Abstract
The recognition and signaling of DNA double strand breaks involves the participation of multiple proteins, including the protein kinase ATM (mutated in ataxia-telangiectasia). ATM kinase is activated in the vicinity of the break and is recruited to the break site by the Mre11-Rad50-Nbs1 complex, where it is fully activated. In human cells, the activation process involves autophosphorylation on three sites (Ser367, Ser1893, and Ser1981) and acetylation on Lys3016. We now describe the identification of a new ATM phosphorylation site, Thr(P)1885 and an additional autophosphorylation site, Ser(P)2996, that is highly DNA damage-inducible. We also confirm that human and murine ATM share five identical phosphorylation sites. We targeted the ATM phosphorylation sites, Ser367 and Ser2996, for further study by generating phosphospecific antibodies against these sites and demonstrated that phosphorylation of both was rapidly induced by radiation. These phosphorylations were abolished by a specific inhibitor of ATM and were dependent on ATM and the Mre11-Rad50-Nbs1 complex. As found for Ser(P)1981, ATM phosphorylated at Ser367 and Ser2996 localized to sites of DNA damage induced by radiation, but ATM recruitment was not dependent on phosphorylation at these sites. Phosphorylation at Ser367 and Ser2996 was functionally important because mutant forms of ATM were defective in correcting the S phase checkpoint defect and restoring radioresistance in ataxia-telangiectasia cells. These data provide further support for the importance of autophosphorylation in the activation and function of ATM in vivo.
Keywords: DNA Damage, Mass Spectrometry (MS), Post-translational Modification, Protein Phosphorylation, Serine Threonine Protein Kinase, Signal Transduction, ATM Kinase, Ataxia-telangiectasia
Introduction
DNA double strand breaks (DSB)4 arise during normal physiological cell processes, such as immunoglobulin and T cell receptor gene rearrangements (1), but also arise in response to exposure to a variety of DNA-damaging agents that, if left unrepaired, lead to cell death or genomic instability, cancer, and other pathologies (2). DNA DSB are repaired in mammalian cells by non-homologous end joining and homologous recombination that predominate at different stages of the cell cycle (3). The Mre11-Rad50-Nbs1 (MRN) complex is a primary sensor of DNA DSB, and there is evidence that it participates in both forms of DSB repair (4–7). Hypomorphic mutations in members of this complex give rise to disorders characterized by sensitivity to agents that generate DNA DSB, cell cycle checkpoint defects, genome instability, and neurological abnormalities (8–10). These syndromes overlap in their clinical and cellular phenotypes with ataxia-telangiectasia (A-T) defective in ATM kinase (11).
ATM is a member of the phosphoinositide 3-kinase-like kinase (PIKK) family of proteins and is primarily activated by DNA DSB to signal to both the DNA repair machinery and the cell cycle checkpoints (12, 13). Considerable progress has been made in understanding the mechanism of ATM activation (14–17). Although the nature of the initial stimulus for activation remains undefined, it seems likely that relaxation of chromatin structure is sufficient to initiate activation (14). ATM is constitutively present as an inactive dimer that monomerizes as part of the activation process, facilitated by both autophosphorylation and acetylation (14, 18). More recently, Sun et al. (19) have shown that after DNA damage, casein kinase 2 phosphorylates and releases HP1β from chromatin, which enables the recruitment of the ATM-Tip60 complex to MRN at the break site. This facilitates interaction between the chromodomain of Tip60 and the unbound histone H3 K9me3, leading to acetylation and activation of the kinase activity of ATM. The initial activation of ATM appears to be incomplete because it relies on recruitment to the site of the break by the MRN complex for full activation (15). This is supported by a variety of in vitro and in vivo studies that demonstrate MRN dependence on the activation of ATM (20–23). In addition, although ATM can be activated in cells deficient in Nbs1, it fails to localize to nuclear foci at sites of DNA damage in these cells (24). Once activated, ATM can then phosphorylate a multitude of substrates that participate in different cellular processes, including DNA repair and cell cycle control (25). A cascade of reactions is initiated, starting with the phosphorylation of H2AX (γH2AX), leading to chromatin modification that assists in signaling reactions and DNA repair (26). Recent data suggest that dephosphorylation at Tyr142 is a prerequisite for H2AX phosphorylation and that this alteration plays a central role in the assembly of the DNA damage response complex (27). Assembly of DNA damage response proteins at the break site is consolidated by phosphorylation and ubiquitylation (13). ATM phosphorylates a number of these proteins, including MDC1, Nbs1, 53BP1, and BRCA1. Phosphorylation of MDC1 by a second enzyme, casein kinase-2, is responsible for the retention of the MRN complex on chromatin (28, 29). Phosphorylation of MDC1 at ATM consensus sites facilitates interaction with the RING finger ubiquitin ligase, RNF8, which in turn ubiquitylates H2A and H2AX, leading to the accumulation of 53BPI, BRCA1, and other proteins at the site of damage (30–32). It is now evident that this accumulation is consolidated by the involvement of a second ubiquitin ligase, RNF168, that also ubiquitylates H2A and H2AX (33, 34). In addition to activation by DNA DSB recent data show that ATM is also an important sensor of reactive oxygen species in human cells (35). Oxidation directly activates ATM, there is no dependence on the MRN complex for activation, and the protein is present as a disulfide-linked dimer under these conditions.
As for other protein kinases, autophosphorylation is observed at several sites in ATM (14, 36). These sites have been described in human cells employing mass spectrometry and include Ser367, Ser1893, and Ser1981. Mutants in these sites fail to correct the extent of radiation-induced cell death and cell cycle defects in A-T cells (36). On the other hand, the dependence on autophosphorylation for ATM activation is not observed in mutant Atm mice generated by BAC recombineering (37, 38). In these experiments, BAC constructs with S367A, S1899A, and S1987A were introduced into an Atm−/− background. ATM signaling and downstream events were normal in these animals. At this stage, these differences between human and murine ATM activation remain unresolved. In this study, we compared ATM phosphorylation in human and murine cells, describing additional sites present in both. We also confirmed ATM Ser367 and ATM Ser2996 autophosphorylation in human cells using phosphospecific antibodies against these sites and showed that they localize to sites of DNA DSB and play a role in regulating the S phase checkpoint.
EXPERIMENTAL PROCEDURES
Cell Culture and Irradiation
Lymphoblastoid cell lines were established from normal healthy individuals (C3ABR) and A-T patients (AT25ABR and AT1ABR) by Epstein-Barr virus immortalization. AT1ABR is homozygous for the 7636del9 mutation that produces nearly full-length ATM protein that is kinase-dead, and AT25ABR fails to express ATM protein. The NBS patient cell line NBS03LA was obtained from Professor R. Gatti, the ATLD cell line (ATLD 6) was a kind gift of Professors D. Delia and L. Chessa, and the Rad50-deficient cell line (F239hTert) was kindly provided by Dr. Thilo Dörk (10). A-T cell lines GM05823-hTert (39) and GM02052A-hTert (40) were kind gifts of Dr. Tej Pandita and Prof. William K. Kaufmann, respectively. The lymphoblastoid cell lines were cultured in RPMI 1640 medium with 10% fetal calf serum (15% FCS in the case of ATLD and Rad50-deficient cells), 100 units/ml penicillin, and 100 units/ml streptomycin (Invitrogen). All irradiations were performed at room temperature using a Gammacell 40 Exactor research irradiator (1 Gy/min; MDS Nordion, Ottawa, Canada).
Antibodies
Affinity-purified ATM-specific sheep polyclonal antibodies were prepared as described earlier (41). ATM 2C1 monoclonal antibody (Abcam/GeneTex), phospho-Ser1981 ATM rabbit polyclonal (Rockland Immunochemicals), Mre11–2D7 monoclonal (GeneTex), Rad50–2C6 monoclonal (Upstate/Chemicon), Nbs1 polyclonal (GeneTex), and γH2AX (Ser(P)139) polyclonal (Novus Biologicals) antibodies were used for immunoblotting.
