Abstract
Pmel17 is a melanocyte/melanoma-specific protein that traffics to melanosomes where it forms a fibrillar matrix on which melanin gets deposited. Before being cleaved into smaller fibrillogenic fragments the protein undergoes processing by proprotein convertases, a class of serine proteases that typically recognize the canonical motif RX(R/K)R↓. The current model of Pmel17 maturation states that this processing step occurs in melanosomes, but in light of recent reports this issue has become controversial. We therefore addressed this question by thoroughly assessing the processing kinetics of either wild-type Pmel17 or a secreted soluble Pmel17 derivative. Our results demonstrate clearly that processing of Pmel17 occurs during secretion and that it does not require entry of the protein into the endocytic system. Strikingly, processing proceeds even in the presence of the secretion inhibitor monensin, suggesting that Pmel17 is an exceptionally good substrate. In line with this, we find that newly synthesized surface Pmel17 is already quantitatively cleaved. Moreover, we demonstrate that Pmel17 function is independent of the sequence identity of its unconventional proprotein convertase-cleavage motif that lacks arginine in P4 position. The data alter the current view of Pmel17 maturation and suggest that the multistep processing of Pmel17 begins with an early cleavage during secretion that primes the protein for later functional processing.
Keywords: Golgi, Protease, Protein Processing, Protein Secretion, Trafficking, Melanosome, Pmel17, Proprotein Convertase, gp100, Trans-Golgi Network
Introduction
Pmel17 (also called gp100, silver, or ME20) is a melanosomal glycoprotein that forms an extensive fibrillar matrix, on which the pigment melanin gets deposited (1). Moreover, these fibrils likely serve for sequestration of toxic reaction intermediates of the melanin synthesis pathway (2, 3). In line with this, Pmel17-defective melanocytes show reduced viability in vivo (4). Consequently, as the major structural component of melanosomes (5), Pmel17 is a key factor in the process of synthesizing and storing melanin, two of the main functions of melanocytes (6).
After insertion into the endoplasmic reticulum (ER)3 membrane as a type I transmembrane glycoprotein called the P1 form, Pmel17 gets exported to the Golgi apparatus (1). There, maturation of oligosaccharides converts Pmel17 into the so-called P2 form, which has an ∼20 kDa higher apparent molecular mass and thus can be well separated from P1 in a gel (7). Subsequently, the protein migrates through the trans-Golgi network (TGN) and from there is routed either directly or via the plasma membrane to melanosomes (8). Immature Pmel17 may also be at the cell-surface to a low extent (9, 10), although other studies have disputed this finding (11). Within melanosomes Pmel17 gets processed into smaller fibrillogenic fragments, which eventually form the fibrous network for melanin deposition (1). This final step in Pmel17 maturation is probably initiated by a membrane-proximal cleavage carried out by a metalloprotease of the a disintegrin and metalloprotease (ADAM) family (12), which releases a soluble fragment that gets further processed into smaller fibrillogenic subunits. Among these are a set of ∼35–45-kDa fragments derived from the repeat domain (RPT) and reactive with monoclonal antibody HMB45 (11, 13–16) as well as a ∼7-kDa fragment derived from the polycystic kidney disease-like domain (PKD) reactive with antibody I51 (17, 18).
During the course of maturation the P2 form of Pmel17 gets processed by a proprotein convertase (pPC) into an N-terminal Mα and a C-terminal Mβ fragment (7). These fragments, however, remain linked to each other by a disulfide bridge. pPC-mediated cleavage is absolutely essential for fibril formation (7), but post-Golgi trafficking appears to depend only little or not at all on this processing step (7). Surprisingly, the generation of at least the set of HMB45-reactive fragments is maintained in a Pmel17 mutant that cannot be cleaved (11, 12, 14).
Theos and co-workers (19) had previously addressed the question in which compartment Pmel17 undergoes pPC processing by using mutants of the protein lacking either the N-terminal region (NTR) or the PKD domain (Δ29–200 and Δ225–292, respectively). Both of these mutants appeared to be impaired in the delivery to intralumenal vesicles within stage I melanosomes, underwent massive missorting to early endosomes, and failed to be cleaved at the pPC-processing motif (19). Based on this data the authors concluded that pPC-mediated cleavage occurs within early stage melanosomes after or during budding from the limiting membrane into the interior of the organelle (19). If this were the case, pPC-mediated processing would probably immediately precede fibril formation and might even be the activating event allowing (or even driving) an instantly following protease cascade that generates the downstream fibrillogenic fragments. Because Pmel17 fibrils represent amyloid aggregates (2) and are thus potentially very toxic structures to a cell when generated in the wrong place, it is certainly of great interest which process actually activates their formation and how this process is controlled.
However, the promising idea that Pmel17 undergoes pPC-mediated cleavage in melanosomes has become increasingly controversial. First, our own work (18) and the work of others (14) has demonstrated that NTR-deletion mutants (Δ28–208 and Δ25–254, respectively) can actually be cleaved by pPCs. In fact, at least for Δ28–208 (called ΔNTR in Ref. 18), cleavage is even more efficient than for wild-type Pmel17 (wt-Pmel17) (18). However, both mutants show a strikingly similar intracellular mislocalization to early endosomes like the NTR-deletion construct originally reported by Theos et al. (14, 18, 19). Importantly, this suggests that failure to target to melanosomes does not necessarily cause a failure to undergo pPC-mediated processing. Consistent with the fact that the NTR-deletion mutant used by Theos and co-workers (11, 19) largely loses reactivity with conformation-sensitive (18) antibodies HMB50 and NKI-beteb, this suggests that the aberrant behavior of this mutant is partially caused by misfolding rather than that correct localization to melanosomes would be a prerequisite for pPC-mediated processing. Furthermore, recent reports demonstrate that a small amount of Pmel17 gets shed from the cell surface (20) and surprisingly this portion of the protein has been shown to be already pPC cleaved (21). Although, it cannot be excluded that the small fraction of Pmel17 destined to be shed from the cell surface is treated differently in the cell than the major protein pool or that the respective shed protein derives from a recycled population that has formerly been cleaved in melanosomes, this result actually suggests that pPC-mediated cleavage of Pmel17 occurs at an early time point prior to plasma membrane exposure, i.e. during secretion and before entry into the endocytic system.
To clarify these issues and conclusively determine whether Pmel17 is cleaved during or after secretion, we assessed the proteolytic maturation of a stably transfected secreted soluble derivative of Pmel17 in melanoma cells. The respective protein never accesses the endocytic system at any time, but strikingly it gets almost quantitatively cleaved by pPCs and this processing is sensitive to pPC inhibitors. Moreover, cleavage of wt-Pmel17 proceeds even in the presence of the secretion inhibitor monensin, suggesting that it is an exceptionally good pPC substrate. In line with this, using various assays we find all surface Pmel17 to be already quantitatively cleaved and this applies to both newly synthesized Pmel17 as well as the protein present at the cell surface at steady-state. Finally, and consistent with the aforementioned results we demonstrate that the sequence identity of the unconventional pPC-cleavage motif (lacking arginine in P4-position) is neither essential for pPC-mediated cleavage nor for fibril formation and function of Pmel17.
Taken together, our report alters the current picture of Pmel17 maturation and suggests that the protein undergoes early pPC-mediated processing during secretion. This may prime the polypeptide for the later protease cascade that gives rise to fibrillogenic fragments, but does not immediately trigger this process. In this way Pmel17 may be much more similar to other pPC substrates, like e.g. Notch (22) and other receptors, which also get cleaved by pPCs during secretion, but in a manner temporarily and spatially separated from actual ligand-mediated activation.
EXPERIMENTAL PROCEDURES
Cell Lines and Cell Culture
LG2-MEL-220 (Mel220), a human Pmel17-deficient melanoma cell line (23), was grown in Iscove's modified Dulbecco's medium (Sigma), 10% FCS (HyClone) containing non-essential amino acids (Invitrogen), GlutaMax (Invitrogen), and penicillin/streptomycin (Invitrogen). Mel220 cells expressing wild-type or mutant Pmel17-i (1, 24) were grown in medium additionally containing 2 mg/ml of G418 (Invitrogen). Mel220 cells expressing wild type Pmel17-i have been described previously (18).
Antibodies
Pep13h (20) and Pmel-N (25) are peptide antibodies recognizing the C and N terminus of newly synthesized Pmel17, respectively. HMB50 (26), NKI-beteb (Abcam), and HMB45 (NeoMarkers) are mouse monoclonal antibodies (IgG2a, IgG2b, and IgG1, respectively) recognizing the folded PKD domain in a conformation-sensitive manner (HMB50 and NKI-beteb) (11, 18) or a sialylated epitope within the RPT domain (HMB45) (15) of Pmel17. The monoclonal antibodies 148.3 (27), 610823 (BD), H4A3 (IgG1) (Abcam), and 9E10 (IgG1) recognize the ER marker TAP1, Golgi marker GM130, lysosomal marker LAMP1, and myc tag, respectively. The rabbit polyclonal antibodies R.gp48N (28) and ab9106 (Abcam) recognize tapasin and the myc tag, respectively. HRP- or fluorophore-labeled isotype-specific or conventional goat anti-mouse and goat anti-rabbit antibodies were purchased from Molecular Probes or Jackson ImmunoResearch.
Vector Constructs and Pmel17 Expression
Pmel17-i (also termed gp100) (24) within expression vector pBMN-IRES-neo (18) served as a template for a standard QuikChange mutagenesis using primer pair 5′-GCCACCTTAAGGCCCAGCCGAAAAAGACGCAGTCTGGATTGTGTTCTG-3′/5′-CAGAACACAATCCAGACTGCGTCTTTTTCGGCTGGGCCTTAAGGTGGC-3′ for generation of construct IR. Construct sPmel17-myc was generated in a similar fashion using a two-step QuikChange procedure with primer pair 5′-GGCAGGTTCCGGAGCAAAAGCTTATCGTCTGAGCGGCG-3′/5′-CGCCGCTCAGACGATAAGCTTTTGCTCCGGAACCTGCC-3′ followed by primer pair 5′-CGGAGCAAAAGCTAATCTCAGAGGAGGACCTCTGAGCGGCG-3′/5′-CGCCGCTCAGAGGTCCTCCTCTGAGATTAGCTTTTGCTCCG-3′. All pBMN vectors containing mutant or wild-type Pmel17 were sequenced from both directions before retroviral transduction into Mel220 cells (29). Mel220 transfectants expressing wild type or mutant Pmel17 were selected in medium containing 2 mg/ml of G418 (Invitrogen) for 3 weeks and expression of Pmel17 was assessed by Western blot.
