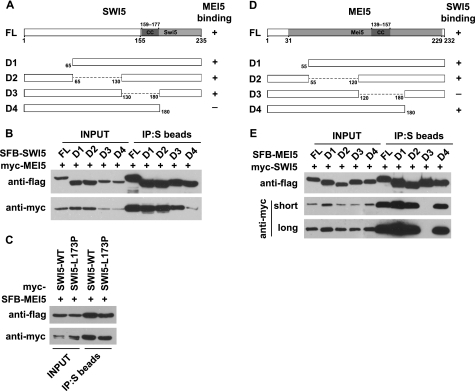
We selected several Swi5 and Mei5 homologues from different organisms and performed multiple sequence alignment using the ClustalW2 program (see Fig. 1A and supplemental Figs. 1 and 2). A conserved coiled-coil motif was identified in both Swi5 and Mei5 from different species (Fig. 1A and supplemental Figs. 1 and 2). As we will show below in Fig. 2, the coiled-coil motif of MEI5 is required for its interaction with SWI5, but the role of coiled-coil motif in SWI5 remains to be determined.
FIGURE 2.
The C-terminal Swi5 domain of SWI5 and the middle coiled-coil region of MEI5 mediate the formation of this protein complex. A, shown is a schematic representation of full-length SWI5 and the mutants used in this study. Their abilities to bind to MEI5 are indicated. FL, full-length. B, the binding of SWI5 to MEI5 is greatly impaired with the deletion of the C-terminal Swi5 domain. C, a point mutation (L173P) within the coiled-coil motif of SWI5 does not affect its binding to MEI5. D, shown is a schematic representation of full-length MEI5 and the mutants used in this study. Their SWI5-binding abilities are indicated. E, a fragment containing coiled-coil motif of MEI5 is required for the binding of MEI5 to SWI5. 293T cells were transfected with plasmids encoding Myc-tagged and SFB-tagged proteins. Co-immunoprecipitation (IP) was performed as described under “Experimental Procedures.”
The SWI5-MEI5 counterpart in budding yeast, Sae3-Mei5, is only expressed during meiosis (20, 21, 30). When we searched human expressed sequence tag (EST) database using UniGene entry Hs.259594 for SWI5 and Hs.93667 for MEI5, we found that both SWI5 and MEI5 are expressed in testis and ovary, but they are also expressed in many other tissues (data not shown). Indeed, the expression of SWI5 and MEI5 was easily detected by RT-PCR in all the nonmeiotic cell lines tested so far (data not shown), suggesting that the expression of human SWI5 and MEI5 is not restricted to meiosis.
The SWI5 and MEI5 homologues in yeasts have been shown to form a stable complex in vivo and in vitro. To check whether human SWI5 and MEI5 also interact with each other, we first performed co-immunoprecipitation experiments using epitope-tagged SWI5 and MEI5. We found that SFB-tagged SWI5 strongly interacted with Myc-tagged MEI5, and the reverse experiment confirmed this result (Fig. 1B). C1orf57 is another uncharacterized protein in the database, and we used it here as an unbiased control. It did not bind to either SWI5 or MEI5 (Fig. 1B). Interestingly, we found that the expression of tagged SWI5 or MEI5 was greatly enhanced when they were co-expressed in the cell (Fig. 1B), which indicates that SWI5 and MEI5 form a stable complex in vivo and are mutually interdependent for their stability. To verify a direct interaction between SWI5 and MEI5, we expressed and purified MBP-tagged and GST-tagged SWI5 or MEI5 from E. coli. Pulldown assay demonstrated that SWI5 and MEI5 bind directly to each other (Fig. 1C). Together, these data support that human SWI5 and MEI5 form a stable complex.
C-terminal Swi5 Domain of SWI5 and Middle Coiled-coil Region of MEI5 Dictate Conserved Association of This Complex
To further determine the interaction between SWI5 and MEI5, we generated a series of truncation or internal deletion mutants of SWI5 and MEI5 (Fig. 2, A and D). As mentioned above, there are conserved coiled-coil motifs found in both Swi5 and Mei5 domains (supplemental Figs. 1 and 2). The coiled-coil motif is often involved in dimerization, oligomerization, and protein-protein interaction. However, we did not detect any homo-dimer or homo-oligomer formation for SWI5 or MEI5 (data not shown). The coiled-coil motif in SWI5 is not required for the binding of SWI5 to MEI5 because the SWI5 mutant deleted of this coiled-coil motif still associated with MEI5 (Fig. 2B), and a point mutation of a conserved residue in this coiled-coil domain of SWI5 (L173P) also failed to disrupt the binding of SWI5 to MEI5 (Fig. 2C). The MEI5-binding domain is located at the C terminus of SWI5, which constitutes the most conservative part of Swi5 domain (Fig. 2, A and B, and supplemental Fig. 1).