Phosphospecific Antibody Production
All peptides were synthesized by Mimotopes (Clayton, Australia). Phosphopeptides were conjugated to keyhole limpet hemocyanin, and phospho-Ser367 and phospho-Ser2996 ATM were prepared by immunizing rabbits with a phospho-Ser367 peptide conjugate of keyhole limpet hemocyanin-SLEIpSQSYTT (where pS represents phosphoserine) and with a phospho-Ser2996 peptide conjugate of keyhole limpet hemocyanin-CKRNLpSDIDQSF (Institute of Medical and Veterinary Science, Salisbury, Australia) followed by affinity purification on affinity columns prepared by coupling of the corresponding phosphorylated and non-phosphorylated ATM peptides to Sulfolink resin (Pierce).
Cell Extracts, Immunoblotting, and ATM Immunoprecipitation
Cell extracts were prepared by lysing cells in ATM kinase lysis buffer containing phosphatase and protease inhibitors (41). For immunoblotting, 50 μg of cell lysate was electrophoresed and transferred to nitrocellulose membranes (Amersham Biosciences). Membranes probed with antibodies were visualized with the ECL kit (PerkinElmer Life Sciences). ATM protein was immunoprecipitated using sheep polyclonal ATM antibodies and resolved on SDS gels (5% acrylamide) as described (41). After transfer to nitrocellulose membranes, they were probed consecutively with phospho-Ser367 ATM, phospho-Ser1981 ATM, phospho-Ser1893 ATM, phospho-Ser2996 ATM, and ATM2C1 antibodies with stripping between each step. Scaled up ATM immunoprecipitations were used to obtain ATM protein for mass spectrometry analysis. A portion of the ATM immunoprecipitations was used for in vitro autophosphorylation reactions with 20 μCi of [γ-32P] ATP to produce 32P-labeled ATM to facilitate analysis by mass spectrometry.
Identification of ATM Phosphorylation Sites
Immunoprecipitated ATM from untreated or irradiated cells was separated by SDS-PAGE. Each ATM band (estimated at 0.5–1 μg) was destained, digested with trypsin, and extracted as described previously (42). Human or mouse ATM tryptic peptides from control or irradiated cells, from three SDS-PAGE bands, were enriched for phosphopeptides using titanium dioxide as described (42). The phosphopeptide-enriched fraction or the non-binding non-phosphorylated peptides were loaded onto an HPLC system (LC Packings Ultimate HPLC system, Dionex) for analysis by a QSTAR XL quadrupole-TOF MS (MDS Sciex) as described (42). One-third of the sample was analyzed in information-dependent acquisition mode where the three most abundant precursors in a 1-s MS scan were selected for MS/MS sequencing in subsequent 2-s scans. Two-thirds of the sample was used to obtain improved spectra of each of the detected phosphopeptides by fixing the precursor at a particular m/z to obtain multiple 2-s scans of the same phosphopeptide across the elution peak, which were later summed together.
Quantitative Analysis Using Isotope Labeling
Human ATM tryptic peptides were labeled with light or heavy stable isotopes for quantitative analyses prior to phosphopeptide enrichment. The dimethylation was done similarly to methods described previously (43, 44). In a 100-μl final reaction volume, 10 μl of 1 m triethylammonium bicarbonate and 10 μl of 1% formaldehyde (0.1% final concentration) were added to ATM tryptic peptides from untreated cells in 60 μl of water and vortexed for 5 min. 10 μl of 50 mm sodium cyanoborohydride was added, and the solution was vortexed for another 10 min. ATM tryptic peptides from irradiated cells were labeled in the same way, except formaldehyde with the “heavy” atoms, 2H and 13C, was used such that the net increase in mass compared with the “light” formaldehyde was at least 6 Da (plus an additional 6 Da for each Lys present in the sequence). The light and heavy labeled peptides were mixed together before titanium dioxide phosphopeptide enrichment. The monoisotopic peak, ±0.15 units of m/z, for each light and heavy phosphopeptide signal was used to extract ion chromatograms, and the peaks were integrated (44). The intensities obtained were normalized for differences in the amount of protein in each of the pair of bands. This was done by comparing the intensity between light and heavy non-phosphopeptide signals from the non-binding titanium dioxide chromatography. Three non-phosphopeptides were used for normalization of protein: 54YLNWDAVFR63, 452TPYVLR457, and 1647LVVNLLQLSK1656. These peptide identities were confirmed in the information-dependent acquisition mode analysis (data not shown). Peptides were identified by exporting the MS/MS spectra from Analyst QS 1.1 to Mascot 2.2 using the Mascot.dll v16b23 script to search the IPI_human 3.57 data base (76,541 entries). All MS/MS sequence data were manually validated.
Expression Vectors and Transfection
Construction of full-length ATM cDNA expression vector pMAT1 was described previously (45). ATM cDNA was placed under the control of a heavy metal-inducible mehallothionein promoter in pMEP4 vector (Invitrogen). Site-directed mutagenesis of the YFP-tagged ATM in pcDNA3 (provided by David Chen) was performed using the QuikChange mutagenesis kit essentially as described by the manufacturer (Stratagene) to generate the S367A and S2996A autophosphorylation site mutants. Transfection of hTERT-immortalized A-T fibroblasts was achieved using the Amaxa Nucleofector system.
Immunofluorescence Microscopy
Lymphoblastoid cells were collected onto glass slides by cytocentrifugation and stained by indirect immunofluorescence. The sheep phospho-Ser1981 ATM antibody was used at 1:100 dilution, and γH2AX antibody (Cell Signaling Technology) was used at 1:300 dilution. Anti-sheep and anti-rabbit secondary antibodies labeled with Alexa dyes (Molecular Probes) were used to visualize binding of primary antibodies. Coverslips were mounted with the Vectashield antifade solution containing DAPI (Molecular Probes) as DNA counterstain. Images were collected using a digital camera attached to a Zeiss Axioskop (Zeiss) fluorescence microscope. Initial digital image processing was performed using Zeiss software. Digital images were prepared for publication using Adobe Photoshop CS software.
Heavy Ion Irradiation and Microscopy
AG1522D human fibroblasts were subjected to beams of 12C, 64Ni, 129Xe, 238U, and 197Au ions at the UNILAC facility at GSI, as described previously (46). Beam parameters were as follows: linear energy transfer (LET), 12C 170 keV/μm; energy on target (Et), 9.8 MeV/unit; 64Ni LET, 3430 keV/μm; Et, 6.0 MeV/unit; 129Xe LET, 8800 keV/μm; Et, 4.4 MeV/unit; 197Au LET, 12,815 keV/μm; Et, 6.2 MeV/unit). Cells were fixed at the indicated time points after irradiation, and immunofluorescence microscopy was performed with antibodies to Ser(P)367 ATM, Ser(P)1981 ATM, Ser(P)2996 ATM, ATM, and γH2AX proteins. DNA was visualized with TOPRO3 or DAPI stain. GM05823-hTert or GM02052A-hTERT A-T cell lines were transfected with wild type YFP-ATM or S367A, S1981A, and S2996A mutants using the Amaxa Nucleofector I system (Amaxa GmbH). HeLa cells were transfected with the same ATM plasmids using Lipofectamine 2000 reagent (Invitrogen). Cells were cultured in a Petri dish on glass coverslips or 25-μm PTFE foil (In Vitro Systems & Solutions, Göttingen, Germany). A-T cells expressing YFP-tagged ATM kinase were irradiated with 238U ions (LET, 14350 keV/μm; Et, 3.8 MeV/unit), 197Au ions (LET, 12,815 keV/μm; Et, 6.2MeV/unit), and 56Fe ions (LET, 3050 keV/μm; Et, 6.1 MeV/unit) and fixed, and immunofluorescence microscopy was performed with antibodies to γH2AX to visualize tracks of DNA damage. Recruitment of YFP-ATM proteins to the sites of ion traversals was visualized using a beamline microscope as described (47).
