Abstract
Bcl-2 homology domain-3 (BH3) peptides are potent cancer therapeutic reagents that target regulators of apoptotic cell death in cancer cells. However, their cytotoxic effects are affected by different expression levels of Bcl-2 family proteins. We recently found that the amphipathic tail-anchoring peptide (ATAP) from Bfl-1, a bifunctional Bcl-2 family member, produced strong pro-apoptotic activity by permeabilizing the mitochondrial outer membrane. Here, we test whether the activity of ATAP requires other cellular factors and whether ATAP has an advantage over the BH3 peptides in targeting cancer cells. Confocal microscopic imaging illustrates specific targeting of ATAP to mitochondria, whereas BH3 peptides show diffuse patterns of cytosolic distribution. Although the pro-apoptotic activities of BH3 peptides are largely inhibited by either overexpression of anti-apoptotic Bcl-2 or Bcl-xL or nullification of pro-apoptotic Bax and Bak in cells, the pro-apoptotic function of ATAP is not affected by these cellular factors. Reconstitution of synthetic ATAP into liposomal membranes results in release of fluorescent molecules of the size of cytochrome c from the liposomes, suggesting that the membrane permeabilizing activity of ATAP does not require additional protein factors. Because ATAP can target to the mitochondrial membrane and its pro-apoptotic activity does not depend on the content of Bcl-2 family proteins, it represents a promising candidate for anti-cancer drugs that can potentially overcome the intrinsic apoptosis-resistant nature of cancer cells.
Keywords: Anticancer Drug, Apoptosis, Cancer Therapy, Mitochondrial Apoptosis, Peptides
Introduction
Recent advances in apoptosis research allow for the possibility of the rational design of cancer therapeutics that selectively activates apoptosis in cancer cells or reduces their apoptotic threshold to other cytotoxic treatments. Extensive efforts have focused on the mitochondrial apoptosis pathway to increase the permeability of the mitochondrial outer membrane (MOM)4 to allow release of cytochrome c, apoptosis-inducing factor, and other mitochondrial proteins that trigger caspase- and nuclease-mediated cell death (1–4). Proteins of the Bcl-2 family regulate permeabilization of the MOM during apoptosis (5–9). All these proteins contain the canonical Bcl-2 homology (BH) domains and are divided into three subfamilies based on the number of BH domains present and the function of the protein. Pro-apoptotic BH3 domain-only proteins convey diverse death signals by either activating pro-apoptotic multi-BH domain proteins such as Bax and Bak, which induce permeabilization of the MOM by forming oligomeric pores in the membrane, or inhibiting anti-apoptotic multi-BH proteins such as Bcl-2 and Bcl-xL, which prevent the membrane permeabilization by neutralizing the activity of Bax, Bak, and/or BH3-only proteins (10–18). Therefore, the pro- and anti-death activities of Bcl-2 family proteins are dictated by their interactions through various BH domains.
Recent studies showed that peptides derived from the BH3 domain of pro-apoptotic Bcl-2 family members, such as BH3-only proteins Bid (an activator of Bax and Bak) and Bad (an inhibitor of Bcl-2 and Bcl-xL), as well as multi-BH pore-forming proteins Bax and Bak, can induce apoptosis of cancer cells by either directly binding and activating Bax and/or Bak or indirectly releasing Bax, Bak, and/or BH3-only proteins from inhibition by anti-apoptotic Bcl-2 family proteins (12, 19, 20). Similar pro-apoptotic activity was observed for small chemical BH3 mimetics, some of which have been advanced into clinical trials and show promising anti-cancer effects (21–28). However, the efficacy of currently available BH3 peptides or chemical mimetics seems to be limited by the contents of Bcl-2 family proteins in cancer cells. The pro-apoptotic activity of BH3 peptides was significantly inhibited by the presence of Bcl-2 or Bcl-xL protein (10, 16, 19, 29, 30). ABT-737, a BH3-mimicking chemical antagonist of Bcl-2, Bcl-xL, and Bcl-w, displayed strong pro-apoptotic activity in small cell lung cancer models but failed to induce apoptosis in acute myeloid leukemia cells expressing anti-apoptotic Mcl-1 due to its weak binding affinity to Mcl-1 (21, 31). Furthermore, overexpression of both Bfl-1 and Bcl-xL caused a significant resistance to ABT-737 in chronic lymphocytic leukemia (32). In addition, the pro-apoptotic activity of BH3 peptides and their mimetics was dependent on other cellular factors as the activity was abrogated in the absence of Bax and Bak (19, 33, 34). The down-regulation of pro-apoptotic and/or the up-regulation of anti-apoptotic Bcl-2 family proteins is commonly observed in cancer cells and often causes resistance to BH3 peptide-induced cell death, and thus the application of BH3 peptides and their mimetics in cancer therapy may be limited by these intrinsic cellular factors.
Previously, we reported that ATAP, a novel amphipathic tail-anchoring peptide of Bfl-1 (amino acids 147–175), targeted specifically to mitochondria and triggered potent apoptotic cell death in the absence of Bax and Bak (35). Synthetic ATAP increased the permeability of lipid bilayer membranes and induced cytochrome c release from isolated mitochondria. Thus, ATAP represents a unique pro-apoptotic peptide with a potential as a therapeutic agent for treatment of cancer cells. However, the molecular mechanism of ATAP action at mitochondria and in cells requires further elucidation. In particular, it is important to determine whether ATAP can form the cytochrome c-releasing pore in the MOM in the absence of other cellular proteins and whether the pro-apoptotic activity of ATAP is sensitive to anti-apoptotic Bcl-2 family proteins. It is also of interest to determine the relative efficacy of ATAP compared with BH3 peptides.
In this study, we analyzed the pore forming activity ATAP in a MOM-mimicking liposomal membrane and its pro-apoptotic activity in cancer cells, and we compared the activities of ATAP and BH3 peptides in these systems. We found that ATAP, unlike BH3 peptides, formed large pores in the liposomal membrane in the absence of any other proteins and induced apoptosis in cells in a Bax/Bak-independent and Bcl-2-insensitive manner. These results suggest that ATAP is potentially a better therapeutic lead for cancer treatment than BH3 peptides and mimetics, as it can bypass the resistance mechanism resulting from dysregulation of Bcl-2 family proteins in cancer cells.
EXPERIMENTAL PROCEDURES
Plasmid Construction and Protein Preparation
Plasmids pFLAG-Bcl-xL and pcDNA-Bcl-2 were described previously (36). To construct pFLAG-Bcl-2, cDNA was amplified using pcDNA-Bcl-2 as template and primers 5′-AAAGAATTCATGGCGCACGCTGGGAGAAC and 5′-AAAGGTACCTCACTTGTGGGACAGATAGGCAC. The PCR product was digested with EcoRI and KpnI and then ligated with pFLAG-ATAP (35) treated with XhoI and BamHI. Expression vectors for GFP-Bak BH3, GFP-Bid BH3, and GFP-Bad BH3 were constructed by the PCR subcloning method as described previously (35) using the following oligonucleotides: 5′-AAAAATCTCGAGCTATGGGGCAGGTGGGACGGCAGCTCGCCATCATCGGGGACG and 5′-AAAAAGGATCCTGAGTCATAGCGTCGGTTGATGTCGTCCCCGATGATGGCGAG for Bak BH3; 5′-AAAAATCTCGAGCTCAAGAAGACATCATCCGGAATATTGCCAGGCACCTCGC and 5′-AAAAAGGATCCACGGTCCATGCTGTCCCCGACCTGGGCGAGGTGCCTGGCAATATTC for Bid BH3; and 5′-AAAAATCTCGAGCTAACCTCTGGGCAGCACAGCGCTATGGCCGCGAGCTCCGGAGGATG and 5′-AAAAAGGATCCCTTCTTAAAGGAGTCCACAACTCGTCACTCATCCTCCGGAGCTCGCGG for Bad BH3. Each paired oligonucleotide contains complementary sequences at the 3′-end (underlined), which were annealed, filled in by PCR, digested with XhoI and BamHI, and then subcloned into the XhoI and BamHI sites of pEGFP-C1. The digested PCR products were also used to generate pFLAG-Bak BH3, pFLAG-Bid BH3, and pFLAG-Bad BH3 constructs by ligating them with pFLAG-ATAP vector (35) treated with XhoI and BamHI. Construction for pGFP-Bax was described previously (35). Expression and purification of His6-tagged Bcl-2ΔTM, tBid, and Bax proteins were conducted as described previously (17, 37).