Immunofluorescence and Flow Cytometry
Mel220 transfectants were seeded overnight on glass coverslips. The next day, cells were washed with PBS containing 0.9 mm CaCl2 and 0.5 mm MgCl2 (PBS+/+) and fixed with 2% formaldehyde (15 min at room temperature). After quenching with PBS+/+, 10 mm glycine followed by a wash with PBS+/+, 0.5% BSA, cells were permeabilized for 1 h in staining buffer (PBS+/+, 0.5% BSA, 0.5% saponin). Staining was performed in a humidity chamber for 1 h with the indicated primary antibodies at concentrations recommended by the manufacturer or 1:50 for 148.3, 1:100 for 9E10, and 1:100 for HMB50. After three washes with staining buffer, Alexa 647-, Alexa 546-, or Alexa 488-conjugated secondary antibodies (Molecular Probes) were applied at a 1:100 dilution in the same buffer, before cells were washed again three times, cells were mounted in ProLong Gold reagent (Invitrogen) and analyzed by confocal fluorescence microscopy using a Leica TCS SP2 Confocal Microscope (Leica Microsystems).
Flow cytometry was performed as described (18) using antibody NKI-beteb at a concentration of 1:10 followed by Alexa 647-conjugated secondary antibodies. Life-gating was performed using YO-PRO-1 iodide (Invitrogen). All data were acquired on a FACSCalibur flow cytometer and analyzed using FlowJo 6.4.7 software (Tree Star).
Electron Microscopy
For conventional Epon embedding of cell samples, Mel220 transfectants were fixed in 2.5% glutaraldehyde, 2% sucrose in 0.1 m sodium cacodylate buffer, pH 7.4 (NaCaCo buffer), for 30 min at room temperature (RT) followed by another 30 min in the same fixation solution at 4 °C. Subsequently, cells were rinsed with NaCaCo buffer and further processed as described (30).
For cryo-immunoelectron microscopy, samples were fixed in 2% paraformaldehyde, 0.1% glutaraldehyde in PBS for 15 min at RT followed by another 15 min in the same fixation solution at 4 °C. Subsequently, cells were rinsed with PBS and further processed as described (30). For immunolabeling, cells were stained with Pmel17-specific antibodies Pep13h or HMB50 at 1:25 followed by protein A-gold (University of Utrecht, Netherlands) or gold anti-mouse conjugate (Jackson ImmunoResearch Laboratories), respectively.
For embedding of cell samples in LR-gold resin (London Resin Gold), cells were fixed in 4% paraformaldehyde, 0.1% glutaraldehyde, 2% sucrose in 0.1 m HEPES buffer for 30 min at RT followed by another 90 min in the same fixation solution, but lacking glutaraldehyde at 4 °C. Subsequently, cells were rinsed with PBS and 50 mm NH4Cl, 100 mm glycine, 3% sucrose for 15 min to quench free aldehydes, scraped in 1% gelatin, and transferred to 5% agar. Once set of samples were placed in 0.5% tannic acid in 0.1 m HEPES for 30 min, rinsed twice in Tris, 50 mm maleate, 3% sucrose (sucrose-maleate buffer), and stained with 2% uranyl acetate in sucrose-maleate buffer. Samples were dehydrated through a graded series of ethanol (50 to 95%) at −20 °C and embedded in LR-gold resin (EMS) at −20 °C. For immunolabeling cells were stained with Pmel17-specific antibody HMB50 at 1:10 followed by gold anti-mouse conjugate (Jackson ImmunoResearch Laboratories).
Samples were viewed using an FEI Tencai Biotwin TEM at 80 Kv. Images were taken using Morada CCD and iTEM (Olympus) software.
Pulse-Chase Analysis, Immunoprecipitation, and Western Blotting
Radiolabeling was performed as described (31). Briefly, 1.5 × 107 starved Mel220 cells expressing Pmel17 derivatives were pulse-labeled at 37 °C with [35S]methionine/cysteine (PerkinElmer Life Sciences) at 0.5 mCi/ml in 1.5 ml for 30 min and subsequently chased in Iscove's modified Dulbecco's medium, 10% FCS containing an excess of cold l-methionine/l-cysteine (both at 0.45 mg/ml) for up to 4 h. Following this, cells were harvested and frozen at −80 °C until the next day or immediately lysed in 2% Triton X-100 (containing protease inhibitor mixture (Roche Applied Science)) and pre-cleared overnight using protein A. Supernatants were always immediately pre-cleared overnight using protein A.
For immunoprecipitation, antibody HMB50-coupled protein A-Sepharose was used. If the pre-clear had not been performed overnight, the frozen cell pellets were thawed, lysed in 2% Triton X-100 (containing protease inhibitor mixture (Roche)) at 107 cells/ml, and precleared using protein A-Sepharose beads. Subsequently, the supernatant was applied to the HMB50-coupled beads and immunoprecipitation was carried out as described (31). After separation of immunoprecipitates by SDS-PAGE, gels were dried, exposed to PhosphorImager screens, and analyzed with ImageQuant 5.2 (GE Healthcare).
Where pharmacological inhibitors were used in a pulse-chase experiment they were included during both labeling and chase period. The pPC-inhibitor Dec-RVKR-CMK (32) (Calbiochem) was used at 100 μm, whereas the secretion inhibitor monensin (33) (eBioscience) was used at 10 μm if not otherwise indicated. Brefeldin A (34) was used at 10 μg/ml. For a specific surface immunoprecipitation, intact cells were incubated with antibody HMB50 for 1 h on ice, before lysis in 2% Triton X-100 (containing protease inhibitor mixture (Roche Applied Science)) and addition of protein A to the postnuclear supernatant to immunoprecipitate labeled surface proteins.
Surface biotinylation was carried out using the cell-impermeable EZ-Link Sulfo-NHS-SS-Biotin reagent (Pierce) according to the protocol of the manufacturer or performing the biotinylation step at 4 °C where indicated. After cell lysis biotinylated proteins were isolated with Monomeric Avidin-agarose (Pierce). Western blotting was carried out as described (31).
RESULTS
The Sequence Identity of the Endogenous Proprotein Convertase Cleavage Motif in Pmel17 Is Not Essential for Fibril Formation
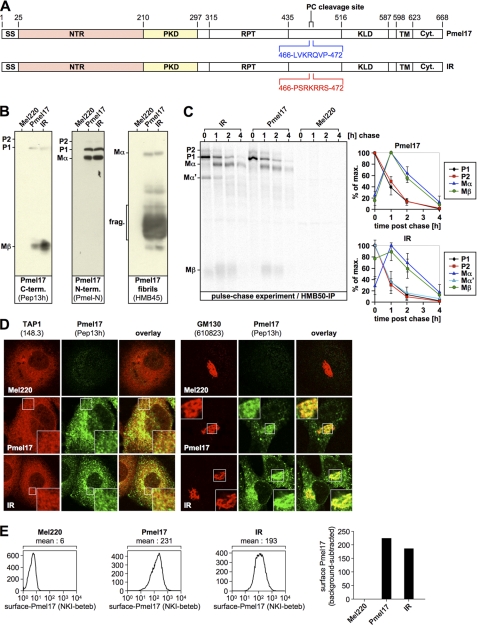
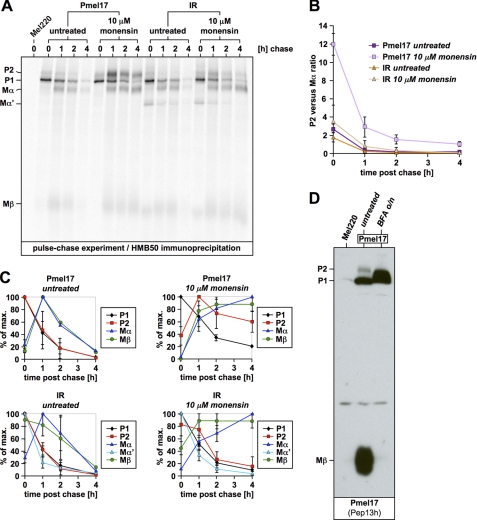
It has been claimed by others that pPC-mediated processing of Pmel17 only takes place in melanosomes (19), but not in the pPC-rich secretory compartments (e.g. the TGN) that the protein traverses before entering the endocytic system (11, 35). If that were the case, one would have to postulate a mechanism that protects Pmel17 from cleavage along the secretory route until proper delivery to melanosomes occurs. One possible way to achieve this would be if Pmel17 were equipped with an unusual pPC-processing motif refractory to TGN-resident pPCs, but susceptible to a specialized enzyme localized only to melanosomes (we call this the “specificity scenario” hereafter). Indeed, Pmel17 has a quite unconventional pPC-cleavage site (LRLVKR↓) (7) that deviates from the consensus processing motif (RX(R/K)R↓) (32, 36, 37) by the lack of an arginine residue in the P4 position. In fact, for most pPCs including furin, an arginine in this position is very critical and often essential for substrate cleavage (37–42). Therefore, to test, whether the sequence identity of this motif is necessary for Pmel17 function or whether it can be replaced by another pPC-processing element, we engineered a Pmel17 mutant, containing an optimal (36) cleavage motif derived from the human proinsulin receptor (43) (PSRKRR↓S) (Fig. 1A). This mutant (hereinafter IR) was expressed in the Pmel17-deficient melanoma cell line LG2-MEL-220 (Mel220) (23) and analyzed by Western blot (Fig. 1B). As expected, newly synthesized IR, which reacts with antibodies Pep13h and Pmel-N, was efficiently cleaved by pPCs as evidenced by the presence of P2-cleavage products Mα and Mβ (Fig. 1B). With regard to the steady-state levels of Mα and Mβ, we observed, if anything, more efficient cleavage, which is consistent with the fact that IR contains an “optimal” pPC motif. When the processing kinetics of IR were assessed in a pulse-chase experiment, we found a very similar profile and rate for the generation of Pmel17 cleavage fragments as for the wild type protein, with the very interesting exception that at early time points IR temporarily gave rise to one additional fragment, Mα′. Because, this fragment appears together with Mβ at the beginning of the chase (Fig. 1C, lane 1), whereas neither this fragment nor Mβ are observed this early for wt-Pmel17 (Fig. 1C, lane 5), it almost certainly corresponds to a cleaved P1 (instead of P2) form. This conclusion is, furthermore, supported by the fact that Mα′ is endoglycosidase H-sensitive (supplemental Fig. S1) and thus ER associated. We note that pPC-mediated processing does not normally occur in the ER (44). However, exceptionally favored substrates have been reported to be able to undergo cleavage even in this compartment (45) if they are able to displace the inhibitory prosegment, which usually keeps pPCs inactive (44). In particular this phenomenon has been observed for a derivative of the human proinsulin receptor (46), whose pPC motif was employed for generation of the IR construct. Thus, that IR is partially cleaved at a very early time point (Fig. 1C) is not entirely unexpected. However, later maturation seems to be very similar for IR and wt-Pmel17, if the levels and decay kinetics of Mα and Mβ at 1, 2, and 4 h post chase are considered (Fig. 1C, compare second to fourth lanes to sixth to eighth lanes). Consistent with a relatively normal (18) behavior of IR, we also observed by immunofluorescence a normal distribution pattern of newly synthesized protein between the ER (Fig. 1D, left panel) and the Golgi apparatus (Fig. 1D, right panel). Moreover, wt-Pmel17 and IR were present to a similar extent at the cell surface (Fig. 1E) and gave rise to similar levels of HMB45-reactive fibrillogenic fragments (Fig. 1B, right panel).
FIGURE 1.