Monitoring Intra-S Phase Checkpoint
Stable cell lines were established in the AT1ABR (A-T) lymphoblastoid cell line expressing wild type ATM (pMAT1), ATM S367A, and ATM S2996A in pMEP4 vector (Invitrogen) (36). AT1ABR cell line alone or with the wild type or ATM S367A or ATM S2996A mutant form was induced with 5 μm cadmium chloride for 16 h prior to the fork labeling. Induced replicating cells were labeled consecutively with 25 μm chlorodeoxyuridine for 20 min, including a mock or 5-Gy treatment, and then for a further 20 min with 100 μm iododeoxyuridine. DNA fibers were made following the approach of Parra and Windle (48). Briefly, 2 × 106 cells were mixed with lysis solution (0.5% SDS, 200 mm Tris-HCl, pH 7.4, and 50 mm EDTA), and then after 10 min, slides were tilted to 20° to spread the DNA fibers. Slides were air-dried both pre- and postfixation in methanol/acetic acid (3:1). Immunolabeling of the denatured DNA fibers (2.5 m HCl for 60 min) followed the approach of Aten et al. (49), using first a rat monoclonal anti-BrdU (Abcam) to label chlorodeoxyuridine and a goat anti-rat Alexa 488-conjugated secondary antibody (Molecular Probes), followed by a mouse monoclonal anti-BrdU (BD Biosciences) to label iododeoxyuridine and donkey anti-mouse Alexa 594-conjugated secondary antibody (Molecular Probes). Fibers were imaged on a Zeiss AxioSkop microscope using Axiovision version 3.0 (Zeiss). DNA forks were scored (400–600) for each cell line for mock (unirradiated) and 5-Gy treatments. The percentage of new initiations was calculated as ((new initiations/(continuing + new initiations)) × 100, and the S.D. value was calculated from three independent experiments.
Statistical Analysis
Statistical analysis was performed using Microsoft Excel. In experiments with YFP-ATM protein recruitment to sites of DNA damage, the error bars represent the 95% confidence interval of the mean. In S phase checkpoint assays, values were expressed as mean ± S.D. In cell survival experiments, error bars represent S.E.
RESULTS
Identification of ATM Autophosphorylation Sites
We have previously identified three autophosphorylation sites in ATM in response to DNA DSB using tandem MS/MS (36). We employed a more sensitive approach here to identify additional autophosphorylation sites. Not surprisingly, we detected phosphopeptides containing the three original sites (Ser(P)367, Ser(P)1893, and Ser(P)1981). A new phosphorylation site was identified on Thr1885 adjacent to the previously identified Ser1893 site. Supplemental Fig. 1, A and B, shows the sequencing of the phosphopeptide containing the novel Thr1885 site. We also confirmed the presence of Ser(P)2996 (supplemental Fig. 1F), close to the kinase domain of ATM, which was recently detected by Daub et al. (50). A summary of the five phosphorylation sites detected in human ATM appears in Table 1.
TABLE 1.
Identification of phosphorylation sites in human and mouse ATM by tandem MS/MS
| Human |
Mouse |
||||
|---|---|---|---|---|---|
| Sequence | Sites | Phosphorylation sites | Sequence | Sites | Phosphorylation sites |
| 363SLEIpSQSYTTTQR375 | 1 | Ser(P)367 | 363SVEIpSQSYVTQR374 | 1 | Ser(P)367 |
| 1883STpTPANLDSEpSEHFFR1898 | 2 | Thr(P)1885 | 1889SApTPANSDSEpSENFLR1904 | 2 | Thr(P)1891 |
| Ser(P)1893 | Ser(P)1999 | ||||
| 1974SLAFEEGpSQSTTISSLSEK1992 | 1 | Ser(P)1981 | 1980SPTFEEGpSQGTTISSLSEK1998 | 1 | Ser(P)1987 |
| 2994NLpSDIDQSFNaK3004 | 1 | Ser(P)2996 | 3003QSLpSDTDQSFNK3014 | 1 | Ser(P)3006 |
a We detected human ATM with the Asp3003 → Asn polymorphism (74).
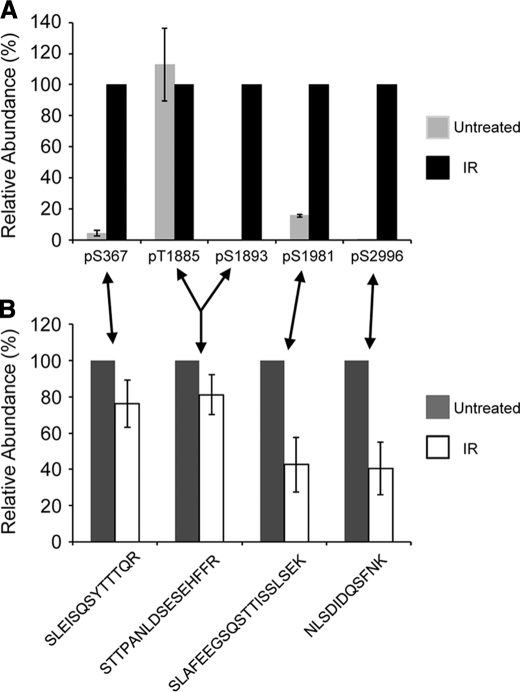
We next quantified the change in phosphorylation at each of these five detectable phosphorylation sites when human cells were irradiated. ATM immunoprecipitated from untreated and irradiated cells was labeled by dimethylation with light and heavy stable isotopes, respectively, as described under “Experimental Procedures.” Analysis of the signal intensity for the labeled peptides allowed relative quantification of the phosphopeptides. Thus, the relative level of phosphorylation at each site was measured. The phosphopeptides containing either Thr1885 or Ser1893 were separable using reverse phase chromatography by an ∼60-s difference in retention time, allowing separate quantification. Four of the five detectable phosphosites, Ser367, Ser1893, Ser1981, and Ser2996, showed increased phosphorylation after irradiation (Fig. 1A). It was not possible to describe the -fold change for most phosphosites, because the phosphopeptide signal from the untreated cells was below the threshold of detection, indicating a low or zero basal phosphorylation. However, there was a measurable 6-fold increase in Ser(P)1981 and a 20-fold increase in Ser(P)367. The proline-directed (Ser/Thr with Pro in the +1-position) phosphosite, Thr(P)1885, did not change significantly, indicating that it does not respond to DNA damage. Further insight was gained by measuring the relative change in the amount of each non-phosphopeptide that can harbor a phosphorylation site (Fig. 1B). An ∼60% drop in the level of 1974SLAFEEGSQSTTISSLSEK1992 and 2994NLSDIDQSFNK3004 indicated that Ser(P)1981 and Ser(P)2996 had a similar response to DNA damage, and there was a large change in the stoichiometry of phosphorylation at these sites. The change in stoichiometry at Ser(P)367 (measured as a decrease in the amount of 363SLEISQSYTTTQR375) was modest in comparison. This peptide may also be influenced by the reported Ser(P)373 site (51). The decrease in the amount of 1883STTPANLDSESEHFFR1898 was very slight. However, the inducible Ser(P)1893 shares this tryptic peptide with the non-inducible Thr1885 (Fig. 1A). Overall, there are large increases in the level of phosphorylation at Ser367, Ser1893, Ser1981, and Ser2996 following irradiation of human cells but not at the new ATM phosphosite Thr1885.
FIGURE 1.