Gene Transfection and Analyses of Cell Death
Apoptotic cell death was monitored after transfection of expression plasmids into HeLa, BMK-W2, and BMK-D3 cells (38). 2 × 105 cells were cultured in 35-mm wells for 24 h in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Cells were transfected with 1 μg (unless specified otherwise) of the indicated expression plasmids using Gene Jammar reagent (Stratagene) and further cultured in the absence or presence of 50 μm pan-caspase inhibitor, QVD-OPH (Enzyme Systems Products). For morphological assessment of apoptotic cell death, cells were plated onto LabTek II chamber slides (Nalgen Nunc International) at densities of 5 × 104 cells per well. 24 h after transfection, cells were washed with phosphate-buffered saline (PBS) prior to fixation with 4% formaldehyde. Subsequently, cells were stained with a Vectasheild mounting solution (Vector Laboratories) containing 1 μg/ml DAPI and visualized under an Axiovert 100 inverted epifluorescence microscope (Carl Zeiss). Nuclei with rippled contours and chromatin condensation were considered to represent the apoptotic cell death. Cell viability was measured by DAPI exclusion. After transfection with GFP fusion plasmids, a total of 1 μg/ml DAPI (Molecular Probes) was added to the cell culture. Cells were observed and photographed with a fluorescence microscope at three different fields containing ∼200 GFP-positive cells. GFP and DAPI double-positive cells were counted as dead cells. Quantitative analysis of cell viability was also determined by β-galactosidase reporter assay according to the procedures described previously (35, 39). Briefly, cells were co-transfected with 1 μg of tested plasmid plus 0.1 μg of pCMV-β plasmid expressing β-galactosidase (Sigma). 24 h following transfection, cells were harvested, and β-galactosidase activity was measured using β-galactosidase enzyme assay system (Promega). In every experiment, each construct was tested in triplicate, and experiments were repeated at least three times.
Confocal Microscopy Study
To observe intracellular localization of GFP fusion proteins, fixed HeLa cells were used for confocal microscopy. HeLa cells were transfected as described above on LabTek II chamber slides and cultured in the presence of 50 μm QVD-OPH. 24 h after transfection, cells were washed with PBS followed by fixation with 4% formaldehyde. Cells finally were washed, mounted, and analyzed with a confocal Zeiss LSM 510 microscope (Carl Zeiss Microscopy, Jena, Germany) equipped with a 63× objective lens. Image acquisition was performed at room temperature.
Immunoprecipitation and Immunoblotting
For co-immunoprecipitation experiments, ∼2 × 105 cells cultured in a 35-mm well were transfected with the indicated plasmids in the presence of 50 μm QVD-OPH. Cells were harvested 24 h after transfection and lysed in Nonidet P-40 lysis buffer, 1% Nonidet P-40 lysis buffer with 20 mm Tris-HCl, pH 7.4, 150 mm NaCl, and protease inhibitor mixture (Roche Applied Science), on ice. Postnuclear lysates were normalized for protein content, precleared with Sepharose 6B (Sigma) for 1 h at 4 °C, and subjected to immunoprecipitation with anti-FLAG-M2-agarose beads (Sigma) for at least 2 h at 4 °C on a orbital shaker. Immunoprecipitates were washed with the Nonidet P-40 lysis buffer and eluted from the beads by boiling in SDS-sample buffer. For immunoblotting analysis, 20 μg of protein was subjected to SDS-PAGE and transferred onto a PVDF membrane, which was blocked with 5% skimmed milk, probed with primary antibodies, and visualized using an ECL chemiluminescence kit (GE Healthcare). Monoclonal anti-GFP antibody and anti-goat horseradish peroxidase (HRP) antibody were purchased from Santa Cruz Biotechnology. Anti-β-actin antibody was purchased from Sigma. Monoclonal anti-FLAG 9E10.2 antibody was purchased from Invitrogen. Anti-mouse HRP antibody was purchased from Amersham Biosciences.
Genomic DNA Isolation and Genotyping PCR
Total genomic DNA was isolated from cultured BMK-W2 or BMK-D3 cells using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. 100 ng of purified genomic DNA was analyzed by PCR using primers 5′-GTTTCATCCAGGATCGAGCAGG and 5′-CCTCTGCAGCTCCATATTGCTG for Bax and 5′-CCTATTTAAGAGTGGCATCAGCTG and 5′-CCAACCGCCTCTCTGTGCGATC for Bak. Each primer pair was designed from a single exon sequence to amplify short PCR products of 149 bp for Bax and 181 bp for Bak. PCR products were then analyzed by electrophoresis on 2% agarose gel and ethidium bromide staining.
Pore Forming Activity Assay
The 29-mer ATAP peptide corresponding to the C terminus of Bfl-1 were synthesized as described previously (35). The BH3 peptides of Bid (QEDIIRNIAR-HLAQVGDSMDR) and Bad (NLWAAQRYGRELRRMSDEFVDSFKK) were synthesized by Abgent (San Diego) with 99% purity as measured by HPLC and mass spectrometry. The peptides were dissolved in dimethyl sulfoxide (DMSO) to make a 10 mm stock. All phospholipids and lipid analogs were purchased from Avanti Polar Lipids (Alabaster, AL). Fluorescent dye Cascade Blue (CB), CB-labeled dextran of 10 kDa, and rabbit anti-CB polyclonal antibody were obtained from Molecular Probes. Liposomes were made with the Xenopus MOM characteristic lipids in the membrane and fluorescent dye CB-labeled dextran (10 kDa) in the lumen by an extrusion method as described previously (16). The Ni2+-chelating liposomes were prepared similarly except that the Ni2+-chelating lipid analog, 1,2-dioleoyl-sn-glycero-3-([N(5-amino-1-carboxypentyl) iminodiacetic acid]-succinyl) (nickel salt), was included in the membrane as described previously (15). The regular or Ni2+-chelating liposomes (12.5 μm) loaded with CB dextran were mixed with 6 μg/ml anti-CB antibodies in 250 μl of buffer A (100 mm NaCl, 51 mm Na2HPO4, and 3.8 mm citric acid, pH 7.4). The initial emission intensity (F0) was determined after equilibrating the sample at 25 °C for 5 min. ATAP, BH3 peptide, His6-tagged Bcl-2ΔTM, tBid, and/or Bax protein were then added. The first fluorescence intensity measurement was started exactly 20 s after the addition of the peptide, chemical, and/or protein(s) and was followed by multiple measurements in a predetermined time interval for 5 h, resulting in multiple intermediate intensities (F). At the end of the time course, 0.1% Triton X-100 was added, and the final measurement was taken, resulting in the final intensity (Ft). The extent of CB dextran release is proportional to the extent of fluorescence quenching that is equal to ΔFProtein/ΔFTriton, where ΔFProtein = F0 − F and ΔFTriton = F0 − Ft. All fluorescence intensities were measured using the SLM-8100 spectrometer as described previously (16).
Cross-linking Assay
Synthetic ATAP (17 μm) was incubated with 1.3 mm regular liposome made with the Xenopus MOM characteristic lipids in 120 μl of buffer A at 22 °C for 1 h. 4 μm BMH (Pierce) was then added to 1 of the 2 aliquots, and the cross-linking reaction was carried out at 22 °C for 30 min and then quenched by 50 mm β-mercaptoethanol for 15 min. The resulting samples were subjected to a step sucrose gradient centrifugation as described previously (37). Top half, bottom half, and pellet fractions were collected, which contain liposome-bound, soluble, and aggregated peptides, respectively. All fractions were precipitated using Cl3CCOOH and analyzed by 16.5% Tris-Tricine SDS-PAGE under reducing conditions and silver staining.