The binding assays performed between SWI5 and various mutants of MEI5 indicate that a fragment of MEI5 (residues 120–180), which contains the coiled-coil motif of MEI5, is required for the binding of MEI5 to SWI5 (Fig. 2, D and E), suggesting that the coiled-coil motif of MEI5 may be involved in the formation of this conserved SWI5-MEI5 complex in humans.
SWI5-MEI5 Is Important for Homologous Recombination Repair in Human Cells
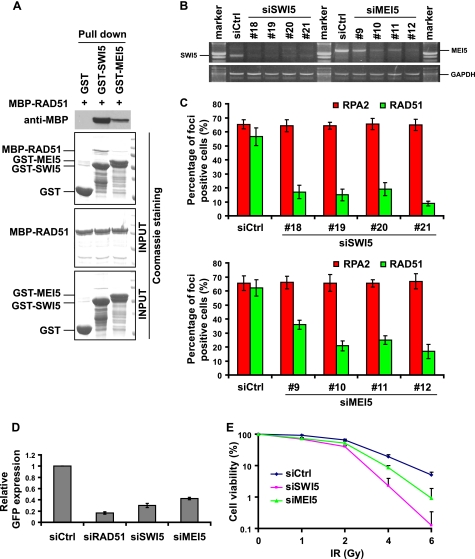
Studies in budding and fission yeasts have revealed that the Swi5 and Mei5 are necessary for HR-mediated DNA repair and meiotic recombination, probably through interacting with the recombinases Rad51 and Dmc1 (20, 22). The broad expression profile of human SWI5 and MEI5 in various tissues and cell lines suggests a potential role of human SWI5-MEI5 in HR mediated DNA repair in mitotic cells. We first checked the interaction between SWI5-MEI5 and human RAD51. GST-tagged SWI5 or MEI5 and MBP-tagged RAD51 were expressed in E. coli, purified, and used in GST pull-down assays. We repeatedly detected the binding of SWI5 or MEI5 with RAD51, with the interaction between SWI5 and RAD51 much stronger than that of MEI5 and RAD51 (Fig. 3A).
FIGURE 3.
SWI5-MEI5 directly interacts with RAD51 and is involved in homologous recombination repair. A, SWI5 and MEI5, especially SWI5, can directly bind to RAD51. B, the efficiency of down-regulating the expression of their target genes by various siRNAs was evaluated by RT-PCR. C, down-regulation of SWI5 or MEI5 greatly impairs IR-induced RAD51 foci formation without influencing RPA foci formation. Immunostaining was performed 6 h after IR treatment with the indicated antibodies. D, decreasing of SWI5 or MEI5 impairs HR repair. U2OS direct repeat GFP cells were electroporated with pCBASce plasmids (see “Experimental Procedures” for details). The percentage of GFP-positive cells was determined by flow cytometry 48 h after electroporation. The data were normalized to those obtained from cells transfected with control siRNA (set as 1.0). Means and S.E. (error bars) shown are obtained from three independent experiments. E, cells with SWI5 or MEI5 down-regulation display increased IR sensitivity. Cell survival assays were performed as described under “Experimental Procedures.” Data were presented as means and S.D. (error bars) from three different experiments. Ctrl, control; Gy, gray.
The conserved SWI5-MEI5 complex formation and the direct binding of this complex to RAD51 suggest that the human SWI5-MEI5 complex may play a role in HR repair. To test whether SWI5 and MEI5 are required in HR mediated DNA repair, sets of siRNAs specifically targeting SWI5 or MEI5 were synthesized and introduced into U2OS cells. All four SWI5-specific siRNAs worked well because they led to significant down-regulation of SWI5 transcripts as detected by RT-PCR. The down-regulation of MEI5 by its siRNAs was also noticeable, although it was not as efficient as that of SWI5 siRNAs (Fig. 3B).