The relative abundance of ATM phosphosites after irradiation using dimethyl labeling with stable isotopes. ATM tryptic peptides from untreated cells and irradiated (IR) cells were labeled with light and heavy isotopes and mixed before quantitative MS analysis. A, the relative change in the abundance of each specific phosphorylation site was determined from the abundance of the phosphopeptides, corrected for the amount of protein in each band (see “Experimental Procedures”). The abundance in the irradiated sample was normalized to 100% for each phosphorylation site. Note that the monophosphorylated peptide that contained Thr1885 eluted ∼1 min before the peptide monophosphorylated at Ser1893, with the same sequence and molecular mass, allowing separate quantification. The data are represented as the mean ± S.E. (error bars), n = 3. B, the relative change in the abundance of the non-phosphopeptides that harbor phosphorylation sites was determined. The abundance in the untreated sample was normalized to 100% for each peptide (ATM 363–375, 1883–1898, 1974–1992, and 2994–3004, where amino acid residue 3003 is an Asp, not a Gln). The data are represented as the mean ± S.E., n = 4.
We also determined the phosphorylation sites on murine ATM in irradiated B16 cells. We confirmed that all of the detectable phosphorylation sites corresponding to those in human ATM are also phosphorylated in mice (see Table 1 and supplemental Fig. 1, C–F). The close correlation with human sites indicates similar usage of sites in both organisms for phosphoregulation. Other autophosphorylation sites have been detected on ATM in large scale proteomic studies (51, 52) (see the PhosphoSitePlus Web site); however, none of these have been validated. All of the sites identified appear in supplemental Table 1.
The Radiation-inducible Phosphorylation of Ser367 and Ser2996
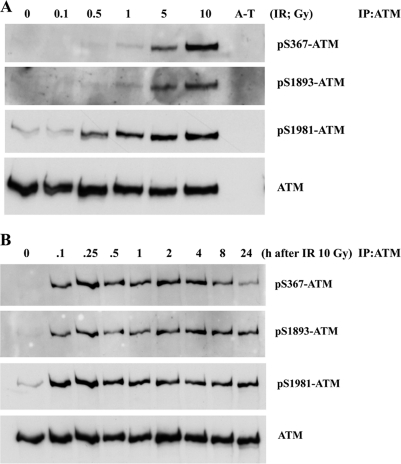
We previously generated phosphospecific antibodies to investigate the importance of the Ser(P)1893 and Ser(P)1981 ATM autophosphorylation sites (36). The same approach was employed here in generating antibodies against the ATM Ser(P)367 site. The specificity of this antibody was demonstrated by detection of a peptide containing Ser(P)367 and no reactivity against the corresponding unphosphorylated peptide (supplemental Fig. 2A). Furthermore, no cross-reactivity was observed against Ser1893- or Ser1981-phosphorylated peptides. The effect of increasing doses of radiation on ATM Ser(P)367 was initially checked. As is evident from the results in Fig. 2A, phosphorylation is detected after 5- and 10-Gy radiation exposure. This was comparable with the level of induction of ATM Ser(P)1893. A more intense signal was observed with anti-ATM Ser(P)1981, but this could be due to the sensitivity of the antibody. After exposure of cells to 10 Gy, ATM Ser(P)367 was detected as early as 6 min postirradiation, comparable with the kinetics of appearance of ATM Ser(P)1893 and Ser(P)1981 (Fig. 2B). In all cases, the level of phosphorylation remained largely unchanged up to 24 h postirradiation.
FIGURE 2.
ATM is autosphosphorylated on Ser367 in response to radiation exposure. A, effect of increasing radiation dose on ATM autophosphorylation on Ser367, Ser1893, and Ser1981. Cells were irradiated (IR) with the indicated doses and incubated for 1 h prior to preparation of extracts, immunoprecipitation (IP) with anti-ATM antibodies, and immunoblotting with phosphospecific and anti-ATM antibodies. AT25ABR, which expresses no ATM protein, was used as a control. B, time course of ATM autophosphorylation on Ser367, Ser1893, and Ser1981 in response to 10 Gy of radiation. Conditions were the same as described in A.
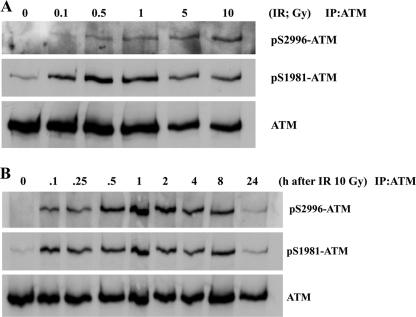
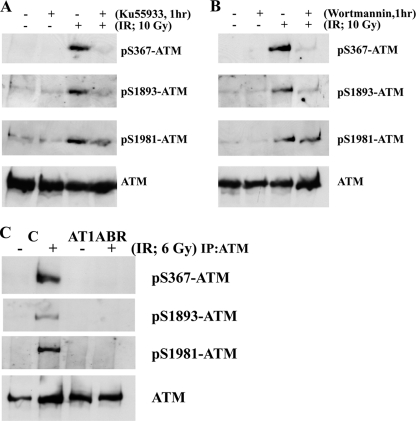
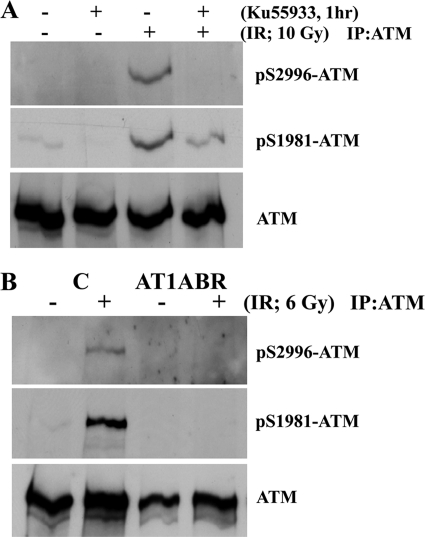
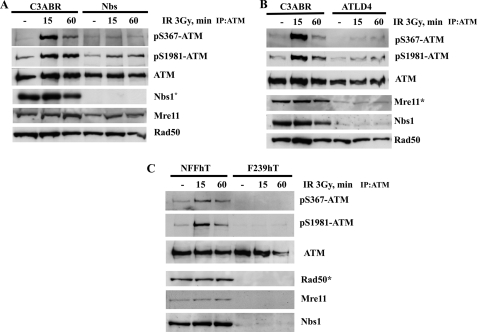
We also raised phosphospecific antibodies against ATM Ser2996 and demonstrated its specificity against phosphorylated peptide (supplemental Fig. 2B). As in the case of ATM ser367 phosphorylation, we also observed time and dose dependence for ATM Ser(P)2996 after exposure to radiation (Fig. 3, A and B). To establish that Ser(P)367 and Ser(P)2996 were ATM-dependent, experiments were carried out with A-T cells and with ATM inhibitors. Exposure of cells to the specific ATM inhibitor KU55933 abolished radiation-induced phosphorylation at Ser367 (Fig. 4A). This was also observed with the more general PIKK inhibitor, wortmannin (Fig. 4B). Under these conditions, the Ser1893 and Ser1981 phosphorylations were also inhibited. The results in Fig. 4C show that there is no induction of ATM Ser(P)367 in AT1ABR cells postirradiation. This was also the case for Ser(P)1893 and Ser(P)1981. Again, as observed for ATM Ser(P)367, the ATM-specific inhibitor (KU55933) abolished ATM Ser2996 phosphorylation (Fig. 5A), and phosphorylation at this site was not observed in AT1ABR cells (Fig. 5B). Previous results reveal a dependence on the MRN complex for ATM autophosphorylation and activation (14, 19, 35). Here we employed NBS cells to demonstrate a marked decrease in DNA damage-induced ATM Ser(P)367 (Fig. 6A). Again, Ser(P)1981 was also defective. This was also observed with ATLD4 cells hypomorphic for Mre11 mutations (Fig. 6B). Similarly, we did not detect any appreciable Ser367 phosphorylation in a cell line, derived from a patient with hypomorphic Rad50 mutations (10) (Fig. 6C). Overall, these results confirm radiation-induced phosphorylation of ATM on Ser367 and Ser2996. They reveal that they are dependent on the presence of active ATM and also demonstrate a dependence of Ser367 phosphorylation on the MRN complex.
FIGURE 3.