RESULTS
Pro-apoptotic Activity of ATAP Is Similar to or Stronger than That of BH3 Peptides
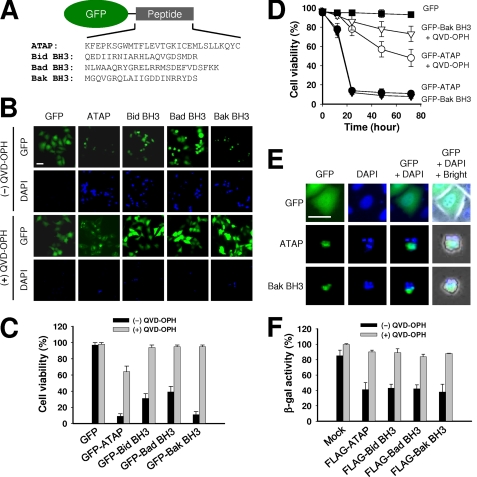
To compare the cellular function of ATAP and BH3 peptides derived from pro-apoptotic proteins Bid, Bad, and Bak, we fused these peptides with GFP and expressed the fusion proteins in HeLa cells for live cell imaging with a confocal microscope (Fig. 1A). Dying cells were observed by staining their nuclei with a cell-impermeable DNA dye, DAPI, to detect late stage apoptotic cells. As shown in Fig. 1, B and C, strong pro-apoptotic activity was observed in cells expressing GFP-ATAP or GFP-Bak BH3, as a similar degree of cell death was observed with GFP-ATAP and GFP-Bak BH3 at 24 h after transfection (Fig. 1C). Transient expression of GFP-Bid BH3 and GFP-Bad BH3 also caused apoptosis in HeLa cells, although to a less extent than GFP-ATAP and GFP-Bak BH3 (Fig. 1, B and C). To test if apoptosis induced by these peptides involves activation of caspase, a pan-caspase inhibitor, QVD-OPH, was added to cells during transfection. As shown in Fig. 1, B and C, the pro-apoptotic activity of ATAP as well as BH3 peptides was strongly inhibited by treatment with 50 μm QVD-OPH at an early stage of the experiment. At 24 h after transfection, more than 78% of cells expressing GFP-ATAP were alive in the presence of QVD-OPH. A stronger inhibitory effect of QVD-OPH was observed against GFP-BH3 peptides because more than 90% of cells transfected with GFP-BH3 peptides survived by treatment with QVD-OPH. At 48 h after transfection, ∼40% of cells expressing GFP-ATAP underwent apoptosis, whereas more than 80% of cells expressing GFP-Bak BH3 were still alive (Fig. 1D). These results show that ATAP can induce apoptosis by both the caspase-dependent apoptotic pathway at an early stage and the caspase-independent pathways at a late stage, whereas BH3 peptides induce apoptosis mainly by activation of the caspase-dependent pathway. When fixed cells were observed by DAPI staining to compare nuclei among transfected cells, most of the cells transiently expressing GFP-ATAP or GFP-Bak BH3 displayed condensed and fragmented chromatin structure, a hallmark of apoptotic cell death (Fig. 1E).
FIGURE 1.
Transient expression of GFP-ATAP or GFP-BH3 peptides induces apoptosis of HeLa cells. A, schematic diagram of constructs expressing ATAP or BH3 peptides fused with GFP protein. B and C, HeLa cells were transfected with 1 μg of plasmids expressing GFP-ATAP- or GFP-BH3-peptide fusion proteins in the absence or presence of the pan-caspase inhibitor QVD-OPH (50 μm). At 24 h after transfection, cells were stained with DAPI and observed under a fluorescence microscope. B, representative photographs of GFP- or DAPI-positive cells transfected with indicated plasmids. The two images in each column were from the same field. Bar, 10 μm. C, percentage of surviving cells was determined by the ratio of GFP-positive cells without DAPI staining to total GFP-positive cells. About 200 cells from three different fields were scored. Data are expressed as the means ± S.E. D, time course effect of GFP-ATAP and GFP-Bak BH3 on HeLa cells. Cell survival was measured by DAPI exclusion in the HeLa cells transfected with 1 μg of plasmids expressing GFP-ATAP or GFP-Bak BH3. E, transient expression of GFP-ATAP- or GFP-Bak BH3-induced apoptotic nuclear morphology. HeLa cells were transfected with GFP-ATAP or GFP-Bak BH3. At 24 h of transfection, cells were fixed, stained with DAPI, and observed under a fluorescence microscope. Bar, 10 μm. F, cytotoxic activity of FLAG-tagged ATAP or BH3 peptides. HeLa cells were co-transfected with 1 μg of the indicated plasmid expressing FLAG peptide and 0.1 μg of pCMV-β reporter plasmid either in the presence or absence of 50 μm QVD-OPH. At 24 h after transfection, β-galactosidase activity was measured. Cell viability is shown as the relative β-galactosidase activity of the FLAG peptide-expressing cells to the control cells.
As fusion with GFP may affect the pro-apoptotic activity of these peptides, we generated FLAG-tagged peptides to test the effect of different tags. The cytotoxic activity of the FLAG peptides was quantified using a β-galactosidase reporter assay 24 h after co-transfection of plasmids encoding β-galactosidase and FLAG peptides in HeLa cells. The decrease in β-galactosidase activity quantitatively reflects the loss of cell viability (35, 39). Compared with cells co-transfected with the mock plasmid, an ∼60% decrease in β-galactosidase activity was observed in cells transfected with the FLAG-ATAP plasmid, which could be prevented by the addition of QVD-OPH (Fig. 1F). The cytotoxic activity of FLAG-BH3 peptides was comparable with that of FLAG-ATAP. Taken together, the data indicate that ATAP exhibits strong pro-apoptotic activity that is comparable with or stronger than that of the tested BH3 peptides.
Bcl-2 or Bcl-xL Cannot Block Apoptosis Induced by ATAP although They Potently Inhibit the Pro-apoptotic Activity of BH3 Peptides
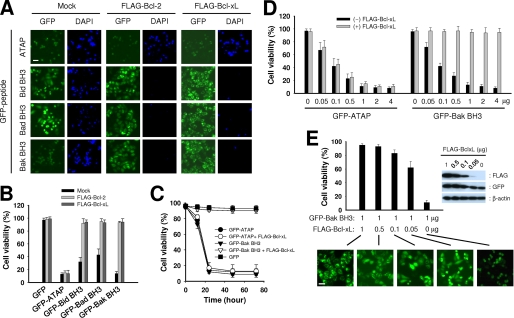
Although published studies established that BH3 peptides can induce apoptosis of cancer cells by antagonizing the anti-apoptotic activity of Bcl-2 and Bcl-xL or by agonizing the pro-apoptotic activity of Bax and Bak, several studies have shown controversial results regarding the effect of Bcl-2 or Bcl-xL overexpression on the cytotoxic effects of BH3 peptides (10, 16, 19, 29, 30). To compare the effect of Bcl-2 and Bcl-xL on the apoptotic activity of ATAP and BH3 peptides, 1 μg of plasmid expressing GFP-ATAP or GFP-BH3 peptides was co-transfected with either 1 μg of FLAG-Bcl-2 or FLAG-Bcl-xL plasmid in HeLa cells. As shown in Fig. 2, A and B, neither FLAG-Bcl-2 nor FLAG-Bcl-xL appeared to have a significant effect on apoptosis induced by GFP-ATAP, because more than 85% of the GFP-positive cells were dead and stained by DAPI no matter whether they expressed the anti-apoptotic proteins or not. In contrast, both FLAG-Bcl-2 and FLAG-Bcl-xL showed complete inhibition of apoptosis induced by all GFP-BH3 peptides in HeLa cells. The inhibitory activity of Bcl-xL on GFP-BH3 peptides could continue for at least 3 days without a significant decrease (Fig. 2C).