It is believed that at least a fraction of HR repair is initiated via DSB end resection, which generates ssDNA overhangs rapidly bound by RPA. Subsequently, the central recombinase enzyme RAD51, with the help of its accessory factors, displaces RPA from ssDNA to form a RAD51 filament, which starts homology search and HR repair. Thus, RPA and RAD51 foci formation can be used as readouts for two different steps during HR repair. Although RPA foci formation indicates the generation of ssDNA regions after DSB induction, the formation of RAD51 foci can be used as an indicator of actively ongoing HR repair process (26, 31). We checked both RPA and RAD51 foci formation after IR treatment in the control, SWI5- or MEI5-depleted U2OS cells. We did not detect any obvious changes in RPA foci formation following SWI5 or MEI5 down-regulation; however, RAD51 foci formation was greatly impaired in cells transfected with SWI5 or MEI5 siRNAs (Fig. 3C, also see supplemental Fig. 3 for representative immunostaining images).
Moreover, we examined HR efficiency using the established direct repeat GFP reporter system (28). In agreement with the results of reduced RAD51 foci formation, the efficiency of HR repair was clearly decreased in cells with SWI5 or MEI5 down-regulation (Fig. 3D). Consistently, down-regulation of SWI5 or MEI5 by siRNAs also resulted in increased cellular sensitivity to IR (Fig. 3E). Taken together, these data support an important role of the human SWI5-MEI5 complex in HR-mediated DNA repair.
DISCUSSION
In this study, we report the identification and characterization of the human SWI5-MEI5 complex. This complex directly binds to RAD51 and promotes RAD51 focus formation following DNA damage. Together, these data support that SWI5-MEI5 is another mediator involved in homologous recombination repair in humans.
During the preparation of our article, the mouse homologues of SWI5-MEI5 (or Swi5-Sfr1) was identified (32). In that study, mouse Swi5-Sfr1 was identified as a complex required for genomic integrity with a specific role in the repair of DNA strand breaks (32). However, HR defects in Swi5−/− and Sfr1−/− embryonic stem cells were relatively mild (32). The variation in the severity of HR defect between human and mouse when SWI5-MEI5 is depleted could be due to different cells used in these studies. Alternatively, it may reflect the difference in the utilization of the SWI5-MEI5 complex in HR repair in embryonic stem cells versus adult somatic cells. Future experiments are needed to distinguish these possibilities.
The possible function of human SWI5-MEI5 in meiosis also needs to be investigated. It remains to be determined whether the human SWI5-MEI5 complex acts in meiosis and, if it does, whether it acts with DMC1, RAD51, or both. Considering that SWI5-MEI5 is the only human counterpart of the yeast Sae3-Mei5 complex, we speculate that SWI5-MEI5 may play important roles both in mitotic and meiotic homologous recombination reactions. Further experimentation on SWI5-MEI5 will reveal molecular mechanisms underlying HR process in humans, which is critical for the prevention of many human diseases including infertility and cancer.
Supplementary Material
Acknowledgments
We thank all members of the Chen laboratory, especially Wenqi Wang and Kelsey Lau, for advice and technical assistance.
This work was supported in part by grants from the National Institutes of Health (to J. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- DSB
- double-strand break
- HR
- homologous recombination
- MBP
- maltose binding protein
- IR
- ionizing radiation
- RPA
- replication protein A.