The sequence identity of the endogenous proprotein convertase-cleavage motif within Pmel17 is not essential for proper early maturation. A, schematic representation of the IR construct. B, IR is processed by pPCs to give rise to Mα, Mβ, and HMB45-reactive fibrillogenic fragments. A total membrane fraction derived from the indicated stable Mel220 transfectants was lysed in 1% SDS, 1% β-mercaptoethanol + protease inhibitors (Complete, Roche Applied Science) and analyzed by Western blot using Pmel17-specific antibodies. C, IR displays a relatively normal early maturation. Cells from B were pulse-labeled for 30 min with 35S and subsequently chased for the indicated times. 2% Triton X-100 lysates were immunoprecipitated with Pmel17-specific antibody HMB50, eluted with 0.5% SDS by boiling for 5 min, and analyzed by autoradiography (left panel). Quantitative PhosphorImager analysis of the pulse-chase data with maximal levels for each band set to 100% is shown (right panel). Error bars reflect the standard deviation from the mean of two independent experiments. D, newly synthesized IR localizes to the ER and Golgi apparatus. Cells from B were analyzed by immunofluorescence using antibodies against newly synthesized Pmel17 (Pep13h) and organelle markers TAP1 (148.3) (ER) or GM130 (610823) (Golgi). A higher magnification of the indicated area is shown as an inset within each image. E, IR is expressed at the cell surface. Cells from B were surface labeled with antibody NKI-beteb against folded Pmel17 and analyzed by flow cytometry (histograms on the left). After background subtraction (untransfected Mel220 cells) data are represented as a bar diagram (right panel).
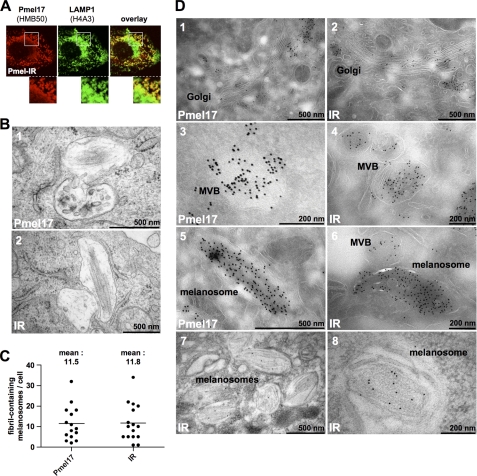
When the distribution of mature protein was assessed, we found IR mostly in a “horseshoe profile” that only weakly co-labeled for the lysosomal marker LAMP1 (Fig. 2A). We have recently described this pattern for wt-Pmel17 in Mel220 cells and determined that it reflects a melanosomal distribution (18). In contrast, the perinuclear LAMP1high-compartments, which we had previously shown to be lysosomes (18), were only low in IR protein, which mimics our observations with wt-Pmel17 (data not shown) (18). When electron microscopy (EM) was employed we clearly observed fibril formation for IR (Fig. 2B) and strikingly, quantification of these results demonstrated an identical number of fibril-containing melanosomes in cells expressing either wt-Pmel17 or the mutant (Fig. 2C). Moreover, when the subcellular distribution of the HMB50-reactive IR was assessed by cryo-immuno-EM, we found an identical pattern as for wt-Pmel17, in that low levels of protein were detected in the Golgi apparatus (Fig. 2D, compare panels 1 and 2), some of the protein localized to multivesicular bodies (Fig. 2D, compare panels 3 and 4), whereas ellipsoid melanosomes showed extensive labeling (Fig. 2D, compare panels 5 and 6). Finally, when we immunolabeled LR-gold embedded EM samples, a procedure, which in our hands substantially improves the preservation of Pmel17 fibrils, we found most gold particles distributed along these fibrils (Fig. 2D, panels 7 and 8).
FIGURE 2.
The sequence identity of the endogenous proprotein convertase-cleavage motif within Pmel17 is not essential for fibril formation. A, most IR in the cell is distributed in a melanosomal pattern, distinct from lysosomes. Mel220 cells stably expressing the IR mutant were analyzed by immunofluorescence using antibodies against folded Pmel17 (HMB50) and LAMP1 (H4A3). A higher magnification of the indicated area is shown at the bottom of each image. B, Mel220 cells expressing IR harbor fibril-containing melanosomes. Electron microscopic analysis of Epon-embedded Mel220 transfectants stably expressing wt-Pmel17 (upper panel) or IR (lower panel). C, IR and wt-Pmel17 are equally efficient in fibril formation. Fibril-containing melanosomes were counted in Epon-embedded samples of Mel220 cells stably expressing wt-Pmel17 or mutant IR. D, IR mostly localizes to fibrils. Mel220 cells expressing wt-Pmel17 or mutant IR were fixed and examined by cryo-immuno-EM (panels 1–6) or by immunolabeling of LR gold-embedded samples (panels 7 and 8) with antibody HMB50. Shown are the Golgi apparatus (panels 1 and 2), multivesicular bodies (MVB) (panels 3 and 4), or melanosomes (panels 5–8).
Altogether, this demonstrates that IR is a fully functional mutant and hence the unusual pPC cleavage site of Pmel17 is not essential for fibril formation. Consequently, if the specificity scenario were correct, protection of Pmel17 from cleavage during secretion would not be necessary for its function.
Entry into the Endocytic System Is Not Necessary for Proprotein Convertase-mediated Cleavage of Pmel17
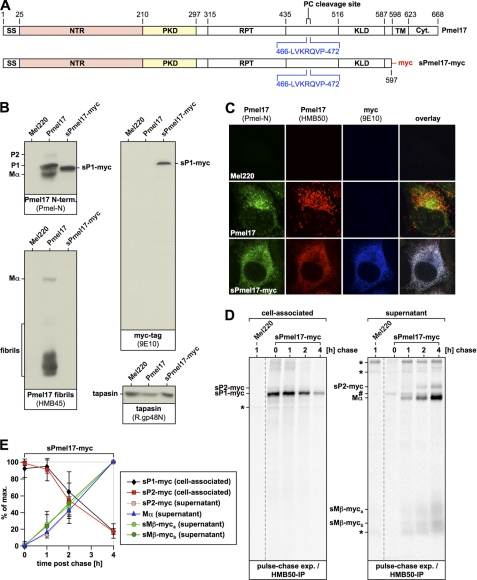
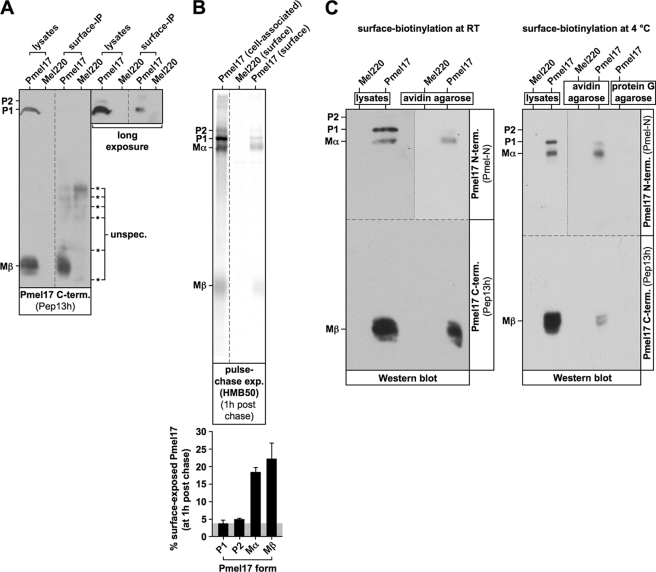
As an alternative to the specificity scenario there is another possible mechanism by which melanosome-restricted cleavage of Pmel17 could be achieved. In this alternative scenario the pPC-processing motif of Pmel17 would be in principle susceptible to all pPCs in the cell, but the protein would fold into a conformation that limits access of proteases to the cleavage site. This conformation would then relax only once Pmel17 enters the melanosome (either via acidic pH or with the assistance of specific melanosomal factors), thus allowing a subcellularly localized cleavage to occur specifically in this compartment (we call this the “conformation scenario” hereafter). If either the specificity scenario or the conformation scenario were correct, access of Pmel17 to the endocytic system would be essential for pPC-mediated processing. To examine whether this is the case, we constructed a soluble, secreted C-terminal deletion mutant of Pmel17 (truncated after amino acid residue proline 597) that contains all lumenal domains, but lacks both the transmembrane region and the cytosolic tail (herein after sPmel17-myc). Instead the protein is tagged at the C terminus with a myc epitope (Fig. 3A). When stably expressed in Mel220 cells, the only cell-associated sPmel17-myc species that could be detected at steady-state by Western blotting was the soluble P1 form (sP1-myc), reactive with antibodies Pmel-N (anti-Pmel17 N terminus) (Fig. 3B, left upper panel) and 9E10 (anti-myc) (Fig. 3B, right upper panel). This form migrated between wt-Pmel17-derived full-length P1 and Mα in a gel (Fig. 3B, left upper panel) and did not react with antibody Pep13h (anti-Pmel17-C terminus) (data not shown). As expected for a secreted mutant of Pmel17, we did not observe any HMB45-reactive fibrillogenic fragments in sPmel17-myc-expressing cells (Fig. 3B, left lower panel). Consistent with our Western blotting results, all cell-associated protein detected at steady-state by immunofluorescence was localized to the ER (Fig. 3C). This was the case no matter whether we stained sPmel17-myc with antibodies that for the wild-type protein in this setting detect almost only newly synthesized protein (Pmel-N) (Fig. 3C, first column), mature fibril-associated protein (HMB50) (Fig. 3C, second column), or with anti-myc antibodies that should label all sPmel17-myc in the cell (Fig. 3C, third column).
FIGURE 3.
A soluble Pmel17 mutant gets secreted from cells in a proprotein convertase-cleaved form. A, schematic representation of the sPmel17-myc construct. B, only the ER-associated P1 form can be detected for sPmel17-myc inside the cells at steady-state. Membrane lysates of Mel220 transfectants stably expressing sPmel17-myc were prepared as in Fig. 1B and analyzed by Western blot using Pmel17-specific antibodies, myc-specific antibodies, or tapasin-specific antibodies for control. C, almost all intracellular sPmel17-myc is localized to the ER. Cells from B were analyzed by immunofluorescence using antibodies against newly synthesized Pmel17 (Pmel-N), mature Pmel17 (HMB50), or the myc-tag (9E10). D, soluble sPmel17-myc gets secreted into the culture medium. Mel220 transfectants stably expressing sPmel17-myc were pulse-labeled for 30 min with 35S and subsequently chased for the indicated times. 2% Triton X-100 lysates (left panel) or culture supernatants (right panel) were immunoprecipitated with Pmel17-specific antibody HMB50, eluted with 0.5% SDS by boiling for 5 min, and analyzed by autoradiography. The dashed lines indicate positions where irrelevant lanes have been removed from the image. The pound symbol indicates background levels of the precipitated P1 form. The asterisks indicate nonspecifically precipitated proteins. E, quantitative PhosphorImager analysis of the pulse-chase data in D and a second independent experiment with maximal levels for each band set to 100%. Error bars reflect the mean ± S.D. of these two independent experiments.