FIGURE 2.
Bcl-xL or Bcl-2 does not inhibit ATAP-induced apoptosis but efficiently inhibits BH3 peptide-induced apoptosis. A and B, HeLa cells were co-transfected with 1 μg of plasmids expressing GFP-ATAP or GFP-BH3 peptides along with 1 μg of mock vector, pFLAG-Bcl-2, or pFLAG-Bcl-xL. At 24 h after transfection, cells were treated with DAPI and observed under a fluorescence microscope. A, representative photographs show GFP- or DAPI-positive cells in the same field. Bar, 10 μm. B, percentage of surviving cells was determined by the ratio of GFP-positive cells without DAPI staining to total GFP-positive cells. About 200 cells from three different fields were scored. Data are expressed as the means ± S.E. C, time course effect of FLAG-Bcl-xL on the apoptosis induced by GFP-ATAP or GFP-Bak BH3 in HeLa cells. Cell survival was measured by DAPI exclusion in the HeLa cells transfected with 1 μg of plasmids expressing GFP-ATAP or GFP-Bak BH3 with or without 1 μg of FLAG-Bcl-xL plasmid. D, HeLa cells were transiently co-transfected with 1 μg of FLAG-Bcl-xL plasmid and various amounts of GFP-ATAP or GFP-Bak BH3 construct as indicated. The percentage of surviving cells was determined by DAPI exclusion assay as described previously above. Data are expressed as the means ± S.E. E, HeLa cells were transiently co-transfected with 1 μg of GFP-Bak BH3 construct and various amounts of FLAG-Bcl-xL construct as indicated. The percentage of surviving cells was determined by the DAPI exclusion assay as described above. Data are expressed as the means ± S.E. Right panel, protein expression in the transfected cells was assessed by immunoblotting the cell lysates using antibodies specific for FLAG, GFP, or β-actin. Lower panel, representative photographs show cellular distribution of GFP-Bak BH3 fusion protein in the transfected cells. Bar, 10 μm.
To examine the dose-dependent effect of Bcl-xL on the apoptosis induced by GFP-ATAP or GFP-BH3 peptides, we co-transfected HeLa cells with 1 μg of FLAG-Bcl-xL and different amounts of GFP-ATAP or GFP-Bak BH3 plasmid, which showed the strongest pro-apoptotic activity among the three constructs for GFP-BH3 peptides (Fig. 1, B and C). As shown in Fig. 2D, GFP-ATAP and GFP-Bak BH3 showed clearly different pro-apoptotic activities in the presence of FLAG-Bcl-xL. Although the pro-apoptotic activity of GFP-ATAP was not affected by FLAG-Bcl-xL, the activity of GFP-Bak BH3 was dramatically blocked by FLAG-Bcl-xL. Even with a 4-fold amount (4 μg) of GFP-Bak BH3 co-transfected with FLAG-Bcl-xL, no apoptosis was observed. These results clearly show that ATAP-induced apoptosis is insensitive to Bcl-2 and Bcl-xL, whereas BH3 peptide-induced apoptosis is strongly inhibited by Bcl-2 and Bcl-xL.
For further examination of the effect of Bcl-xL on BH3-induced apoptosis, we co-transfected HeLa cells with a fixed amount (1 μg) of GFP-Bak BH3 plasmid together with different amounts of FLAG-Bcl-xL plasmid. The transfected cells were analyzed by Western blotting to check the expression level of GFP-Bak BH3 and FLAG-Bcl-xL (Fig. 2E, upper right panel). Clearly, more dead cells were detected at higher GFP-Bak BH3 to FLAG-Bcl-xL ratios (Fig. 2E, graph), indicating that the BH3 peptide induces cell death by saturating Bcl-xL, and thus by preventing Bcl-xL from inhibiting other endogenous pro-apoptotic proteins. Of interest, co-transfection with 20-fold less FLAG-Bcl-xL than GFP-Bak BH3 still showed significant inhibition of apoptosis in HeLa cells compared with cells that were not transfected by FLAG-Bcl-xL, suggesting that cancer cells displaying up-regulation of anti-apoptotic protein expression would be more resistant to the BH3 peptide-based therapeutics than the normal cells if the cells contain a larger pool of free anti-apoptotic proteins. Interestingly, even at the high dose ratio, Bcl-xL did not inhibit GFP-ATAP-induced apoptosis (1 μg of FLAG-Bcl-xL to 0.05 μg of GFP-ATAP in Fig. 2D), suggesting that elevated level of anti-apoptotic Bcl-2 proteins in cancer cells may not affect the cytotoxic activity of ATAP-based therapeutics.
ATAP Targets Mitochondria to Induce Apoptosis without Interaction with Bcl-2 and Bcl-xL in HeLa Cells
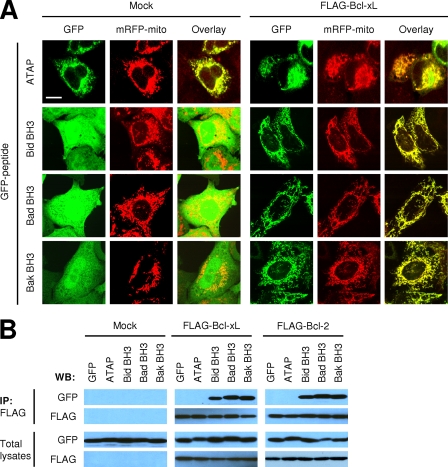
Notably, we found that the subcellular localization of GFP-BH3 peptides seemed to be changed from cytosolic distribution to mitochondrial or perinuclear distribution by co-transfection with Bcl-2 or Bcl-xL (Fig. 2A). Furthermore, more GFP-Bak BH3 protein was localized in the cytosol as less FLAG-Bcl-xL plasmid was used in co-transfection (Fig. 2E, lower panel), suggesting that a function of the anti-apoptotic protein is to sequester the pro-apoptotic BH3 peptides to intracellular membranes. To assess the subcellular distribution of these GFP-peptide fusion proteins, we used confocal microscopy and treated the transfected cells with QVD-OPH to prevent morphological changes caused by apoptosis. As shown in Fig. 3A, GFP-ATAP was principally localized in the mitochondria as it co-localized with mRFP-mito, a mitochondrial marker, confirming our previous findings that ATAP has the intrinsic property of a specific mitochondrial target signal (35). The mitochondrial localization of GFP-ATAP was not altered by co-expression of FLAG-Bcl-xL. In contrast, co-expression of FLAG-Bcl-xL dramatically changed the localization of GFP-BH3 peptides from the cytosol to mitochondria (Fig. 3A). Similarly, the co-expression of FLAG-Bcl-2 also led the GFP-BH3 peptides to the intracellular membranes such as the mitochondrial or perinuclear compartment (data not shown).
FIGURE 3.
GFP-ATAP constitutively localizes in mitochondria and does not interact with Bcl-xL, although GFP-BH3 peptides were translocated from cytosol to mitochondria by physical association with Bcl-xL. A, subcellular localization of GFP peptides. HeLa cells were co-transfected with 1 μg of plasmids expressing GFP-ATAP or GFP-BH3 peptides along with 0.5 μg of pmRFP-mito and with 1 μg of mock vector or pFLAG-Bcl-xL in the presence of 50 μm QVD-OPH. At 24 h after transfection, cells were fixed using 4% paraformaldehyde and observed under a confocal microscope. Bar, 5 μm. B, GFP peptides were co-expressed with FLAG-Bcl-xL, FLAG-Bcl-2, or mock vector in HeLa cells in the presence of 50 μm QVD-OPH. Cell lysates were immunoprecipitated (IP) with anti-FLAG M2-agarose and detected with anti-GFP antibody. WB, Western blot.