REFERENCES
- 1. Sancar A., Lindsey-Boltz L. A., Unsal-Kaçmaz K., Linn S. (2004) Annu. Rev. Biochem. 73, 39–85 [DOI] [PubMed] [Google Scholar]
- 2. Branzei D., Foiani M. (2008) Nat. Rev. Mol. Cell Biol. 9, 297–308 [DOI] [PubMed] [Google Scholar]
- 3. Yuan J., Ghosal G., Chen J. (2009) Genes Dev. 23, 2394–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jeggo P. A., Löbrich M. (2007) Oncogene 26, 7717–7719 [DOI] [PubMed] [Google Scholar]
- 5. McKinnon P. J., Caldecott K. W. (2007) Annu. Rev. Genomics Hum. Genet. 8, 37–55 [DOI] [PubMed] [Google Scholar]
- 6. Ogawa T., Yu X., Shinohara A., Egelman E. H. (1993) Science 259, 1896–1899 [DOI] [PubMed] [Google Scholar]
- 7. Sung P., Robberson D. L. (1995) Cell 82, 453–461 [DOI] [PubMed] [Google Scholar]
- 8. Lord C. J., Ashworth A. (2007) Nat. Struct. Mol. Biol. 14, 461–462 [DOI] [PubMed] [Google Scholar]
- 9. Pellegrini L., Venkitaraman A. (2004) Trends Biochem. Sci. 29, 310–316 [DOI] [PubMed] [Google Scholar]
- 10. Thorslund T., West S. C. (2007) Oncogene 26, 7720–7730 [DOI] [PubMed] [Google Scholar]
- 11. Xia B., Sheng Q., Nakanishi K., Ohashi A., Wu J., Christ N., Liu X., Jasin M., Couch F. J., Livingston D. M. (2006) Mol. Cell 22, 719–729 [DOI] [PubMed] [Google Scholar]
- 12. Sy S. M., Huen M. S., Chen J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7155–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang F., Ma J., Wu J., Ye L., Cai H., Xia B., Yu X. (2009) Curr. Biol. 19, 524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Takata M., Sasaki M. S., Tachiiri S., Fukushima T., Sonoda E., Schild D., Thompson L. H., Takeda S. (2001) Mol. Cell Biol. 21, 2858–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Deans B., Griffin C. S., O'Regan P., Jasin M., Thacker J. (2003) Cancer Res. 63, 8181–8187 [PubMed] [Google Scholar]
- 16. West S. C. (2003) Nat. Rev. Mol. Cell Biol. 4, 435–445 [DOI] [PubMed] [Google Scholar]
- 17. Meindl A., Hellebrand H., Wiek C., Erven V., Wappenschmidt B., Niederacher D., Freund M., Lichtner P., Hartmann L., Schaal H., Ramser J., Honisch E., Kubisch C., Wichmann H. E., Kast K., Deissler H., Engel C., Müller-Myhsok B., Neveling K., Kiechle M., Mathew C. G., Schindler D., Schmutzler R. K., Hanenberg H. (2010) Nat. Genet. 42, 410–414 [DOI] [PubMed] [Google Scholar]
- 18. Vaz F., Hanenberg H., Schuster B., Barker K., Wiek C., Erven V., Neveling K., Endt D., Kesterton I., Autore F., Fraternali F., Freund M., Hartmann L., Grimwade D., Roberts R. G., Schaal H., Mohammed S., Rahman N., Schindler D., Mathew C. G. (2010) Nat. Genet. 42, 406–409 [DOI] [PubMed] [Google Scholar]
- 19. Okada T., Keeney S. (2005) Curr. Biol. 15, R200–202 [DOI] [PubMed] [Google Scholar]
- 20. Hayase A., Takagi M., Miyazaki T., Oshiumi H., Shinohara M., Shinohara A. (2004) Cell 119, 927–940 [DOI] [PubMed] [Google Scholar]
- 21. Tsubouchi H., Roeder G. S. (2004) Genetics 168, 1219–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Akamatsu Y., Dziadkowiec D., Ikeguchi M., Shinagawa H., Iwasaki H. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 15770–15775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ellermeier C., Schmidt H., Smith G. R. (2004) Genetics 168, 1891–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ferrari S. R., Grubb J., Bishop D. K. (2009) J. Biol. Chem. 284, 11766–11770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jia S., Yamada T., Grewal S. I. (2004) Cell 119, 469–480 [DOI] [PubMed] [Google Scholar]
- 26. Yuan J., Chen J. (2010) J. Biol. Chem. 285, 1097–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yuan J., Chen J. (2009) J. Biol. Chem. 284, 31746–31752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Weinstock D. M., Nakanishi K., Helgadottir H. R., Jasin M. (2006) Methods Enzymol. 409, 524–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Richardson C., Moynahan M. E., Jasin M. (1998) Genes Dev. 12, 3831–3842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McKee A. H., Kleckner N. (1997) Genetics 146, 817–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang J., Huen M. S., Kim H., Leung C. C., Glover J. N., Yu X., Chen J. (2009) Nat. Cell Biol. 11, 592–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Akamatsu Y., Jasin M. (2010) PLoS Genet 6, e1001160. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.