To assess whether sPmel17-myc was in fact secreted into the supernatant, we performed a pulse-chase experiment monitoring the accumulation of Pmel17-associated radioactivity in the culture medium (Fig. 3D, right panel). As expected, whereas cell-associated sPmel17-myc decayed over time during the course of the experiment (Fig. 3D, left panel), secreted protein accumulated in the culture supernatant (Fig. 3D, right panel). Interestingly, this secreted protein was already cleaved by pPCs, as evidenced by the presence of Mα and sMβ-myc. The latter appeared in the form of two fragments (termed sMβ-myca and sMβ-mycb), which probably differ only in their N terminus, because both forms can be immunoprecipitated with anti-myc antibodies (see Fig. 4D). We speculate that the smaller form is generated by N-terminal processing of the larger form subsequent to the action of pPCs. Strikingly, pPC-mediated cleavage of sPmel17-myc was almost quantitative, resulting in a 10-fold excess of pPC-generated cleavage fragments (Mα, sMβ-myca, and sMβ-mycb) over the uncleaved secreted precursor (sP2-myc) in the supernatant.
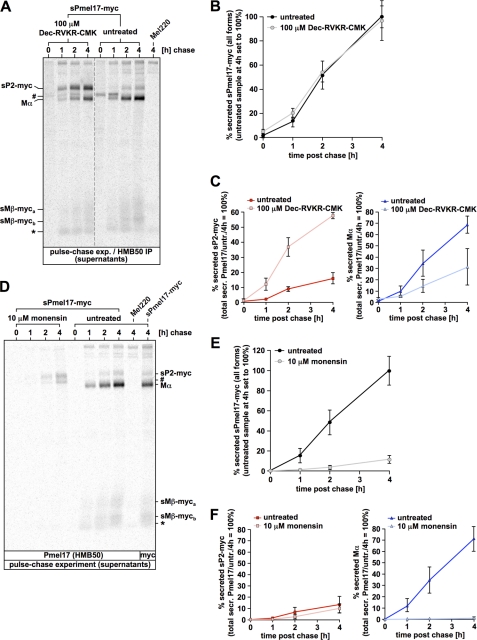
FIGURE 4.
Proprotein convertase-mediated cleavage of sPmel17-myc is inhibited by Dec-RVKR-CMK or monensin. A, cleavage of sPmel17-myc is sensitive to proprotein convertase inhibitor Dec-RVKR-CMK. Mel220 transfectants stably expressing sPmel17-myc were pulse-labeled for 30 min with 35S and subsequently chased for the indicated times. During both labeling and chase, 100 μm Dec-RVKR-CMK was included (four left lanes) or no inhibitor was included at all (fifth to eighth lanes). Culture supernatants were immunoprecipitated with Pmel17-specific antibody HMB50, eluted with 0.5% SDS by boiling for 5 min, and analyzed by autoradiography. The dashed line indicates a position where irrelevant lanes have been removed from the image. The pound symbol indicates background levels of the precipitated P1 form. The asterisk indicates a nonspecifically precipitated protein. B, total secretion is not affected by Dec-RVKR-CMK. Quantitative PhosphorImager analysis of the pulse-chase data in A is shown. The figure displays the total amount of all secreted sPmel17-myc forms (total released protein under untreated conditions at chase time point 4 h set to 100%). Error bars reflect the mean ± S.D. of two independent experiments. C, treatment with Dec-RVKR-CMK impairs proprotein convertase-mediated cleavage of the P2 form of sPmel17-myc. Quantitative PhosphorImager analysis of the pulse-chase data in A is shown. The figure displays the amount of secreted sP2-myc (left panel) or Mα (right panel) (total released protein under untreated conditions at chase time point 4 h set to 100%). Error bars reflect the mean ± S.D. of two independent experiments. D, cleavage of sPmel17-myc is sensitive to the secretion inhibitor monensin. Mel220 transfectants stably expressing sPmel17-myc were pulse labeled for 30 min with 35S and subsequently chased for the indicated times. During both labeling and chase, 10 μm monensin was included (four left lanes) or no inhibitor was included at all (five right lanes). Culture supernatants were immunoprecipitated with Pmel17-specific antibody HMB50 (nine left lanes) or myc-specific antibody ab9106 (last lane on the right), eluted with 0.5% SDS by boiling for 5 min and analyzed by autoradiography. The pound symbol indicates background levels of the precipitated P1 form. The asterisk indicates a nonspecifically precipitated protein. E, total secretion is almost completely suppressed by monensin. Quantitative PhosphorImager analysis of the pulse-chase data in D is shown. The figure displays the total amount of all secreted sPmel17-myc forms (total released protein under untreated conditions at chase time point 4 h set to 100%). Error bars reflect the mean ± S.D. of two independent experiments. F, treatment with monensin impairs proprotein convertase-mediated cleavage of the P2 form of sPmel17-myc. Quantitative PhosphorImager analysis of the pulse-chase data in D is shown. The figure displays the amount of secreted sP2-myc (left panel) or Mα (right panel) (total released protein under untreated conditions at chase time point 4 h set to 100%). Error bars reflect the mean ± S.D. of two independent experiments.
To confirm that cleavage of sPmel17-myc was indeed carried out by a pPC, we analyzed its processing in the presence of the specific pPC-inhibitor Dec-RVKR-CMK (32, 47), which has been shown by others to block Pmel17 cleavage (12, 21) (Fig. 4A). Indeed, the drug substantially impaired processing of secreted sPmel17-myc (Fig. 4A and supplemental Fig. S6, first and second panels), without affecting secretion as such (if the inhibitor was only included during the labeling and the chase) (Fig. 4B). In the presence of Dec-RVKR-CMK the uncleaved P2 form (sP2-myc) accumulated in the supernatant (Fig. 4C, left panel, and supplemental Fig. S6, second panel), whereas only low levels of Mα were generated (Fig. 4C, right panel, and supplemental Fig. S6, second panel).
In summary, these results show that a secreted Pmel17 mutant can be efficiently cleaved by pPCs. Thus, entry into the endocytic system is not a prerequisite for pPC processing of Pmel17. We note that this data clearly argues against the specificity scenario and is also very difficult to reconcile with the conformation scenario.
Pmel17 Cleavage in the Presence of Secretion Inhibitor Monensin Suggests It Is an Exceptionally Good Proprotein Convertase Substrate
Cleavage of most pPC substrates, e.g. pro-bone morphogenetic protein-1, β-site APP cleaving enzyme, transforming growth factor-β (TGFβ), metalloprotease disintegrin cysteine-rich 15 or procarboxypeptidase E, is blocked when cells are treated with the secretion inhibitor monensin (48–52), a drug that arrests intra-Golgi transport (33). This may be due in part by pH neutralization of cleavage compartments by monensin, which acts as an ionophore (33) and perturbs the pH that is necessary for efficient pPC activation (i.e. inhibitory prosegment removal) (53). Additionally, low pPC content within the monensin-arrested Golgi compartment (monensin prevents pPC substrates from access to the pPC-rich TGN) is likely to contribute to cleavage inefficiency, because concomitant furin overexpression can drive processing of substrates that would fail to be cleaved by endogenous pPCs in the presence of the drug (54). Nevertheless, despite these issues a small subset of substrates like a proliferation-inducing ligand (APRIL) persist to be cleaved even in the presence of monensin (55), suggesting that some substrates are particularly susceptible to pPC-mediated processing even under very unfavorable conditions.
To assess whether active secretion is a requirement for Pmel17 cleavage, we first determined the effect of the drug on sPmel17-myc processing (Fig. 4D). As expected, 10 μm monensin drastically impaired the release of this protein from the cell (Fig. 4E), whereas levels of initially labeled sP1-myc were not affected (supplemental Fig. S2). No pPC cleavage was observed in the presence of the drug (Fig. 4F, right panel, and supplemental Fig. S2). Rather the P2 form (sP2-myc) displayed a mild accumulation without any evidence for generation of Mα- or sMβ-myc fragments. However, we note that sPmel17-myc is likely to be a more disadvantaged substrate than wt-Pmel17, because it lacks a membrane anchor that restricts its movement within the plane of the membrane (i.e. within the Golgi apparatus the protein can move in three dimensions rather than two). Thus, association with membrane-bound pPCs, like e.g. furin, PC7, or PC5/6B, would be predictably less favored.
We therefore decided to investigate how the processing of full-length wt-Pmel17 or the IR mutant would be affected at a monensin concentration that is sufficient to fully block the cleavage of the otherwise cleavable sPmel17-myc (Fig. 4, D and F). Surprisingly, cleavage of wt-Pmel17 was remarkably efficient in the presence of the drug (Fig. 5, A and C, and supplemental Fig. S5), although it was reduced when compared with an untreated control (Fig. 5B). That pPC-mediated cleavage occurred in monensin-treated cells was shown by the vigorous appearance of Mα and Mβ fragments even at early time points after synthesis, like 1 h post chase (Fig. 5A, compare lanes 7 and 3 for wt-Pmel17 and lanes 15 and 11 for IR, and supplemental Fig. S5), whereas the P2 form decayed over time (Fig. 5C, right panels). The ratio of P2 versus Mα continuously dropped during the chase period (Fig. 5B) indicating that the former was converted into the latter. Similar observations were also made using monensin at a lower concentration of 2 μm, which the manufacturer suggests to block secretion (supplemental Figs. S3 and S4). Interestingly, however, whereas the monensin-arrested Golgi compartment obviously provided an environment that allowed pPC-mediated cleavage of Pmel17 to occur, the ER did not, as shown by the complete block of processing upon brefeldin A treatment (Fig. 5D). This is consistent with the pPC-processing profile of other pPC substrates that can be cleaved in the presence of monensin, but not in the presence of brefeldin A (54, 55).
FIGURE 5.
Brefeldin A, but not monensin treatment abrogates proprotein convertase-mediated processing of Pmel17. A, monensin treatment does not abrogate proprotein convertase-mediated processing of Pmel17. Mel220 transfectants stably expressing wt-Pmel17 or IR were pulse-labeled for 30 min with 35S and subsequently chased for the indicated times. During both labeling and chase, 10 μm monensin was included (sixth to ninth and 14th to 17th lanes 6–9) or no inhibitor was included at all (first to fifth lanes and 10th to 13th). 2% Triton X-100 lysates were immunoprecipitated with Pmel17-specific antibody HMB50, eluted with 0.5% SDS by boiling for 5 min, and analyzed by autoradiography. B, quantitative PhosphorImager analysis of the pulse-chase data in A and a second independent experiment (supplemental Fig. S5) is shown. The figure displays the ratio of the P2 form versus the Mα fragment at the indicated time points of chase. Error bars reflect the mean ± S.D. of the two independent experiments. C, quantitative PhosphorImager analysis of the pulse-chase data in A and a second independent experiment (supplemental Fig. S5) with maximal levels for each band set to 100% is shown. Error bars reflect the mean ± S.D. of two independent experiments. D, Mel220 transfectants stably expressing wt-Pmel17 were treated or not with 10 μg/ml of brefeldin A (BFA) overnight and subsequently analyzed by Western blot using the Pmel17-specific antibody Pep13h.