Because FLAG-Bcl-xL predominantly localized to mitochondria (35), the mitochondrial localization of GFP-BH3 peptides by Bcl-xL implies that Bcl-xL physically interacts with them on the mitochondrial membrane to inhibit their pro-apoptotic activity. To examine whether GFP-ATAP and GFP-BH3 peptides interact with Bcl-xL or Bcl-2, we performed co-immunoprecipitation assays. HeLa cells were co-transfected with individual GFP fusion plasmid and either mock, FLAG-Bcl-xL, or FLAG-Bcl-2 plasmids. Cell lysates were immunoprecipitated with anti-FLAG antibody and then immunoblotted with anti-GFP antibody. As shown in Fig. 3B, GFP-ATAP did not co-precipitate with any of the anti-apoptotic proteins, whereas all GFP-BH3 fusion proteins co-precipitated with Bcl-2 or Bcl-xL. Taken together, these results indicate that anti-apoptotic Bcl-2 and Bcl-xL proteins inhibit the pro-apoptotic activity of BH3 peptides through physical interaction with these peptides at mitochondria, whereas these proteins do not interact with ATAP and cannot inhibit its pro-apoptotic activity.
Bax and Bak Do Not Contribute to ATAP Function but Are Required for BH3 Peptide-induced Apoptosis
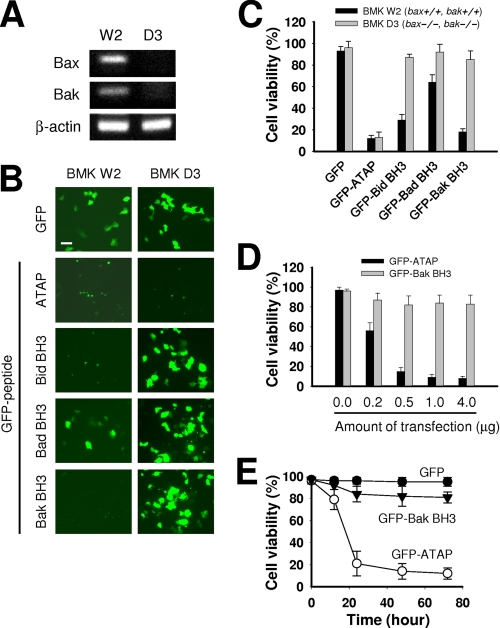
Previously, we reported that ATAP can induce apoptosis without Bax or Bak proteins (35). In contrast, apoptosis induced by pro-apoptotic BH3 peptides and mimetics requires the activation of pro-apoptotic Bax or Bak proteins (10, 19, 21, 30, 31). To compare the effect of Bax and Bak on the pro-apoptotic activity of ATAP and BH3 peptides, the corresponding GFP fusion proteins were transiently expressed in BMK-D3, a baby mouse kidney cell line derived from the bax−/− bak−/− mice (38) and in BMK-W2, a cell line derived from wild type littermates. PCR analysis on genomic DNA isolated from BMK-W2 and BMK-D3 cells confirmed ablations of Bax and Bak genes in BMK-D3 cells (Fig. 4A). Counting dead cells among GFP-positive cells indicated that GFP-ATAP induced apoptosis in BMK-D3 cells at a similar level as in BMD-W2 cells, which is consistent with our previous observation (Fig. 4, B and C) (35). In contrast, all GFP-BH3 peptides failed to induce apoptosis in BMK-D3 cells, although they induced apoptosis in BMK-W2 cells to various degrees (Fig. 4, B and C). These data demonstrate that ATAP-induced apoptosis is independent of Bax and Bak, although BH3 peptide-induced apoptosis requires the presence of either Bax or Bak. The pro-apoptotic effect of GFP-ATAP showed a dose- and time-dependent manner, whereas GFP-Bak BH3 could not induce apoptosis at all doses and time points tested (Fig. 4, D and E).
FIGURE 4.
ATAP does not require either Bax or Bak activity to induce apoptosis although BH3 peptides do. A, detection of Bax and Bak genes by PCR. 100 ng of genomic DNA isolated from cultured BMK-W2 (bax+/+, bak+/+) and BMK-D3 (bax−/−, bak−/−) cells was analyzed by PCR using primers designed to amplify an exon sequence of 149 bp in Bax gene and of 181 bp in Bak gene. PCR products were then analyzed by agarose gel electrophoresis and visualized by ethidium bromide staining. B, viability of BMK W2 or BMK D3 cells transfected with 1 μg of GFP-ATAP or GFP-BH3 expression plasmid. At 24 h after transfection, cells were stained with DAPI and observed under a fluorescence microscope. Representative photographs show GFP-positive cells after 24 h of transfection. Bar, 10 μm. C, percentage of surviving cells was determined by the ratio of GFP-positive cells without DAPI staining to total GFP-positive cells. About 200 cells from three different fields were scored. Data are expressed as the means ± S.E. D, dose effect of GFP-ATAP and GFP-Bak BH3 on apoptosis of BMK D3 cells. Cells were transiently transfected with the indicated amounts of GFP-ATAP or GFP-Bak BH3 construct. Cell survival was measured by DAPI exclusion assay. Data are expressed as the means ± S.E. E, time course study of apoptosis induced by GFP-ATAP and GFP-Bak BH3 in BMK D3 cells. Cells were transiently transfected with 1 μg of GFP-ATAP or GFP-Bak BH3 construct. Cell survival was measured by DAPI exclusion assay. Data are expressed as the means ± S.E.
Together, our data show that BH3 peptides require Bax or Bak for their apoptotic function, which is inhibited by Bcl-2, Bcl-xL, or probably other anti-apoptotic family members, whereas ATAP induces Bax/Bak-independent apoptosis that cannot be inhibited by Bcl-2 and Bcl-xL anti-apoptotic proteins.
Synthetic ATAP Peptide Releases Large Molecules from Liposomes
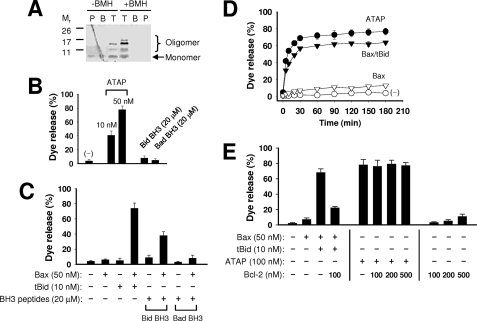
Previously, we reported that ATAP could permeabilize MOM causing the loss of mitochondrial membrane potential and release of cytochrome c to induce apoptosis in cultured cells (35). We also showed that synthetic ATAP could induce release of cytochrome c from mitochondrial preparations isolated from BMK D3 cells (see Fig. 6C in our previous report published by Ko et al. (35)). Thus, we postulated that oligomerization of ATAP at MOM could lead to formation of a large pore for release of cytochrome c and other pro-apoptotic factors to trigger apoptosis. To test whether ATAP could oligomerize in the membrane, we performed chemical cross-linking assay after reconstitution of synthetic ATAP to the liposomal membrane composed of the MOM characteristic lipids (MOM-liposome) (15, 16). Taking advantage of the two cysteine residues present in ATAP, we used bis-maleimidohexane (BMH), a thio-specific bifunctional cross-linking agent, to examine the potential oligomerization of ATAP reconstituted in the MOM-liposome. The membrane-bound ATAP was separated from the soluble and precipitated peptides by floating the liposome and associated ATAP to the top of a three-step sucrose gradient using a centrifugation procedure described previously (37). The resulting fractions were analyzed by SDS-PAGE and silver staining. As shown in Fig. 5A, complexes larger than ATAP monomer (Mr 3.4) were detected in the liposome fraction even in the absence of BMH treatment, suggesting that ATAP can form stable oligomers in the membrane. Cross-linking further stabilized and enlarged the ATAP oligomers as more and larger complexes were detected in the presence of BMH. Interestingly, the oligomeric ATAP was only detected in the liposome fraction, suggesting that membrane targeting could promote oligomerization of ATAP and pore formation activities.