ATM is autosphosphorylated on Ser2996 in response to radiation exposure. A, effect of increasing radiation dose on ATM autophosphorylation on Ser2996 and Ser1981. Cells were irradiated (IR) with the indicated doses and incubated for 1 h prior to preparation of extracts, immunoprecipitation (IP) with anti-ATM antibodies, and immunoblotting with phosphospecific and anti-ATM antibodies. B, time course of ATM autophosphorylation on Ser2996 and Ser1981 in response to 10 Gy of radiation. Conditions were the same as described in A.
FIGURE 4.
ATM S367 autophosphorylation is dependent on ATM protein kinase. A, the ATM-specific inhibitor KU55933 prevents ATM autophosphorylation on Ser367, Ser1893, and Ser1981 sites after exposure to 10 Gy. B, the effect of the PIKK inhibitor wortmannin on ATM autophosphorylation at the three sites. C, ATM autophosphorylation on Ser367 is not detected in AT1ABR cells. AT1ABR is an A-T cell line expressing a mutant, less stable, and kinase-dead form of ATM (7636del9). C3ABR is a control cell line. ATM was immunoprecipitated from extracts prepared 1 h after exposure of cells to 6 Gy, and immunoblotting was carried out as described in Fig. 2. IR, irradiation; IP, immunoprecipitation.
FIGURE 5.
ATM Ser2996 autophosphorylation is dependent on ATM protein kinase. A, the ATM-specific inhibitor KU55933 prevents ATM autophosphorylation on Ser2996 and Ser1981 sites after exposure to 10 Gy. B, ATM autophosphorylation on Ser29967 is not detected in AT1ABR cells, expressing a mutant, less stable, and kinase-dead form of ATM. C, C3ABR is a control cell line. ATM was immunoprecipitated from extracts prepared 1 h after exposure of cells to 6 Gy, and immunoblotting was carried out as described in Fig. 2. IR, irradiation; IP, immunoprecipitation.
FIGURE 6.
Mutations in members of the MRN complex reduce the efficiency of ATM autophosphorylation on Ser367. A, C3ABR (control) and NBS (Nijmegen breakage syndrome) cells were exposed to 3 Gy of radiation and incubated for 15 or 60 min prior to preparation of extracts. ATM was immunoprecipitated, and immunoblotting was carried out with the phosphospecific antibodies (top). Separated extracts were also immunoblotted with Nbs1, Mre11, and Rad50 antibodies. B, autophosphorylation in control cells (C3ABR) and ATLD4 cells (deficient in Mre11) in response to 3 Gy of radiation. As in A, ATM was immunoprecipitated, followed by blotting with phosphospecific antibodies. Mre11, Nbs1, and Rad50 were detected by immunoblotting. C, NFFhTert (control) and F239hTert fibroblasts (deficient in Rad50) were exposed to 3 Gy of radiation and incubated for 15 or 60 min prior to preparation of extracts. ATM was immunoprecipitated, and immunoblotting was carried out with the phosphospecific antibodies (top). Extracts were also immunoblotted with Nbs1, Mre11, and Rad50 antibodies. Note that Rad50 mutation in F239hTert cells also affects levels of Mre11 and Nbs1 (10). *, defective protein in the specific cell line.
ATM Ser(P)367 and ATM Ser(P)2996 Localize to Sites of DNA Damage
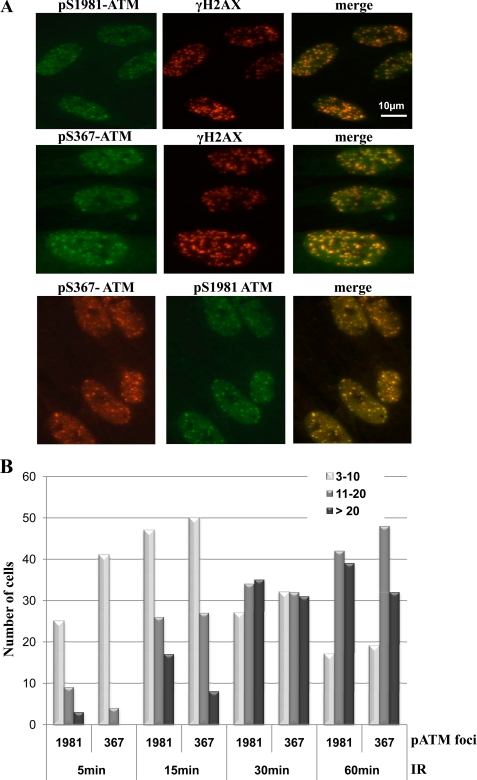
Exposure of cells to ionizing radiation leads to the accumulation of DNA damage recognition and repair proteins at the sites of DNA damage in the form of nuclear foci (53, 54). It has also been shown that ATM Ser(P)1981 co-localizes with γH2AX in nuclear foci postirradiation (14, 36). We employed two different qualities of radiation (low LET and high LET), both of which give rise to DNA DSB, to investigate ATM Ser(P)367 localization at short times after radiation exposure. ATM Ser(P)367 foci were detected at 5 min after 3 Gy of γ-rays, and these coincided with γH2AX foci indicative of DNA DSB (Fig. 7A, middle). γH2AX foci also colocalized with ATM Ser(P)1981 foci at the same time point (Fig. 7A, top), and there was co-localization between ATM Ser(P)367 and ATM Ser(P)1981 (bottom). We also employed co-localization with 53BP1 to demonstrate the presence of ATM Ser(P)367 at sites of DNA damage (supplemental Fig. 3). Quantitation of Ser(P)367 and Ser(P)1981 foci showed similar kinetics of accumulation (Fig. 7B).
FIGURE 7.
Localization of ATM Ser367 to sites of DNA damage. A, control fibroblasts were exposed to 3 Gy of radiation, fixed, and stained for γH2AX foci as indicators of DNA DSB using immunofluorescence microscopy. The same cells were also stained for Ser(P)367 or Ser(P)1981 ATM foci, and co-localization was carried out using green and red Alexa dye labeling. Co-localization of Ser(P)367 and Ser(P)1981 ATM was also investigated. B, quantitative analysis of Ser(P)367 and Ser(P)1981 foci formation after 3 Gy of radiation. Control fibroblasts were exposed to 3 Gy of radiation, fixed at the indicated time points, and stained with Ser(P)367 and Ser(P)1981 antibodies. Ser(P)367 and Ser(P)1981 foci were analyzed by immunofluorescence microscopy using the DeltaVision deconvolution microscope system (Applied Biosciences). Semiautomated foci counting was performed using macros written for ImageJ software (National Institutes of Health). A histogram of foci distribution per cell nucleus is shown. Combined foci counts from two independent experiments are shown. Counts were obtained from 50 nuclei, and nuclei with two or less foci were excluded from analysis.