Altogether, these results demonstrate that at a concentration sufficient to completely block pPC-mediated processing of sPmel17-myc (Fig. 4, D and F) and to almost completely shut off secretion (Fig. 4E), monensin fails to efficiently suppress processing of wild-type Pmel17 (Fig. 5, A–C, and supplemental Fig. S5) and its effect on IR is only marginal (Fig. 5B). Consistent with our finding that entry into the endocytic system is not a prerequisite for Pmel17 cleavage (Figs. 3, D and E, and 4, A and C), this shows that the protein can be cleaved in the Golgi apparatus even prior to exposure to the pPC-rich TGN. Moreover, we note that our monensin experiments are inconsistent with the conformation scenario, which implies Pmel17 to exist in an uncleavable conformation during its passage through secretory compartments. This becomes even more obvious in light of mutant IR, which is even less affected by monensin (Fig. 5B), but given its full functionality (Fig. 2) and reactivity with conformation-sensitive antibodies (Figs. 1E and 2A) is very likely folded appropriately.
All Surface Pmel17 Is in an Already Proprotein Convertase-cleaved State
Our results in Figs. 3 and 5 strongly suggest that Pmel17 is cleaved during and not after secretion. If this were correct, all surface Pmel17 would be expected to exist in a state already pre-cleaved by pPCs. Indeed, that is what we find when we specifically immunoprecipitate the surface population of Pmel17 and analyze this fraction by Western blotting (Fig. 6A). In particular, we surface-labeled Mel220 transfectants on ice with antibody HMB50, extensively washed the cells before lysis, and added protein A-Sepharose to specifically recover surface Pmel17. Strikingly, the only Pmel17 form detectable by antibody Pep13h above background in this experiment was Mβ (Fig. 6A, lane 3) and thus “cleaved” protein, whereas the “uncleaved” P2 form was completely absent (Fig. 6A, lanes 3 and 7). We note that this experiment is internally controlled by the ER form P1, which is clearly present inside the cells (Fig. 6A, lane 1), but not to a significant extent at the cell surface (Fig. 6A, lane 3) at steady-state. Our results are consistent with the work of Valencia and colleagues (9) who showed that cleaved Pmel17 (Mβ), but not the uncleaved P2 form, can be detected at the cell surface. However, our results differ from this study with respect to recovery of immature Pmel17 from the plasma membrane, which we precipitate at most at marginal levels (Fig. 6A). We note that in our experience surface immunoprecipitations can sometimes pull down large contaminating amounts of the respective ER species of the same protein if the ER population is much more abundant than the surface population in the cell (56). Indeed, some melanoma cell lines appear to contain vastly more P1 than any other Pep13h-reactive Pmel17 form at steady-state under the conditions used in the respective publications (e.g. MNT-1 cells used in Ref. 9 and SK-MEL-28 cells used in Ref. 14). That P1 is present at the cell surface to a significant extent is also called into question by a recent study by Hoashi and co-workers (21), demonstrating that all surface-shed Pmel17 exclusively derives from the mature Mα-S-S-Mβ-dimer (itself derived from P2), but not from P1. Therefore, in line with our immunofluorescence results using either antibody Pmel-N or Pep13h (Figs. 1D and 3C, and Ref. 18), we believe that at least in Mel220 cells the P1 form is not expressed at the plasma membrane to a significant extent.
FIGURE 6.
Surface Pmel17 is already in a proprotein convertase-cleaved state. A, all Pmel17 at the cell surface is already in a proprotein convertase-cleaved state. Mel220 transfectants stably expressing wt-Pmel17 were incubated on ice with Pmel17-specific antibody HMB50, then extensively washed and lysed in 2% Triton X-100 before protein A-Sepharose beads were added to specifically immunoprecipitate the surface population of Pmel17. Immunoprecipitates (third, fourth, seventh, and eighth lanes) or corresponding total cell lysates (first, second, fifth, and sixth lanes) were analyzed by Western blot using antibody Pep13h. A shorter (first to fourth lanes) and a longer (fifth to eighth lanes) exposure of the same membrane is shown. The dashed line indicates a position where irrelevant lanes have been removed from the image. The asterisks indicate nonspecifically precipitated proteins. Note that the ER P1 form (present in total cell lysate, but not in the surface-IP sample) serves as an internal control in the experiment. B, newly synthesized Pmel17 at the cell surface is already in a proprotein convertase-cleaved state. Mel220 transfectants stably expressing wt-Pmel17 were pulse-labeled for 30 min with 35S and subsequently chased for 1 h. Antibody HMB50 was added to the intact cells on ice, before extensive washing, lysis in 2% Triton X-100, and addition of protein A-Sepharose beads to specifically immunoprecipitate the surface population of Pmel17 (second and third lanes). In parallel, a 2% Triton X-100 total cell lysate (lane 1) was immunoprecipitated with Pmel17-specific antibody HMB50 as described in the legend to Fig. 1C. Immunoprecipitates were subsequently analyzed by autoradiography (upper panel). For quantification, the ratio of surface-immunoprecipitated (lane 3) versus total cell-associated Pmel17 forms (P1, P2, Mα, and Mβ) (first lane) was determined. Note that this percentage is almost equally low for the P2 form and the ER-located P1 form, which serves as an indicator for background precipitation (gray shaded area). Only Mα and Mβ are precipitated above this background. Error bars reflect the mean ± S.D. of two independent experiments. The dashed line indicates a position where irrelevant lanes have been removed from the image. C, all Pmel17 at the cell surface is already in a proprotein convertase-cleaved state. Cells from A were surface-biotinylated at room temperature according to the protocol of the manufacturer (left panel) or at 4 °C to avoid any residual endocytosis during this step (right panel). Subsequently, biotinylated surface proteins were precipitated using avidin-agarose. Protein G-agarose was used as a specificity control. These samples (third and fourth lanes in left panel and third to fifth in right panel) or total cell lysates (first and second lanes in left and right panels) were analyzed by Western blot using Pmel17-specific antibodies Pep13h and Pmel-N. The horizontal dashed lines separate the regions of the membrane that were incubated with antibody Pmel-N (upper part) and antibody Pep13h (lower part), respectively. The vertical dotted lines separate longer (right part) or shorter (left part) exposures of the same membrane. Note that the ER P1 form (present in total cell lysate, but only marginally in the avidin-precipitation samples) serves as an internal control in the experiment.
A similar surface immunoprecipitation approach like the one shown in Fig. 6A, combined with a pulse-chase experiment, confirmed the exposure of newly synthesized pPC-processed, disulfide-linked Mα-S-S-Mβ dimer at the cell surface, whereas uncleaved newly synthesized P2 was not detected above background (again, some contamination from internal proteins is expected in these kinds of experiments and we consider the low percentage of P1 recovery as the respective background threshold) (Fig. 6B). To provide even more evidence that the surface population of Pmel17 is quantitatively cleaved, we employed another approach, where we specifically biotinylated surface proteins using a membrane-impermeable labeling reagent and subsequently pulled down these proteins using avidin-agarose (Fig. 6C). The surface biotinylation step was either carried out at room temperature as suggested by the manufacturer (Fig. 6C, left panel) or at 4 °C (Fig. 6C, right panel) to rule out any residual endocytosis during labeling. Consistent with our former results, surface proteins subsequently isolated with avidin-agarose contained both Mα and Mβ, and thus cleaved Pmel17, but not uncleaved P2 (Fig. 6C, fourth lane in left and right panels). Again, these results are internally controlled by ER form P1, which is present in a total cell lysate (Fig. 6C, second lane in the left and right panels), but is only marginally precipitated with avidin-agarose (Fig. 6C, fourth lane in the left and right panels). Moreover, we confirmed that precipitation of biotinylated surface Pmel17 was specific (Fig. 6C, fifth lane in the right panel). We note that these results are consistent with a recent report that demonstrated that the small fraction of Pmel17 that gets shed from the cell surface is already cleaved by pPCs (21).
To investigate a possible processing step of Pmel17 at the plasma membrane, we also tried to examine directly whether exogenously applied protein can be cleaved at the cell surface. To this end, we pretreated Mel220 cells expressing the soluble sPmel17-myc mutant with 100 μm Dec-RVKR-CMK overnight, before starving the cells and pulsing them with radioactively labeled 35S again in the presence of inhibitor, but finally chasing for 4 h without inhibitor for collection of supernatant thereafter. Although, removal of the inhibitor may lead to the appearance of newly synthesized “uninhibited” convertases during the chase, the underlying idea was that more radiolabeled uncleaved sP2-myc might be harvested than under a setting where inhibitor is omitted throughout. Indeed, this was the case. The majority of protein secreted within these 4 h of chase was the uncleaved 35S-labeled sP2-myc form (supplemental Fig. S6, third panel, time point 0 h), although some Mα was also present. We harvested this protein from the supernatant and directly applied it exogenously to non-labeled untransfected Mel220 cells, which do not produce any Pmel17 on their own. Interestingly, the labeled sP2-myc decayed only marginally during the experiment and the “P2 to Mα” ratio remained almost constant throughout the whole 4-h chase (supplemental Fig. S6, third panel), arguing that sP2-myc is not converted into Mα at any significant rate when exposed exogenously to potentially plasma membrane-located convertases. We also controlled that the failure to cleave was not due to residual Dec-RVKR-CMK contamination in the harvested supernatant, because when this same supernatant was added to freshly 35S-labeled Mel220(wt-Pmel17) cells, cleavage of full-length wild-type Pmel17 was completely unimpeded when compared with a control sample that had received regular (non-radioactive) chase medium instead of sPmel17-myc-containing supernatant (supplemental Fig. S6, fourth panel). Thus, cleavage of sPmel17-myc does not occur post secretion at any significant rate, but must be carried out while the protein is still cell-associated. Together with our monensin inhibition experiments showing that cleavage can occur in the Golgi (Fig. 5, A—C, and supplemental Figs. S3–5) and our quantitative immuno-EM results suggesting that large amounts of cleaved Pmel17 do reside in the Golgi at steady-state (see Fig. 7), this strongly suggests that, if at all, only a small minority of Pmel17 undergoes pPC-mediated processing at the cell surface (and if the latter happens at all, then probably immediately concomitant to arrival of the protein at the plasma membrane). Consequently, the practically complete cleavage status of plasma membrane-localized Pmel17 in melanoma cells (Fig. 6) can, at least for the bulk of Pmel17, be explained by earlier processing in the secretory pathway.