FIGURE 6.
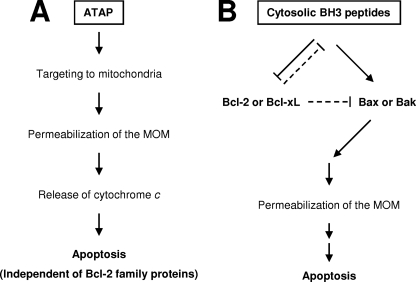
Proposed events for the pro-apoptotic actions of ATAP and BH3 peptides at mitochondria. A, ATAP derived from tail-anchor domain of Bfl-1 can target to mitochondrial membrane by its intrinsic nature of the mitochondrial targeting signal (35). Once at mitochondria, ATAP may insert into the MOM through a translocation complex on the MOM or through a direct interaction with the lipid bilayer. An amphipathic helical feature on the transmembrane segment of ATAP is likely involved in the perturbation of mitochondria membrane integrity by pore formation. The pore formation and pro-apoptotic function of ATAP are independent of Bcl-2 family proteins. B, distinctively, BH3 peptides derived from pro-apoptotic Bcl-2 family proteins accumulate in the cytoplasm, and hence only a small fraction would interact with Bcl-2 family proteins on the MOM. For the pro-apoptotic activity of BH3 peptides, the intracellular content of Bcl-2 family proteins is critical. First, the BH3 peptides must be in excess of anti-apoptotic Bcl-2 proteins such as Bcl-2 or Bcl-xL to inhibit their anti-apoptotic function. Otherwise, the peptides might be sequestered by these anti-apoptotic proteins on the MOM (shown by dashed lines). Second, there must be enough pro-apoptotic Bcl-2 proteins such as Bax or Bak because the pro-apoptotic activity of BH3 peptides was dependent on these proteins. Under this situation, BH3 peptides can induce apoptosis by either directly binding and activating Bax and/or Bak or preventing anti-apoptotic Bcl-2 family proteins from blocking Bax, Bak, and/or BH3-only pro-apoptotic Bcl-2 proteins (12, 19, 20).
FIGURE 5.
ATAP forms large pores in the MOM-like liposomal membrane. A, oligomerization of ATAP in the liposomal membrane was examined by BMH cross-linking. The samples were fractionated using the step sucrose gradient centrifugation. The resulting top (T), bottom (B), and pellet (P) fractions contained liposome-bound, -soluble, and -aggregated peptide and adducts, respectively. All fractions were subjected to Tris-Tricine SDS-PAGE, and the peptide and adducts were visualized by silver staining. B, release of 10-kDa CB dextrans from the MOM-liposome by the indicated concentrations of ATAP peptide or BH3 peptides was monitored using the fluorescence quenching method. Data shown are the average extent of release after 5 h of incubation from three independent experiments with standard deviation. C, release of 10-kDa CB dextrans from the MOM-liposome by BH3 peptides (20 μm) in the presence of Bax (50 nm) was monitored as above. Data shown are averages from three independent experiments with standard deviation. D, release kinetics of 10-kDa CB dextrans by ATAP (50 nm) or Bax (50 nm) together with or without tBid (10 nm) was monitored. A negative control shows an effect by DMSO vehicle without any peptide. Data shown are averages from three independent experiments with standard deviation. E, release of 10-kDa CB dextrans from 12.5 μm Ni2+-chelating liposome by ATAP peptide or tBid-activated Bax in the presence or absence of His6-Bcl-2ΔTM was monitored as above. Data shown are averages from three independent experiments with standard deviation. The concentration of ATAP was doubled compared with that in B–D because the batch of peptide used here is less active than the one used in B–D.
To test the pore-forming potential of ATAP, we used a liposome-based fluorescent dye release assay following our established protocols (15, 16). For this study, dextran molecules of 10 kDa with a size similar to that of cytochrome c were labeled with a fluorescent dye, CB, and encapsulated into the MOM-liposomes. The peptide-induced release of CB dextran from the liposome was monitored by quenching of CB fluorescence by anti-CB antibody presented outside of the liposome. As shown in Fig. 5, B and D, ATAP efficiently released CB dextrans from the liposomes in a dose- and time-dependent manner. With 50 nm ATAP, the release of CB dextrans reached a near maximal level in 30 min. In contrast, neither Bid nor Bad BH3 peptide, which triggered release of cytochrome c from native mitochondrial preparations (10, 19, 20, 40, 41), could release CB dextran from the liposomes even at a much higher dose (20 μm) than we used for ATAP (Fig. 5B). Because Bid-BH3 peptide and truncated Bid (tBid) is known to permeabilize MOM indirectly by activating pro-apoptotic Bax or Bak proteins (10, 12, 15, 19, 20, 40, 41), we tested whether Bid and Bad BH3 peptides could permeabilize the membrane in the presence of the purified Bax protein. As shown in Fig. 5C, the Bid-BH3 peptide displayed potent activity of releasing CB dextrans from liposomes in the presence of Bax, whereas Bad-BH3 showed weaker activity than Bid-BH3 (Fig. 5C). The different activities between Bid and Bad BH3 peptides for the Bax-mediated pore formation is consistent with previous reports (19, 41).
To estimate the potency of the pore forming activity of ATAP, we compared it with tBid-activated Bax. As shown in the Fig. 5D, both ATAP and tBid-activated Bax efficiently generate large pores. The activity of ATAP was higher than that of tBid-activated Bax. Because the size of a hydrated dextran of 10 kDa is similar to that of cytochrome c (42), the ATAP pore formed in the liposomal membrane would allow for cytochrome c release, as we previously observed in isolated mitochondria from BMK D3 cells (35). Moreover, because the ATAP pore in the MOM-liposomal membrane is formed in the absence of any additional proteins, ATAP alone may form a cytochrome c-releasing pore in MOM.
We next tested whether Bcl-2 could inhibit the pore formation by ATAP in the liposomal membrane. Because native Bcl-2 is an integral membrane protein anchored in MOM by its C-terminal transmembrane (TM) sequence, we tethered the purified recombinant Bcl-2 protein lacking the TM sequence (Bcl-2ΔTM) to the MOM-liposomal membrane via interaction of a His6 tag at the C terminus of Bcl-2ΔTM with a Ni2+-chelating lipid analog incorporated into the membrane as described previously (15), and we used this membrane-tethered recombinant Bcl-2 to mimic the native membrane-anchored Bcl-2 for the inhibition test. As shown in Fig. 5E, the pore forming activity of ATAP was not inhibited by Bcl-2 at various concentrations of Bcl-2, whereas the activity of tBid-activated Bax was significantly inhibited by Bcl-2. Parallel experiments with full-length Bcl-xL containing the C-terminal TM sequence and the MOM-liposomes lacking the Ni2+-lipid demonstrated that the pore forming activity of ATAP was also insensitive to Bcl-xL (data not shown). Together, these studies demonstrate that, unlike tBid- or BH3 peptide-activated Bax (16), the pore forming activity of ATAP is not inhibited by Bcl-2 or Bcl-xL, which is consistent with the finding from our cell-based assays (Fig. 2).
DISCUSSION
We conclude that ATAP uses a different mechanism to induce apoptosis than BH3 peptides based on the following evidence. (i) The pro-apoptotic activity of ATAP in cells is insensitive to Bcl-2 and Bcl-xL expression, whereas the pro-apoptotic effect of BH3 peptides is strongly inhibited by these anti-apoptotic proteins. (ii) The cell killing activity of ATAP does not require Bax or Bak, whereas BH3 peptides cause cell death in a strictly Bax- or Bak-dependent manner. These observations were complemented by our in vitro reconstitution studies that demonstrate that ATAP is sufficient to form cytochrome c-permeable pores in a MOM-mimicking liposomal membrane in the absence of other cellular proteins.