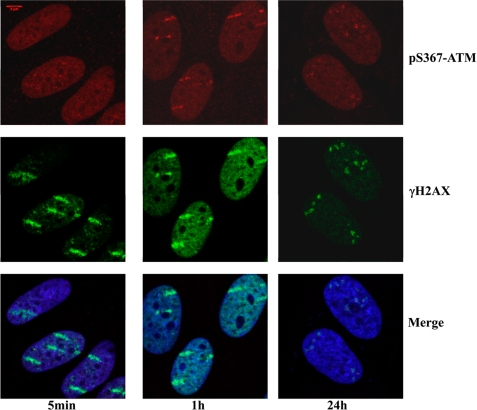
Biological imaging of charged particle tracks represents a useful approach to investigate the association of DNA damage response proteins with chromatin (55–57). The results in Fig. 8 show that fibroblasts exposed to xenon ions accumulate ATM Ser(P)367 along tracks, and these coincide with γH2AX signal. This was also the case for total ATM and ATM Ser(P)1981 (supplemental Fig. 4, A and B). Similar results for Ser(P)367 and Ser(P)1981 were obtained after irradiation with carbon or nickel ions (supplemental Fig. 5, A and B). We have also observed weak accumulation of ATM Ser(P)367 and ATM Ser(P)1981 in Nbs cells after irradiation with 64Ni ions (supplemental Fig. 6, A and B), and pretreatment of normal fibroblasts with ATM inhibitor Ku55933 abolishes ATM Ser(P)367 signal along the tracks of DNA damage (data not shown). Similar experiments were carried out for localization of ATM Ser(P)2996. Exposure of fibroblasts to gold ions led to accumulation of ATM Ser(P)2996 at sites of DNA damage as evidenced by co-localization with γH2AX (Fig. 9). We next determined whether phosphosite mutants in ATM would affect its recruitment to chromatin in transfected A-T cells in response to DNA DSB, using a unique real-time beamline microscopy system as described previously (56, 57). We initially established localization of YFP-tagged ATM proteins to the sites of DNA damage produced by uranium ions (Fig. 10). Cells were fixed 30 min after irradiation and immunostained with antibodies against γH2AX. We observed co-localization of YFP-ATM wild type, YFP-S1981A ATM, and YFP-S367A ATM with γH2AX along “tracks” of trajectories of the uranium particles (Fig. 10, A–C). This correlates well with the results in Fig. 8 and supplemental Fig. 4 showing accumulation of ATM, ATM Ser(P)1981, and ATM Ser(P)367 along tracks produced by heavy ions. No quantitative differences in recruitment of YFP-ATM and ATM phosphorylation site mutants to the sites of DNA damage were evident.
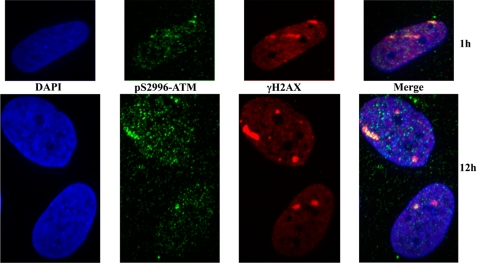
FIGURE 8.
Recruitment of ATM kinase phosphorylated at Ser367 to the sites of DNA damage induced by heavy ions. Normal AG1522D human fibroblasts were subjected to a beam of xenon ions. Cells were fixed at the indicated time points after irradiation, and immunofluorescence microscopy was performed with antibodies to Ser(P)367 ATM and γH2AX proteins. DNA was visualized with TOPRO3 stain. Top panels, Ser(P)367 ATM staining; middle, γH2AX staining; bottom, merged images with TOPRO3 staining.
FIGURE 9.
Recruitment of ATM kinase phosphorylated at Ser2996 to the sites of DNA damage induced by heavy ions. Normal AG1522D human fibroblasts were subjected to a beam of 197Au ions. Cells were fixed at the indicated time points after irradiation, and immunofluorescence microscopy was performed with antibodies to Ser(P)2996 ATM and γH2AX proteins. DNA was visualized with DAPI stain.
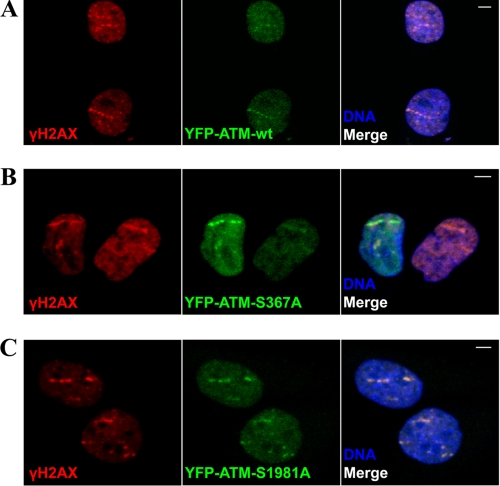
FIGURE 10.
Recruitment of YFP-ATM, YFP-ATM S367A, and YFP-ATM S1981A to sites of DNA damage induced by heavy ions. YFP-ATM WT, YFP-S1981A ATM, and YFP-S367A ATM were transfected into the A-T GM05823hTert cell line, and cells were subjected to heavy ion irradiation (238U). Cells were fixed 30 min after irradiation and stained with antibodies against γH2AX. DNA was visualized with TOPRO3 stain.
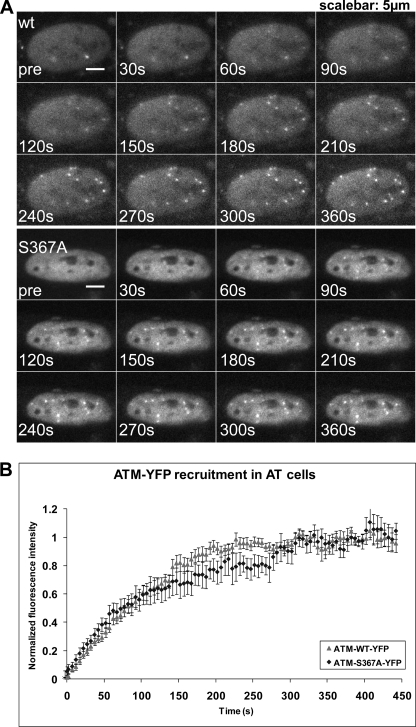
We then analyzed the kinetics of accumulation of different phosphorylated forms of ATM at DSB sites induced by uranium ions. Both ATM and the S367A ATM mutant accumulated at sites of DNA damage with approximately the same kinetics (Fig. 11). Both proteins show time constants of accumulation of ∼120–150 s, comparable with those reported previously for Nbs1 and Mdc1 (58). It was not possible to use this real-time beam microscopy system to analyze binding over longer periods of time because the signal fades. To further analyze the recruitment kinetics of ATM, we transfected HeLa cells with YFP-ATM wild type, S1981A, and S367A and subjected them to irradiation to determine accumulation of ATM in the presence of endogenous ATM protein. Similar to the situation in A-T cells, we did not observe any significant differences in recruitment of wild type ATM and the two mutant forms of ATM to the sites of DNA damage (supplemental Fig. 7). Use of the ATM-YFP S2996A mutant also demonstrated that this form of ATM had accumulation kinetics at sites of DNA damage similar to that of wild type (supplemental Fig. 8).
FIGURE 11.
Time lapse imaging of YFP-ATM-expressing A-T cells before and after irradiation with 238U ions. A, an example of cells expressing YFP-ATM WT. Shown is live cell imaging prior to irradiation and 7 min postirradiation. Recruitment of YFP-ATM to the sites of damage produced by heavy ions is visualized in real time as white dots (foci) appearing in the nucleus. Images were recorded at 1-s intervals and binned afterward to 5-s intervals. The real-time animation is available upon request. B, kinetics of WT ATM versus S367A ATM accumulation at the sites of DNA damage induced by uranium irradiation. The error bars represent the 95% confidence interval of the mean, n = 10.
We have previously shown that the ATM S367A mutant was defective in correcting radiosensitivity in A-T cells (36). Transfection of AT1ABR cells with ATM S2996A also failed to correct radiosensitivity (supplemental Fig. 9A). Under these conditions, the level of induction of the mutant ATM protein was comparable with that of wild type (supplemental Fig. 9B). It is notable that in ATM S2996A transfectants, the induced levels of protein decreased with time of cells in culture (data not shown).
We examined the effect of both the S367A and S2996A mutations on the S phase checkpoint, a hallmark of the A-T phenotype: radioresistant DNA synthesis (59). Radioresistant DNA synthesis was determined by consecutively labeling sites of replication to distinguish new initiations from ongoing forks (49). Both the S367A and S2996A mutants were defective in correcting radioresistant DNA synthesis in A-T cells (Fig. 12). Under these conditions, the control cells showed marked inhibition of DNA synthesis.
FIGURE 12.