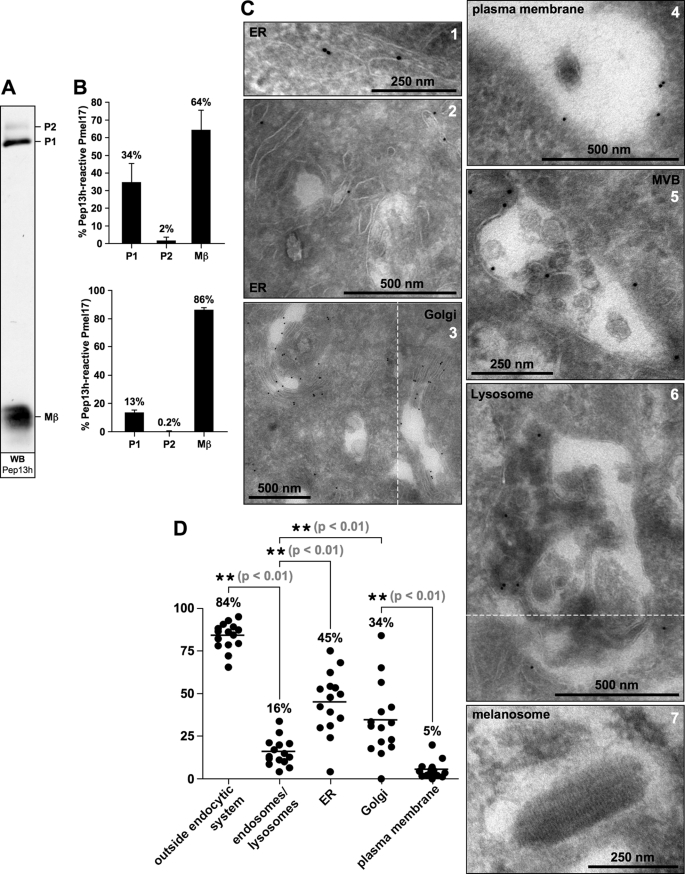
FIGURE 7.
Proprotein convertase-cleaved Pmel17 partially distributes along the secretory route. A, the peptide antibody Pep13h predominantly recognizes cleaved Pmel17 (Mβ) at steady-state. A total cell lysate of Mel220 cells stably expressing wt-Pmel17 was analyzed by Western blot using antibody Pep13h, which recognizes the extreme C terminus of Pmel17. One typical example of numerous experiments is shown. B, quantitative Western blotting was performed on Pep13h-stained membranes like the one shown in Fig. 7A. The intensity of Pep13h-stained, Pmel17-specific bands (P1 + P2 + Mβ = 100%) shows the strongest labeling of pPC-cleaved Pmel17 (Mβ) followed by lower levels of ER-associated Pmel17 (P1) and only negligible levels of P2. Two independent experiments are shown (top and bottom panels). C, Pep13h-specific labeling of wt-Pmel17 in Mel220 cells is predominantly found along the secretory route and only to a minor extent in the endocytic system. Mel220 cells expressing wt-Pmel17 were fixed and examined by cryo-immuno-EM using antibody Pep13h. Shown is Pep13h-specific labeling in the ER (panels 1 and 2), in the Golgi apparatus (panel 3), on the plasma membrane (panel 4), and in multivesicular bodies (MVB) (panel 5). Only occasionally is labeling observed in lysosomes (panel 6) and melanosomes are not stained at all (panel 7). D, the majority of Pep13h-reactive Pmel17 distributes along the secretory route. The Pep13h-specific immunolabeling outside (ER, Golgi apparatus, and plasma membrane) and inside the endocytic system (endosomal and lysosomal) was quantified in 15 entire cells and the percentage of labeling associated with a particular organelle is shown. Each dot corresponds to one cell.
Cleaved Pmel17 Is Distributed Along the Secretory Route in Melanoma Cells
Our results in Fig. 5, and supplemental Figs. S3–S5, suggest that pPC-mediated cleavage of wt-Pmel17 can occur as early as in the Golgi apparatus. This, and our results presented under supplemental Fig. S6, suggest that cleaved protein should partially distribute along the secretory route at steady-state instead of being restricted to the plasma membrane (Fig. 6) and the endocytic system. To examine whether this is the case, we first characterized by quantitative Western blotting the specificity of the Pmel17-specific antibody Pep13h, a reagent raised against a short peptide at the extreme C terminus of the protein (20). This antibody is known to recognize only newly synthesized Pmel17, but not mature fibrils (11, 18, 20, 35). We found that Pep13h predominantly detects cleaved Pmel17 (Mβ) in Mel220 transfectants (Figs. 1B, 5D, 6A, and 7A, and Ref. 18) and we confirmed this finding by quantitative Western blotting (Fig. 7B). In fact, ∼64–86% of the total Pep13h-recognized protein is already cleaved (Mβ) at steady-state (Fig. 7B, right bars), whereas the ER-associated P1 form contributes much less to the overall labeling (13–34%). The levels of the P2 form, the immediate precursor for pPC-mediated cleavage, are practically negligible (0.2–2%).
We then examined where in the cell the Pep13h-reactive protein resides using quantitative immuno-EM, a method allowing very high organellar resolution. Consistent with our results shown in Fig. 1D most of the Pmel17 protein was found in the ER (Fig. 7C, panels 1 and 2) and in the Golgi apparatus (Fig. 7C, panel 3). Together these two compartments alone contained at steady-state almost 80% of the total Pep13h-specific labeling in the cell (Fig. 7D), whereas only a minority of the labeling was associated with the endocytic system (Fig. 7C, panels 5–7) (16%) or the plasma membrane (Fig. 7C, panel 4) (5%). However, since the majority of the Pep13h-reactive protein is already pPC-cleaved at steady-state (Figs. 1B, 5D, 6A, and 7, A and B), we conclude that cleaved protein must at least partially distribute along the secretory route prior to the plasma membrane. Because we know that cleavage of wt-Pmel17 (in contrast to mutant IR) does not occur inside the ER (Figs. 1C and 5D) where the P1 form is abundant, it is thus highly likely that the Golgi apparatus contains very significant amounts of pPC-cleaved protein at steady-state. This compartment alone comprises 34% of the Pep13h-specific labeling in the cell (Fig. 7D) and in fact, this number suggests that almost all Golgi-associated protein is already cleaved. This is also consistent with the fact that despite the dense Pep13h-specific Golgi staining (Fig. 7, C, panel 3, and D), only minute levels of the Pmel17 P2 form, a form that arises in the Golgi apparatus through modification of oligosaccharides can be found at steady-state (Figs. 1B, 5D, 6A, and 7, A and B). Furthermore, results obtained using Pmel17-specific peptide antibody Pmel-N point to the same conclusion. Also Pmel-N hardly detects any P2 form, but lots of pPC-cleaved protein (Mα) at steady-state in Mel220 cells (Fig. 1B, middle panel, and Ref. 18), whereas immunofluorescence analysis shows most Pmel-N-specific labeling was concentrated in the ER and Golgi apparatus (18). Thus, altogether our results strongly suggest that pPC-cleaved protein can be found in the Golgi at steady-state and hence, consistent with our monensin-inhibition experiments (Fig. 5, A—C, and supplemental Figs. S3–5), we conclude that proprotein convertases largely process Pmel17 along the secretory route and most of it probably prior to arrival of the protein at the plasma membrane (Fig. 8B).
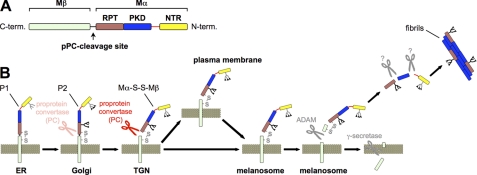
FIGURE 8.
Model of Pmel17 maturation. A, schematic domain structure of Pmel17. The pPC cleavage site is located between the lumenal RPT domain and the transmembrane domain. B, model of Pmel17 maturation based on our results. Pmel17 is synthesized as a ∼100 kDa precursor in the ER (so-called P1 form) that quickly migrates to the Golgi where its oligosaccharides get modified, which leads to a large shift in the apparent molecular weight (so-called P2 form). Proprotein convertase cleavage might occur as early as in this compartment. However, the major fraction of the protein may be cleaved only in the pPC-rich trans-Golgi network, thereby generating an N-terminal Mα- and a membrane-tethered C-terminal Mβ-fragment. The two fragments remain linked to each other via a disulfide bridge (Mα-S-S-Mβ). In this form Pmel17 gets translocated to early stage melanosomes. Some protein may route there directly from the TGN, but a large fraction accesses melanosomes only via the plasma membrane. Part of the protein gets shed there from the cell surface. In melanosomes, an unknown trigger induces cleavage of Pmel17 by a metalloprotease of the ADAM family, which releases a soluble Mα derivative, which gets further cleaved into at least two (probably non-overlapping) sets of fragments: those containing the PKD domain (reactive with antibody I51) and those containing the RPT domain (reactive with antibody HMB45). These fragments eventually assemble into mature fibrils.
DISCUSSION
The main function of Pmel17 is to form a network of fibrillar sheets in melanosomes that serves for deposition of the pigment melanin as well as to concentrate reaction intermediates of the melanin synthesis pathway and keep these potentially toxic compounds from diffusing into the cell (1–3, 57). Because Pmel17 fibrils represent an amyloid structure (2, 16, 17) and the uncontrolled formation of amyloid aggregates may be harmful to cells or tissue it is of interest to comprehend how the assembly of this fibrous matrix in melanosomes is carefully regulated and orchestrated.
Based on the mistargeting of two N-terminal Pmel17 deletion mutants (termed ΔNTR and ΔPKD in Ref. 19) that also proved to be refractory to pPC-mediated cleavage, it had been speculated that correct delivery of the protein to melanosomes is a prerequisite for pPC cleavage and that this processing step takes place in melanosomes (19). However, Theos and co-workers (18) found that at least their cleavage-resistant NTR-deletion mutant also lost to a significant extent reactivity with two conformation-sensitive antibodies, HMB50 and NKI-beteb (11, 19) (in their second mutant, ΔPKD, the domain that these two antibodies recognize is deleted and therefore correct folding of this construct cannot be probed this way). Therefore, it is possible that misfolding rather than mistargeting of the construct is actually causing the processing defect. In line with this, we (18) and others (14) recently reported very similar deletion mutants that, despite sharing the massive mislocalization phenotype, were nevertheless, efficiently cleaved by pPCs. In fact, if anything, cleavage was even more efficient for the trafficking mutant Δ28–208 (named ΔNTR in Ref. 18) than wild-type Pmel17 (18). Additionally, we recently generated a third N-terminal deletion mutant (Δ28–189), which shows the same subcellular misdistribution as Δ28–208 (ΔNTR), and this construct is also cleaved by pPCs (data not shown). Furthermore, Pmel17 surface internalization mutants, which are retained at the plasma membrane (and thus in a preendosomal compartment) do not show a significantly altered rate of Mα and Mβ formation (in fact, both fragments accumulate to high levels in the respective pulse-chase experiments) (58). This is interesting, because the amount of Pmel17 that migrates to melanosomes via the plasma membrane (in contrast to direct trafficking from the TGN) is probably not negligible. Our own results shown in Fig. 6B suggest that 1 h after labeling, i.e. the time when Mα and Mβ levels peak (Figs. 1C and 5, A and C, and supplemental Figs. S3—5), about 25% of the total cellular Mα-S-S-Mβ dimer is detectable at the cell surface. Because it is likely that at this time point some fraction of the cellular pPC-cleaved protein is already in a “post-internalization” state, whereas some protein may still be on its way to the cell surface, the actual percentage of Pmel17 trafficking via the plasma membrane is probably much greater than 25%. Altogether, this shows that much of the published data is at odds with the current model of pPC-mediated Pmel17 processing in melanosomes. In fact, this data points toward cleavage taking place outside these organelles.