Our data provide a model for the pro-apoptotic action of ATAP in cells (Fig. 6A). Once it is delivered into cells, ATAP targets to mitochondria using an intrinsic mitochondrial targeting sequence (35). The specificity of this targeting may be achieved via a direct interaction with the MOM lipids or a protein-mediated process. The amphipathic nature of ATAP is required for the peptide to insert into the MOM lipid bilayer and form aqueous pores (35). Oligomerization of ATAP in the membrane may facilitate the formation of large pores that cause release of cytochrome c and potentially other mitochondrial proteins. Previously, several amphipathic peptides have been shown to induce mitochondrial membrane permeability transition and the release of cytochrome c by modulating the translocases of the outer and inner mitochondrial membrane (43, 44). Future studies are warranted to determine whether other mitochondrial proteins are involved in the pro-apoptotic action of ATAP, although our in vitro reconstitution assays suggest that other proteins are not required. For the pro-apoptotic function of BH3 peptides, there are at least two critical steps controlled by Bcl-2 family proteins (Fig. 6B). First, the cytosolic BH3 peptides must overwhelm the membrane-bound anti-apoptotic proteins such as Bcl-2 or Bcl-xL to antagonize their anti-apoptotic activity. Based on our studies, we speculate that at least 10–20-fold of excess BH3 peptide over Bcl-2 protein might be needed to trigger apoptosis. Sequestration of BH3 peptides to MOM represents a potent mechanism for the anti-apoptotic action of Bcl-2 or Bcl-xL. Second, there must be pro-apoptotic Bax or Bak protein in the cell in order for BH3 peptides to induce apoptosis.
Cancer cells often acquire resistance to apoptotic stimuli, including chemotherapeutics and γ-irradiations, by alteration of the expression levels of Bcl-2 family proteins. For example, anti-apoptotic Bcl-2 is overexpressed in 90% of human follicular lymphoma and causes chemoresistance in such tumors (45). Overexpression of Bcl-xL was detected in certain colorectal adenocarcinomas, which contributes to suppression of apoptosis (46). In contrast, Bax expression is reduced in approximately one-third of the metastatic breast cancers and correlates with poor response to treatment (47). Therefore, development of novel therapeutics that can induce apoptosis by directly targeting the Bcl-2 family proteins has been a major focus in the cancer research field. As an attempt to induce apoptosis by directly targeting the Bcl-2 family proteins, several groups have developed BH3 peptides derived from pro-apoptotic Bcl-2 family proteins or produced chemical compounds that could act as BH3 peptides. Recent biochemical and genetic studies suggest that BH3 peptides from BH3-only proteins such as Bid, Bad, Bim, and Noxa could antagonize the function of anti-apoptotic Bcl-2 family proteins in a very specific way and induce apoptosis by releasing pro-apoptotic Bax and/or Bak protein from the anti-apoptotic proteins (11, 12). Applications of BH3 peptides or their derivatives (peptidomimetics or chemical mimetics) produced some successful outcomes in treating cancers (20–25). However, this approach seems to have limitations when it is applied to cancers with overexpression of Bcl-2-related anti-apoptotic proteins or reduced expression of pro-apoptotic Bax and Bak, because these aberrant expression levels of Bcl-2 family proteins cause resistance to these BH3 peptides and mimetics (31–33, 48–50). Therefore, we must consider alternative strategies that can bypass the regulation by Bcl-2 family proteins in cancer cells, such as the application of ATAP as a treatment for cancer.
The results from this investigation suggest that ATAP can bypass the regulation by the Bcl-2 family to induce apoptosis. ATAP permeabilizes the mitochondrial membrane without the participation of Bax and Bak, which often display compromised function or reduced expression in cancer cells. Additionally, ATAP-induced apoptosis is not inhibited by Bcl-2 and other anti-apoptotic family members, which are frequently overexpressed in cancer cells. Therefore, ATAP represents a promising agent for development as a therapeutic that may escape the Bcl-2-mediated protection mechanisms and effectively kill resistant cancer cells.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants RO1-CA095739 and RO1-AG028614 (to J. M.), GM062964 (to J. L.), and K99/R00-AR054793 (to N. W.).
This article was selected as a Paper of the Week.
- MOM
- mitochondrial outer membrane
- ATAP
- amphipathic tail-anchoring peptide
- BH
- Bcl-2 homology
- BH3
- Bcl-2 homology domain-3
- CB
- Cascade Blue
- QVD-OPH
- Q-Val-Asp-OPH
- BMK cells
- baby mouse kidney cells
- BMH
- bis-maleimidohexane
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- TM
- transmembrane.
REFERENCES
- 1. Labi V., Grespi F., Baumgartner F., Villunger A. (2008) Cell Death Differ. 15, 977–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Meng X. W., Lee S. H., Kaufmann S. H. (2006) Curr. Opin. Cell Biol. 18, 668–676 [DOI] [PubMed] [Google Scholar]
- 3. Benz E. J., Jr., Nathan D. G., Amaravadi R. K., Danial N. N. (2007) Clin. Cancer Res. 13, 7250–7253 [DOI] [PubMed] [Google Scholar]
- 4. Vogler M., Dinsdale D., Dyer M. J., Cohen G. M. (2009) Cell Death Differ. 16, 360–367 [DOI] [PubMed] [Google Scholar]
- 5. Green D. R., Kroemer G. (2004) Science 305, 626–629 [DOI] [PubMed] [Google Scholar]
- 6. Chipuk J. E., Green D. R. (2008) Trends Cell Biol. 18, 157–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Leber B., Lin J., Andrews D. W. (2007) Apoptosis 12, 897–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Youle R. J., Strasser A. (2008) Nat. Rev. Mol. Cell Biol. 9, 47–59 [DOI] [PubMed] [Google Scholar]
- 9. Adams J. M., Cory S. (2007) Curr. Opin. Immunol. 19, 488–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kuwana T., Bouchier-Hayes L., Chipuk J. E., Bonzon C., Sullivan B. A., Green D. R., Newmeyer D. D. (2005) Mol. Cell 17, 525–535 [DOI] [PubMed] [Google Scholar]
- 11. Uren R. T., Dewson G., Chen L., Coyne S. C., Huang D. C., Adams J. M., Kluck R. M. (2007) J. Cell Biol. 177, 277–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Willis S. N., Fletcher J. I., Kaufmann T., van Delft M. F., Chen L., Czabotar P. E., Ierino H., Lee E. F., Fairlie W. D., Bouillet P., Strasser A., Kluck R. M., Adams J. M., Huang D. C. (2007) Science 315, 856–859 [DOI] [PubMed] [Google Scholar]