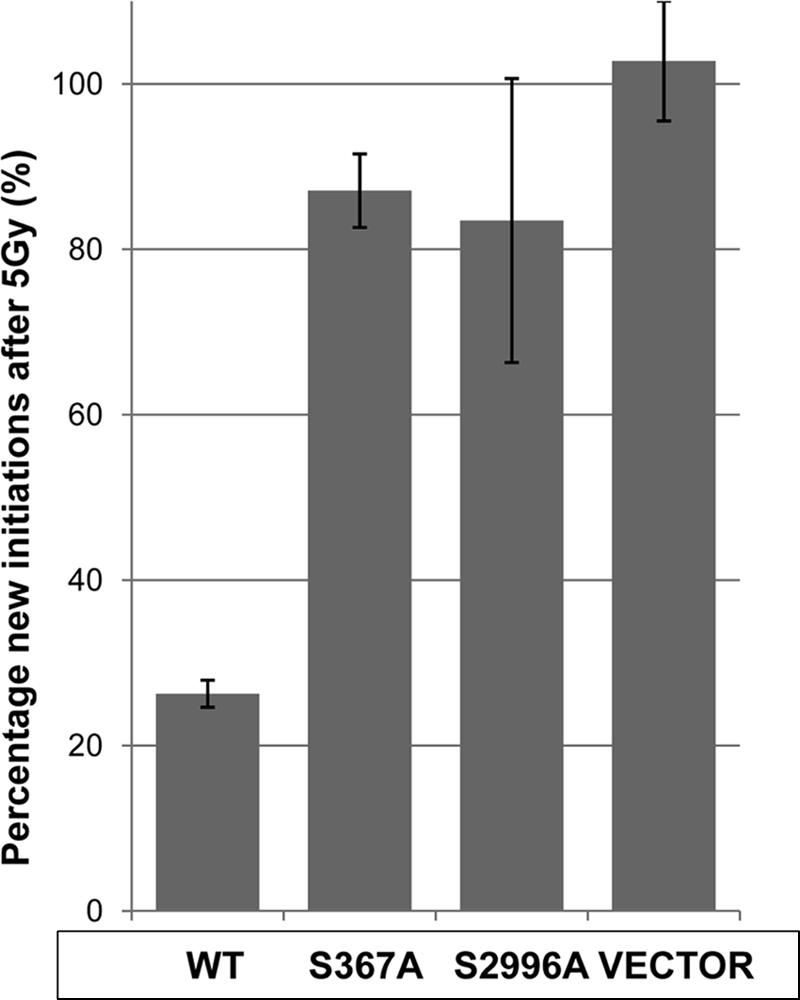
ATM S367A and ATM S2996A are unable to correct radioresistant DNA synthesis seen in A-T cells. Wild type ATM, mutant ATM S367A, or ATM S2996A was transfected into A-T lymphoblastoid cells (AT1ABR) to investigate the role of Ser367/Ser2996 autophosphorylation sites in radioresistant DNA synthesis. DNA forks were consecutively labeled with nucleotide analogs to follow the response of cells to ionizing radiation (5 Gy). The percentage of new initiations after irradiation/unirradiated new initiations is shown. The percentage of new initiations was calculated as ((new initiations/(continuing + new initiations)) × 100. At least 500 DNA forks were scored for each cell line for unirradiated and 5-Gy treatments from three independent experiments. Results are mean ± S.D. (error bars), n = 3.
DISCUSSION
The four ATM autophosphorylation sites (Ser(P)367, Ser(P)1893, Ser(P)1981, and Ser(P)2996) described previously using different approaches (14, 36, 50, 51) were detected, and the response to DNA damage was quantified for each site. In addition, we have described here the existence of Thr(P)1885 on ATM and shown that it is not involved in the DNA damage response. The best described of the four radiation-sensitive sites is ATM Ser(P)1981, which has been associated with ATM monomerization and activation in the vicinity of DNA DSB (14, 15). Phosphorylation at this site is widely used as an indicator of ATM activation. The description of a second site, Ser(P)1893, and the availability of a phosphospecific antibody against that site reveal that this phosphorylation can also be used to monitor activity (36). However, this site is not a consensus (SEpSE) ATM site and could point to the involvement of another protein kinase closely associated with ATM and involved in its activation. However, ATM has been shown to phosphorylate some substrates at non-consensus sites (60). Using a similar approach, we have shown here that the other two sites (Ser367 and Ser2996), are also phosphorylated in response to DNA DSB. As outlined in supplemental Table 1, five of the reported phosphorylation sites are located close to the N terminus of the molecule between amino acids 72 and 373, and several of these are in the substrate binding region (61). Phosphorylation at these sites may alter substrate binding or the interaction of ATM with other proteins.
Three other sites are in the region of Ser1981, and the final site, Ser2996, is located adjacent to the kinase domain in the PIKK regulatory domain. Mordes and Cortez (62) revealed that TopBP1 interacts with ATRIP and facilitates its binding to the PIKK regulatory domain of ATR for activation of that protein kinase. The PIKK regulatory domains of ATM, DNA-PKcs, and mTOR have also been shown to regulate the activities of these PIKK family members (63). We have shown for the first time phosphorylation of ATM on Ser2996 and demonstrated that it is particularly responsive to radiation-induced phosphorylation, similar to Ser1981. It is also of interest that this site has been identified by Shiloh and co-workers (65). Perhaps not by coincidence, the Ser(P)2996 site is located close to an acetylation site (Lys3016) (18). This site is acetylated in parallel to Ser(P)1981 in response to DNA DSB (64). Furthermore, mutation in this site inhibited monomerization of the inactive ATM dimer and prevented induction of ATM activation and phosphorylation of downstream substrates. We have also provided evidence that ATM Ser2996 phosphorylation plays a functional role in radiation signaling. ATM post-translational modifications may influence the interaction of activators with this region of the protein, such as TopBP1 does for ATR (62). Identification of such activators will assist further in unraveling the mechanism of ATM activation. Whereas ATM activation by DNA DSB involves autophosphorylation and formation of an active monomer, activation by oxidative stress occurs by formation of a dimeric form of ATM held together by disulfide linkages (35). A critical site for disulfide bond formation is Cys2991. It will be of interest to determine how the phosphorylation status of Ser2996 impacts on this process. We did not observe here the recently described Cdk5 (cyclin-dependent kinase 5) phosphorylation site on ATM, Ser794 (64). Ser794 is located in a small tryptic peptide and therefore could be easily missed in our MS/MS analysis. This site was identified on ATM in postmitotic neurons by indirect approaches. It is not clear if activation of Cdk5 kinase could be achieved in our cell culture model, lymphoblastoid cells.
As observed previously for ATM Ser1981 and Ser1893, both ATM Ser367 and ATM Ser2996 are rapidly phosphorylated in response to DNA DSB. In the case of ATM Ser(P)1981, this is correlated with dissociation of ATM into active monomers, which is prevented by mutation at this site (14). Mutation at this site did not interfere with phosphorylation on ATM Ser1893, but it reduced ATM-dependent phosphorylation of p53, Nbs1, SMC1, and Chk2 in A-T cells complemented with ATM S1981A cDNA (36). This mutant also failed to correct DNA repair, cell cycle checkpoint defects, and radiosensitivity in A-T cells. These results are at variance with those of Pellegrini et al. (37), who demonstrated that Atm-dependent responses are functional at the organism and cellular levels in mice expressing the corresponding mutant (S1987A) form of Atm. In the case of the human ATM S367A mutant, results similar to these with ATM S1981A were obtained. Mutation at this site was defective in supporting ATM-dependent signaling and did not correct downstream events, such as radiosensitivity and cell cycle checkpoint defects (36). Although no mouse mutant expressing the ATM S367A mutation has been described, a triple autophosphorylation site mutant (S367A/S1893A/S1981A) revealed that all of these sites were dispensable for ATM activation in vivo (38). They propose that these data are consistent with Atm autophosphorylation correlating with DNA damage-induced activation of the kinase but not being required for ATM function in vivo. The difference between human and mouse studies remains unclear at present. However, ATM from these organisms have the same phosphorylation sites, which implies similar usage of sites for phosphoregulation, although important differences may be revealed by future detailed studies of these sites.