These results are also consistent with a recent report that investigated the small fraction of Pmel17 that is shed from the cell surface in melanoma cells (21). In particular, these authors found that shed Pmel17 is already in a post-pPC-processed state under normal conditions. Here we extend these findings and demonstrate that all surface Pmel17 (Fig. 6, A and C), including the newly synthesized population (Fig. 6B) has already been quantitatively pPC-cleaved, whereas no uncleaved P2 form can be detected at the plasma membrane. This strongly suggests that the surface-shed Pmel17 population described by Hoashi et al. (21) is not a “side population” undergoing some special pPC-mediated processing, but follows in this respect the bulk of the protein.
One major problem of those studies that have tried to promote the view that pPCs cleave Pmel17 only in melanosomes is that none so far has come up with a convincing, clear-cut demonstration of how Pmel17 processing during secretion can be so efficiently avoided. This is a serious problem, because according to the current model (8) newly synthesized Pmel17 migrates through several compartments that are known to be very rich in pPCs, particularly the TGN (that may contain furin, PC7, PC5/6B, and PC1) (59–62), post-TGN vesicles (that may contain PC7) (63, 64), and also the plasma membrane (that may contain PC7, PC5/6A, and PACE4) (65, 66). In fact, our own studies have revealed expression of at least furin, PACE4, PC7, and PC1 on the RNA level in human melanoma cells and one melanoma cell line additionally expressed PC5/6 (data not shown). Therefore, if pPC-mediated processing of Pmel17 were restricted only to melanosomes (19), the protein would obviously have to be protected from cleavage in these secretory compartments in some way. This is all the more important, because furin, a convertase predominantly localized to the TGN, is able to cleave Pmel17 in cells (7, 19, 67). Principally, there are two scenarios of how such a protection might be achieved mechanistically. Either the cleavage motif of Pmel17 would be generally refractory to cleavage by all pPCs that the protein encounters during its passage through the secretory route. In that case the existence of a distinct, specific melanosome-located enzyme would have to be predicted that can eventually cleave the site (specificity scenario). Alternatively, Pmel17 might in principle be cleavable by all cellular pPCs, but may fold into a conformation that specifically restricts access of its processing motif during secretion. This conformational protection would then be relaxed only once the protein arrives in melanosomes allowing cleavage to occur (conformation scenario). To examine whether either of these scenarios can explain the Pmel17 processing pattern, we analyzed the cleavage of a soluble, secreted Pmel17 mutant in melanoma cells. This mutant contains all lumenal domains, but lacks the transmembrane region and the cytosolic tail. Unlike wt-Pmel17, this construct does not traffic to melanosomes or access early endosomes (Fig. 3C). However, although the secreted Pmel17 mutant never enters the endocytic system, it is nevertheless, almost quantitatively cleaved by pPCs (Figs. 3, D, right panel, and 4), resulting in a 10-fold excess of pPC-generated cleavage fragments (Mα, sMβ-myca, and sMβ-mycb) over the uncleaved secreted precursor (sP2-myc) in the supernatant. This clearly argues against both the specificity scenario and the conformation scenario, whereas strongly promoting our own new model featuring secretory cleavage (Fig. 8B). This model is also further corroborated by our quantitative immuno-EM analysis (Fig. 7), which indicates that the Golgi apparatus already contains significant amounts of cleaved wt-Pmel17 at steady-state. Moreover, this model is also supported by the behavior of the Pmel17 mutant IR, in that the replacability of the pPC-cleavage motif argues against the specificity scenario whereas, the conformation scenario is inconsistent with the partial cleavage of IR in the ER as well as the only marginal sensitivity of IR cleavage to monensin. Proper folding of this mutant is very likely given because IR appears to be fully functional (Fig. 2) and reacts well with conformation-sensitive antibodies like NKI-beteb (Fig. 1E) and HMB50 (e.g. Fig. 1C).
Only low, transient intracellular levels of sP2-myc are observed in cells expressing the secreted Pmel17 mutant (Figs. 3, D and E, and 4), suggesting that the protein probably traverses the secretory route very rapidly once released from the ER. This, however, implies only very transient exposure of sP2-myc to TGN enzymes and hence, this suggests that Pmel17 is a quite decent pPC substrate. This is also consistent with the ongoing pPC-mediated processing of wt-Pmel17 even in the presence of monensin (Fig. 5 and supplemental Figs. S3–S5), which indicates that Pmel17 may even be an exceptionally good substrate. In this context we emphasize that the monensin-arrested compartment probably provides a much less favorable environment for pPC activity than the cis or medial Golgi would do under normal circumstances, due to the pH-neutralizing properties of monensin as an ionophore (33) (low pH supports displacement of the inhibitory prosegment from pPCs (53)). We also note that in case of wild-type Pmel17 (or mutants IR, ΔNTR (Δ28–208) and Δ28–189) a substantial fraction of the newly synthesized protein localizes to a Golgi compartment at steady-state (Fig. 1D, right panel, and Ref. 18) (data not shown). This high cleavage efficiency comes as a surprise given the atypical deviation of the Pmel17 cleavage motif (LVKR↓) from the consensus cleavage site (RX(R/K)R↓) (32, 36, 37). Many pPC substrates in fact essentially require an arginine in position P4 for efficient cleavage (37–42). However, substrates lacking a basic residue in this position have been described, although unconventional cleavage motifs may affect the processing capacity of the respective target proteins. For instance, wild-type von Willebrand factor has been shown to be efficiently cleaved by furin, PACE4, and PC5/6A at the processing site (RSKR↓), whereas a mutated processing site lacking arginine in position P4 (ASKR↓) permitted cleavage only by furin (68). That the motif in Pmel17 can be easily replaced by a most optimal processing site (PSRKRR↓S) (36) seemingly with no effect on fibril formation whatsoever (Fig. 1B, right panel, and Fig. 2, B–D, panels 6–8) therefore suggests that the original motif does permit efficient cleavage. This is also supported by the rapid cleavage kinetics of the protein in melanoma cells (Fig. 1C and see Refs. 13 and 20).
Finally, our results suggest yet another so far unanticipated similarity between the protease cascade that initiates the activation of fibril formation by Pmel17 (12) and the one that drives activation of a group of cellular receptors like Notch (69) and receptor protein-tyrosine phosphatase κ (RPTPκ) (70). In all these cases, first a pPC-mediated cleavage occurs (S1) (7, 69, 70), followed by the action of a metalloprotease of the ADAM family (S2) (12, 69, 70), and finally intramembrane cleavage by γ-secretase (S3) (12, 69, 70). Interestingly, both Notch and RPTPκ are cleaved by furin early during secretion in the TGN and prior to arrival at the plasma membrane, whereas receptor activation via S2 cleavage follows at the plasma membrane only once the receptor meets its ligand (69, 70). Thus, pPC-mediated priming and S2 cleavage-mediated receptor activation are both temporarily and spatially separated. pPC-mediated processing of these receptors does not trigger immediate activation.
Our results suggest a very similar sequence of events also for Pmel17. In analogy to Notch and RPTPκ, pPC-mediated cleavage appears to be carried out while the protein migrates along the secretory route, before and not after its delivery into the endocytic system (Fig. 8B). Fibril formation, however, occurs in melanosomes and thus S2 cleavage is likely to be restricted to this organelle. Hence, our results suggest that for Pmel17, S1 (pPC-mediated) and S2 cleavage are both spatially and temporarily separated, even though only on a small scale (i.e. S2-cleavage may follow “rapidly” after, but not “immediately” after S1 cleavage). Thus, pPC-mediated cleavage of Pmel17 cannot be the immediate trigger of fibril formation, because otherwise aggregation of the protein into fibrous sheets would have to be observed in secretory compartments as is the case for other multimerizing proteins such as the von Willebrand factor expressed in endothelial cells (71). But if pPC-mediated processing is not the immediate trigger for fibril formation this certainly adds one more point to the already surprisingly long list of events that this processing step has already been shown not to be relevant for, like surface shedding (21), subcellular trafficking (7), S2 cleavage by ADAM10/ADAM17 metalloproteases (12), or generation of the HMB45-reactive fibrillogenic fragments (11, 12, 14). Nevertheless, pPC-mediated processing of Pmel17 is absolutely required for fibril formation (7). Hence, it remains an important goal for the future to determine what this maturation step actually serves for on a molecular level and how fibril formation is affected by it.
Altogether, our results alter the current picture of Pmel17 maturation and demonstrate that pPC-mediated processing of Pmel17 occurs during secretion, but not after transfer of the protein into the endocytic system. Hence, our data add to the understanding of the process of fibril formation by this complex protein and its regulation and thus provide deeper insights into these key events that are so crucial to the biology of melanocytes.
Supplementary Material
Acknowledgments
We are indebted to Drs. M. Marks, A. Gown, and M. Skelly for the kind donation of Pmel17-specific antibodies Pep13h, Pmel-N, and HMB50. We further thank Dr. M. Marks for very helpful discussions and communication of unpublished results. We are grateful to Dr. M. Graham for help with the electron microscopic analysis. Dr. J. Grotzke is acknowledged for critically reading the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R37-AI23081, the Howard Hughes Medical Institute, and Yale SPORE in Skin Cancer Grant 5P50 CA121974 (to P. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- ER
- endoplasmic reticulum
- NTR
- N-terminal region
- PKD
- polycystic kidney disease-like domain
- pPC
- proprotein convertase
- RPT
- repeat domain
- TGN
- trans-Golgi network
- ADAM
- a disintegrin and metalloprotease.