- 13. Billen L. P., Kokoski C. L., Lovell J. F., Leber B., Andrews D. W. (2008) PLoS Biol. 6, e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lovell J. F., Billen L. P., Bindner S., Shamas-Din A., Fradin C., Leber B., Andrews D. W. (2008) Cell 135, 1074–1084 [DOI] [PubMed] [Google Scholar]
- 15. Peng J., Tan C., Roberts G. J., Nikolaeva O., Zhang Z., Lapolla S. M., Primorac S., Andrews D. W., Lin J. (2006) J. Biol. Chem. 281, 35802–35811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tan C., Dlugosz P. J., Peng J., Zhang Z., Lapolla S. M., Plafker S. M., Andrews D. W., Lin J. (2006) J. Biol. Chem. 281, 14764–14775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang Z., Lapolla S. M., Annis M. G., Truscott M., Roberts G. J., Miao Y., Shao Y., Tan C., Peng J., Johnson A. E., Zhang X. C., Andrews D. W., Lin J. (2004) J. Biol. Chem. 279, 43920–43928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Willis S. N., Adams J. M. (2005) Curr. Opin. Cell Biol. 17, 617–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Letai A., Bassik M. C., Walensky L. D., Sorcinelli M. D., Weiler S., Korsmeyer S. J. (2002) Cancer Cell 2, 183–192 [DOI] [PubMed] [Google Scholar]
- 20. Walensky L. D., Kung A. L., Escher I., Malia T. J., Barbuto S., Wright R. D., Wagner G., Verdine G. L., Korsmeyer S. J. (2004) Science 305, 1466–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Oltersdorf T., Elmore S. W., Shoemaker A. R., Armstrong R. C., Augeri D. J., Belli B. A., Bruncko M., Deckwerth T. L., Dinges J., Hajduk P. J., Joseph M. K., Kitada S., Korsmeyer S. J., Kunzer A. R., Letai A., Li C., Mitten M. J., Nettesheim D. G., Ng S., Nimmer P. M., O'Connor J. M., Oleksijew A., Petros A. M., Reed J. C., Shen W., Tahir S. K., Thompson C. B., Tomaselli K. J., Wang B., Wendt M. D., Zhang H., Fesik S. W., Rosenberg S. H. (2005) Nature 435, 677–681 [DOI] [PubMed] [Google Scholar]
- 22. Wang G., Nikolovska-Coleska Z., Yang C. Y., Wang R., Tang G., Guo J., Shangary S., Qiu S., Gao W., Yang D., Meagher J., Stuckey J., Krajewski K., Jiang S., Roller P. P., Abaan H. O., Tomita Y., Wang S. (2006) J. Med. Chem. 49, 6139–6142 [DOI] [PubMed] [Google Scholar]
- 23. Wang J. L., Liu D., Zhang Z. J., Shan S., Han X., Srinivasula S. M., Croce C. M., Alnemri E. S., Huang Z. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 7124–7129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yin H., Lee G. I., Sedey K. A., Kutzki O., Park H. S., Orner B. P., Ernst J. T., Wang H. G., Sebti S. M., Hamilton A. D. (2005) J. Am. Chem. Soc. 127, 10191–10196 [DOI] [PubMed] [Google Scholar]
- 25. Zeitlin B. D., Joo E., Dong Z., Warner K., Wang G., Nikolovska-Coleska Z., Wang S., Nör J. E. (2006) Cancer Res. 66, 8698–8706 [DOI] [PubMed] [Google Scholar]
- 26. Kitada S., Leone M., Sareth S., Zhai D., Reed J. C., Pellecchia M. (2003) J. Med. Chem. 46, 4259–4264 [DOI] [PubMed] [Google Scholar]
- 27. Oliver C. L., Miranda M. B., Shangary S., Land S., Wang S., Johnson D. E. (2005) Mol. Cancer Ther. 4, 23–31 [PubMed] [Google Scholar]
- 28. Xu L., Yang D., Wang S., Tang W., Liu M., Davis M., Chen J., Rae J. M., Lawrence T., Lippman M. E. (2005) Mol. Cancer Ther. 4, 197–205 [PubMed] [Google Scholar]
- 29. Cosulich S. C., Worrall V., Hedge P. J., Green S., Clarke P. R. (1997) Curr. Biol. 7, 913–920 [DOI] [PubMed] [Google Scholar]
- 30. Polster B. M., Kinnally K. W., Fiskum G. (2001) J. Biol. Chem. 276, 37887–37894 [DOI] [PubMed] [Google Scholar]
- 31. van Delft M. F., Wei A. H., Mason K. D., Vandenberg C. J., Chen L., Czabotar P. E., Willis S. N., Scott C. L., Day C. L., Cory S., Adams J. M., Roberts A. W., Huang D. C. (2006) Cancer Cell 10, 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vogler M., Butterworth M., Majid A., Walewska R. J., Sun X. M., Dyer M. J., Cohen G. M. (2009) Blood 113, 4403–4413 [DOI] [PubMed] [Google Scholar]
- 33. Konopleva M., Contractor R., Tsao T., Samudio I., Ruvolo P. P., Kitada S., Deng X., Zhai D., Shi Y. X., Sneed T., Verhaegen M., Soengas M., Ruvolo V. R., McQueen T., Schober W. D., Watt J. C., Jiffar T., Ling X., Marini F. C., Harris D., Dietrich M., Estrov Z., McCubrey J., May W. S., Reed J. C., Andreeff M. (2006) Cancer Cell 10, 375–388 [DOI] [PubMed] [Google Scholar]
- 34. Vogler M., Weber K., Dinsdale D., Schmitz I., Schulze-Osthoff K., Dyer M. J., Cohen G. M. (2009) Cell Death Differ. 16, 1030–1039 [DOI] [PubMed] [Google Scholar]
- 35. Ko J. K., Choi K. H., Pan Z., Lin P., Weisleder N., Kim C. W., Ma J. (2007) J. Cell Sci. 120, 2912–2923 [DOI] [PubMed] [Google Scholar]
- 36. Yang W. S., Ko J. K., Park S. O., Choi H. Y., Kim Y. N., Kim C. W. (2005) J. Cell. Biochem. 94, 1234–1247 [DOI] [PubMed] [Google Scholar]
- 37. Yethon J. A., Epand R. F., Leber B., Epand R. M., Andrews D. W. (2003) J. Biol. Chem. 278, 48935–48941 [DOI] [PubMed] [Google Scholar]
- 38. Degenhardt K., Sundararajan R., Lindsten T., Thompson C., White E. (2002) J. Biol. Chem. 277, 14127–14134 [DOI] [PubMed] [Google Scholar]
- 39. Wood D. E., Newcomb E. W. (2000) Exp. Cell Res. 256, 375–382 [DOI] [PubMed] [Google Scholar]
- 40. Kuwana T., Mackey M. R., Perkins G., Ellisman M. H., Latterich M., Schneiter R., Green D. R., Newmeyer D. D. (2002) Cell 111, 331–342 [DOI] [PubMed] [Google Scholar]
- 41. Walensky L. D., Pitter K., Morash J., Oh K. J., Barbuto S., Fisher J., Smith E., Verdine G. L., Korsmeyer S. J. (2006) Mol. Cell 24, 199–210 [DOI] [PubMed] [Google Scholar]
- 42. Venturoli D., Rippe B. (2005) Am. J. Physiol. Renal Physiol. 288, F605–F613 [DOI] [PubMed] [Google Scholar]
- 43. Kushnareva Y. E., Polster B. M., Sokolove P. M., Kinnally K. W., Fiskum G. (2001) Arch. Biochem. Biophys. 386, 251–260 [DOI] [PubMed] [Google Scholar]
- 44. Muro C., Grigoriev S. M., Pietkiewicz D., Kinnally K. W., Campo M. L. (2003) Biophys. J. 84, 2981–2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Crisan D. (1996) Clin. Lab. Med. 16, 23–47 [PubMed] [Google Scholar]
- 46. Krajewska M., Moss S. F., Krajewski S., Song K., Holt P. R., Reed J. C. (1996) Cancer Res. 56, 2422–2427 [PubMed] [Google Scholar]
- 47. Krajewski S., Blomqvist C., Franssila K., Krajewska M., Wasenius V. M., Niskanen E., Nordling S., Reed J. C. (1995) Cancer Res. 55, 4471–4478 [PubMed] [Google Scholar]
- 48. Chen S., Dai Y., Harada H., Dent P., Grant S. (2007) Cancer Res. 67, 782–791 [DOI] [PubMed] [Google Scholar]
- 49. Deng J., Carlson N., Takeyama K., Dal Cin P., Shipp M., Letai A. (2007) Cancer Cell 12, 171–185 [DOI] [PubMed] [Google Scholar]
- 50. Tahir S. K., Yang X., Anderson M. G., Morgan-Lappe S. E., Sarthy A. V., Chen J., Warner R. B., Ng S. C., Fesik S. W., Elmore S. W., Rosenberg S. H., Tse C. (2007) Cancer Res. 67, 1176–1183 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.