REFERENCES
- 1. Theos A. C., Truschel S. T., Raposo G., Marks M. S. (2005) Pigment Cell Res. 18, 322–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fowler D. M., Koulov A. V., Alory-Jost C., Marks M. S., Balch W. E., Kelly J. W. (2006) PLoS Biol. 4, e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lee Z. H., Hou L., Moellmann G., Kuklinska E., Antol K., Fraser M., Halaban R., Kwon B. S. (1996) J. Invest. Dermatol. 106, 605–610 [DOI] [PubMed] [Google Scholar]
- 4. Quevedo W. C., Fleischmann R. D., Dyckman J. (1981) in Phenotypic Expression in Pigment Cells (Seiji M. ed) pp. 177–184, Tokyo University Press, Tokyo [Google Scholar]
- 5. Yamaguchi Y., Hearing V. J. (2009) BioFactors 35, 193–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Plonka P. M., Passeron T., Brenner M., Tobin D. J., Shibahara S., Thomas A., Slominski A., Kadekaro A. L., Hershkovitz D., Peters E., Nordlund J. J., Abdel-Malek Z., Takeda K., Paus R., Ortonne J. P., Hearing V. J., Schallreuter K. U. (2009) Exp. Dermatol. 18, 799–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Berson J. F., Theos A. C., Harper D. C., Tenza D., Raposo G., Marks M. S. (2003) J. Cell Biol. 161, 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Raposo G., Marks M. S. (2007) Nat. Rev. Mol. Cell Biol. 8, 786–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Valencia J. C., Watabe H., Chi A., Rouzaud F., Chen K. G., Vieira W. D., Takahashi K., Yamaguchi Y., Berens W., Nagashima K., Shabanowitz J., Hunt D. F., Appella E., Hearing V. J. (2006) J. Cell Sci. 119, 1080–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Valencia J. C., Rouzaud F., Julien S., Chen K. G., Passeron T., Yamaguchi Y., Abu-Asab M., Tsokos M., Costin G. E., Yamaguchi H., Jenkins L. M., Nagashima K., Appella E., Hearing V. J. (2007) J. Biol. Chem. 282, 11266–11280 [DOI] [PubMed] [Google Scholar]
- 11. Harper D. C., Theos A. C., Herman K. E., Tenza D., Raposo G., Marks M. S. (2008) J. Biol. Chem. 283, 2307–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kummer M. P., Maruyama H., Huelsmann C., Baches S., Weggen S., Koo E. H. (2009) J. Biol. Chem. 284, 2296–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yasumoto K., Watabe H., Valencia J. C., Kushimoto T., Kobayashi T., Appella E., Hearing V. J. (2004) J. Biol. Chem. 279, 28330–28338 [DOI] [PubMed] [Google Scholar]
- 14. Hoashi T., Muller J., Vieira W. D., Rouzaud F., Kikuchi K., Tamaki K., Hearing V. J. (2006) J. Biol. Chem. 281, 21198–21208 [DOI] [PubMed] [Google Scholar]
- 15. Chiamenti A. M., Vella F., Bonetti F., Pea M., Ferrari S., Martignoni G., Benedetti A., Suzuki H. (1996) Melanoma Res. 6, 291–298 [DOI] [PubMed] [Google Scholar]
- 16. McGlinchey R. P., Shewmaker F., McPhie P., Monterroso B., Thurber K., Wickner R. B. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 13731–13736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Watt B., van Niel G., Fowler D. M., Hurbain I., Luk K. C., Stayrook S. E., Lemmon M. A., Raposo G., Shorter J., Kelly J. W., Marks M. S. (2009) J. Biol. Chem. 284, 35543–35555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leonhardt R. M., Vigneron N., Rahner C., Van den Eynde B. J., Cresswell P. (2010) J. Biol. Chem. 285, 16166–16183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Theos A. C., Truschel S. T., Tenza D., Hurbain I., Harper D. C., Berson J. F., Thomas P. C., Raposo G., Marks M. S. (2006) Dev. Cell 10, 343–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Berson J. F., Harper D. C., Tenza D., Raposo G., Marks M. S. (2001) Mol. Biol. Cell 12, 3451–3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hoashi T., Tamaki K., Hearing V. J. (2010) FASEB J. 24, 916–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kopan R., Ilagan M. X. (2009) Cell 137, 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vigneron N., Ooms A., Morel S., Ma W., Degiovanni G., Van den Eynde B. J. (2005) Tissue Antigens 65, 156–162 [DOI] [PubMed] [Google Scholar]
- 24. Adema G. J., de Boer A. J., Vogel A. M., Loenen W. A., Figdor C. G. (1994) J. Biol. Chem. 269, 20126–20133 [PubMed] [Google Scholar]
- 25. Nichols S. E., Harper D. C., Berson J. F., Marks M. S. (2003) J. Invest. Dermatol. 121, 821–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Esclamado R. M., Gown A. M., Vogel A. M. (1986) Am. J. Surg. 152, 376–385 [DOI] [PubMed] [Google Scholar]
- 27. Meyer T. H., van Endert P. M., Uebel S., Ehring B., Tampé R. (1994) FEBS Lett. 351, 443–447 [DOI] [PubMed] [Google Scholar]
- 28. Sadasivan B., Lehner P. J., Ortmann B., Spies T., Cresswell P. (1996) Immunity 5, 103–114 [DOI] [PubMed] [Google Scholar]
- 29. Barton G. M., Medzhitov R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14943–14945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Carrithers M. D., Chatterjee G., Carrithers L. M., Offoha R., Iheagwara U., Rahner C., Graham M., Waxman S. G. (2009) J. Biol. Chem. 284, 8114–8126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Leonhardt R. M., Keusekotten K., Bekpen C., Knittler M. R. (2005) J. Immunol. 175, 5104–5114 [DOI] [PubMed] [Google Scholar]
- 32. Garten W., Hallenberger S., Ortmann D., Schäfer W., Vey M., Angliker H., Shaw E., Klenk H. D. (1994) Biochimie 76, 217–225 [DOI] [PubMed] [Google Scholar]
- 33. Mollenhauer H. H., Morré D. J., Rowe L. D. (1990) Biochim. Biophys. Acta 1031, 225–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Klausner R. D., Donaldson J. G., Lippincott-Schwartz J. (1992) J. Cell Biol. 116, 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Raposo G., Tenza D., Murphy D. M., Berson J. F., Marks M. S. (2001) J. Cell Biol. 152, 809–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Remacle A. G., Shiryaev S. A., Oh E. S., Cieplak P., Srinivasan A., Wei G., Liddington R. C., Ratnikov B. I., Parent A., Desjardins R., Day R., Smith J. W., Lebl M., Strongin A. Y. (2008) J. Biol. Chem. 283, 20897–20906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vey M., Orlich M., Adler S., Klenk H. D., Rott R., Garten W. (1992) Virology 188, 408–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Watanabe T., Murakami K., Nakayama K. (1993) FEBS Lett. 320, 215–218 [DOI] [PubMed] [Google Scholar]
- 39. Rehemtulla A., Kaufman R. J. (1992) Blood 79, 2349–2355 [PubMed] [Google Scholar]
- 40. Oda K., Ikeda M., Tsuji E., Sohda M., Takami N., Misumi Y., Ikehara Y. (1991) Biochem. Biophys. Res. Commun. 179, 1181–1186 [DOI] [PubMed] [Google Scholar]
- 41. Molloy S. S., Bresnahan P. A., Leppla S. H., Klimpel K. R., Thomas G. (1992) J. Biol. Chem. 267, 16396–16402 [PubMed] [Google Scholar]
- 42. Mark M. R., Lokker N. A., Zioncheck T. F., Luis E. A., Godowski P. J. (1992) J. Biol. Chem. 267, 26166–26171 [PubMed] [Google Scholar]
- 43. Goldfine I. D. (1987) Endocr. Rev. 8, 235–255 [DOI] [PubMed] [Google Scholar]
- 44. de Cicco R. L., Bassi D. E., Benavides F., Conti C. J., Klein-Szanto A. J. (2007) Mol. Carcinog. 46, 654–659 [DOI] [PubMed] [Google Scholar]
- 45. Salvas A., Benjannet S., Reudelhuber T. L., Chrétien M., Seidah N. G. (2005) FEBS Lett. 579, 5621–5625 [DOI] [PubMed] [Google Scholar]
- 46. Bass J., Turck C., Rouard M., Steiner D. F. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 11905–11909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jean F., Stella K., Thomas L., Liu G., Xiang Y., Reason A. J., Thomas G. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 7293–7298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Leighton M., Kadler K. E. (2003) J. Biol. Chem. 278, 18478–18484 [DOI] [PubMed] [Google Scholar]
- 49. Bennett B. D., Denis P., Haniu M., Teplow D. B., Kahn S., Louis J. C., Citron M., Vassar R. (2000) J. Biol. Chem. 275, 37712–37717 [DOI] [PubMed] [Google Scholar]
- 50. Basque J., Martel M., Leduc R., Cantin A. M. (2008) Can. J. Physiol. Pharmacol. 86, 606–612 [DOI] [PubMed] [Google Scholar]
- 51. Lum L., Reid M. S., Blobel C. P. (1998) J. Biol. Chem. 273, 26236–26247 [DOI] [PubMed] [Google Scholar]
- 52. Song L., Fricker L. (1995) J. Neurochem. 65, 444–453 [DOI] [PubMed] [Google Scholar]
- 53. Anderson E. D., VanSlyke J. K., Thulin C. D., Jean F., Thomas G. (1997) EMBO J. 16, 1508–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vey M., Schäfer W., Berghöfer S., Klenk H. D., Garten W. (1994) J. Cell Biol. 127, 1829–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. López-Fraga M., Fernández R., Albar J. P., Hahne M. (2001) EMBO Rep. 2, 945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhu Y., Zhang W., Veerapen N., Besra G., Cresswell P. (2010) J. Biol. Chem. 285, 38283–38292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hurbain I., Geerts W. J., Boudier T., Marco S., Verkleij A. J., Marks M. S., Raposo G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 19726–19731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Theos A. C., Berson J. F., Theos S. C., Herman K. E., Harper D. C., Tenza D., Sviderskaya E. V., Lamoreux M. L., Bennett D. C., Raposo G., Marks M. S. (2006) Mol. Biol. Cell 17, 3598–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Seidah N. G., Mayer G., Zaid A., Rousselet E., Nassoury N., Poirier S., Essalmani R., Prat A. (2008) Int. J. Biochem. Cell Biol. 40, 1111–1125 [DOI] [PubMed] [Google Scholar]
- 60. Molloy S. S., Thomas L., VanSlyke J. K., Stenberg P. E., Thomas G. (1994) EMBO J. 13, 18–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. De Bie I., Marcinkiewicz M., Malide D., Lazure C., Nakayama K., Bendayan M., Seidah N. G. (1996) J. Cell Biol. 135, 1261–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Marcinkiewicz M., Seidah N. G., Chretien M. (1996) Acta Neurobiol. Exp. 56, 287–298 [DOI] [PubMed] [Google Scholar]
- 63. Wouters S., Leruth M., Decroly E., Vandenbranden M., Creemers J. W., van de Loo J. W., Ruysschaert J. M., Courtoy P. J. (1998) Biochem. J. 336, 311–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Leonhardt R. M., Fiegl D., Rufer E., Karger A., Bettin B., Knittler M. R. (2010) J. Immunol. 184, 2985–2998 [DOI] [PubMed] [Google Scholar]
- 65. Mayer G., Hamelin J., Asselin M. C., Pasquato A., Marcinkiewicz E., Tang M., Tabibzadeh S., Seidah N. G. (2008) J. Biol. Chem. 283, 2373–2384 [DOI] [PubMed] [Google Scholar]
- 66. Rousselet E., Benjannet S., Hamelin J., Canuel M., Seidah N. G. (2011) J. Biol. Chem. 286, 2728–2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Raposo G., Marks M. S. (2002) Traffic 3, 237–248 [DOI] [PubMed] [Google Scholar]
- 68. Creemers J. W., Groot Kormelink P. J., Roebroek A. J., Nakayama K., Van de Ven W. J. (1993) FEBS Lett. 336, 65–69 [DOI] [PubMed] [Google Scholar]
- 69. Fortini M. E. (2009) Dev. Cell 16, 633–647 [DOI] [PubMed] [Google Scholar]
- 70. Anders L., Mertins P., Lammich S., Murgia M., Hartmann D., Saftig P., Haass C., Ullrich A. (2006) Mol. Cell. Biol. 26, 3917–3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sadler J. E. (2009) J. Thromb. Haemost. 7, Suppl. 1, 24–27 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.