We also demonstrated here that ATM Ser(P)367 is localized to nuclear foci and to sites of DNA damage induced by both low and high LET-ionizing radiation (Figs. 7 and 8). This behavior is also observed for ATM Ser(P)1981 as reported earlier (14). However, we did not observe ATM Ser(P)2996 localizing to foci in response to γ-radiation, whereas ATM Ser(P)2996 could be detected at higher DSB density damage sites (Fig. 9). More recently, Berkovich et al. (15) showed that although wild type ATM was bound to PpoI-induced DNA DSB, neither kinase-inactive ATM nor ATM S1981A was able to bind, suggesting that autophosphorylation is important for localization to sites of DNA damage. After this initial activation, the MRN complex is instrumental in recruiting ATM to the break site for full activation. In contrast, You et al. (16) showed that ATM activation is dispensable for initial recruitment to DNA fragments added to Xenopus egg extracts. This is substantiated by a recent report showing that ATM Ser(P)1981 is only required for sustained retention of ATM at sites of the breaks (35). This is in agreement with the findings of Lee and Paull (17, 22), who used an in vitro system to demonstrate large increases in ATM kinase activity without a concomitant increase in ATM autophosphorylation. These data suggest that mutant ATM may dissociate following the initial recruitment, whereas wild type is retained on chromatin to amplify signaling. An alternative model of ATM kinase activation suggests a two-step mechanism (23), where the level of DNA damage defines a threshold for MRN complex requirement in the activation process. It is feasible that some discrepancies in the interpretation of the behavior of ATM phosphorylation mutants can be attributed to different doses and types of DNA damage employed.
We have shown here that both the YFP-ATM S367A and YFP-ATM S2996A mutants are recruited to sites of DNA damage induced by heavy ions with kinetics similar to that of wild type YFP-ATM, at least under the experimental conditions studied (Fig. 10 and supplemental Figs. 7 and 8). Similarly, the recruitment of YFP-ATM S1981A mutant did not seem to differ much from wild type YFP-ATM as can be judged from immunofluorescence data. We previously published data demonstrating that an ATM S367A cDNA was defective in correcting radiation-induced signaling, DNA repair, radiosensitivity, and cell cycle checkpoint defects in A-T cells (36). Because the initial recruitment of YFP-S367A ATM to the sites of DNA damage induced by uranium ions was similar to that of wild type YFP-ATM, it seems that functional effects of Ser367 mutation can be explained by defective ATM signaling to downstream substrates (at least at low doses of irradiation). Indeed, moderate variation in kinase activity of ATM phosphorylation site mutants at the sites of DNA DSB could influence the effective concentration of “activated” pools of ATM downstream targets (Nbs1, Chk2, p53, etc.). The transient nature of the interaction of ATM with its substrates (Chk2 and Nbs1) at sites of DNA DSB and the existence of multiple pools of chromatin-bound ATM substrates have been observed previously (58, 66). It is also impossible to exclude the possibility that the differences in expression systems and levels of expressed ATM proteins (constitutive expression via CMV promoter in pcDNA3 vector versus heavy metal-inducible metallothionein promoter in pMEP4 vector) could influence the outcome. However, we have been able again to detect the functional effect of S367A ATM and S2996A mutations in the S phase checkpoint assay (Fig. 12) and their inability to correct radioresistant DNA synthesis in A-T cells. One intriguing possibility is that Ser367 and Ser2996 phosphorylations might play a distinct role in a specific stage of the cell cycle. From our analysis of Ser367 and Ser2996 ATM phosphorylation, it is clear that phosphospecific Ser367 and Ser2996 ATM antibodies can be used as additional and reliable markers of ATM activation.
Curiously, tethering Cherry-LacR-ATM protein, with a truncation of the N terminus (residues 1300–3060) lacking the Ser367 phosphorylation site, to chromatin seems to be sufficient to activate DNA damage signaling (67), even in the absence of DNA damage. In addition, manipulating the N-terminal area of ATM by adding the ATRIP-binding N-terminal region (residues 1–388) of ATR kinase (68) created ATM protein that was capable of binding ATRIP and localizing it to stalled replication forks. Although this chimeric ATM protein retained normal kinase activity in vitro and was phosphorylated on Ser1981, it was nevertheless deficient in checkpoint signaling and did not complement the cellular phenotype of A-T cells, probably due to deficiency in binding to Nbs1. Fernandez et al. (69) showed that ATM kinase with mutations in the N-terminal interaction domain (amino acids 91–97) had kinase activity and was autophosphorylated on Ser1981 but did not correct radiosensitivity in A-T cells. The N-terminal “substrate binding region” of ATM described previously (61, 70, 71) was also identified as a “chromatin association” domain (amino acids 5–224) (72). Importantly, Nbs1 interaction region is also mapped to the N-terminal HEAT repeats of ATM (73). All of these studies underscore the importance of regulatory phosphorylation sites in the N terminus of ATM and unresolved issues related to the role of N-terminal domains of ATM in its activation as well as potential critical role of C-terminal phosphorylation sites in the initiation of DNA damage signaling. Both the N-terminal and C-terminal ATM phosphorylation sites described here will assist in elucidating the precise mechanism of ATM kinase activation after DNA damage.
Finally, it is clear from the data presented here (Table 1 and supplemental Table 1) that ATM kinase is capable of phosphorylation on multiple sites in both human and mouse cells. It is also evident that complex arrays of post-translational modifications are involved in regulation of ATM kinase activation (e.g. see Ref. 13; see the PhosphoSitePlus Web site). The combinations of post-translational modifications might allow precise cell- and damage signal-specific ATM kinase activation. Future studies aimed at detailed mapping of ATM modification sites in different model organisms, especially in tissues of neuronal origin, will undoubtedly lead to a greater understanding of ATM kinase activation.
Supplementary Material
Acknowledgments
We thank Aine Farell and Gudrun Becker for excellent technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants CA050519 and CA92584 (to D. C.). This work was also supported by grants from the National Health and Medical Research Council (to M. F. L.) and GSI Helmholtzzentrum für Schwerionenforschung GmbH (to G. T.-S. and B. J.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–9.
- DSB
- double strand break(s)
- MRN
- Mre11-Rad50-Nbs1
- A-T
- ataxia-telangiectasia
- PIKK
- phosphoinositide 3-kinase-like kinase
- Gy
- gray(s)
- LET
- linear energy transfer.
REFERENCES
- 1. Bredemeyer A. L., Sharma G. G., Huang C. Y., Helmink B. A., Walker L. M., Khor K. C., Nuskey B., Sullivan K. E., Pandita T. K., Bassing C. H., Sleckman B. P. (2006) Nature 442, 466–470 [DOI] [PubMed] [Google Scholar]
- 2. McKinnon P. J., Caldecott K. W. (2007) Annu. Rev. Genomics Hum. Genet. 8, 37–55 [DOI] [PubMed] [Google Scholar]
- 3. Shrivastav M., De Haro L. P., Nickoloff J. A. (2008) Cell Res. 18, 134–147 [DOI] [PubMed] [Google Scholar]
- 4. Stracker T. H., Theunissen J. W., Morales M., Petrini J. H. (2004) DNA Repair 3, 845–854 [DOI] [PubMed] [Google Scholar]
- 5. Helmink B. A., Bredemeyer A. L., Lee B. S., Huang C. Y., Sharma G. G., Walker L. M., Bednarski J. J., Lee W. L., Pandita T. K., Bassing C. H., Sleckman B. P. (2009) J. Exp. Med. 206, 669–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rass E., Grabarz A., Plo I., Gautier J., Bertrand P., Lopez B. S. (2009) Nat. Struct. Mol. Biol. 16, 819–824 [DOI] [PubMed] [Google Scholar]
- 7. Xie A., Kwok A., Scully R. (2009) Nat. Struct. Mol. Biol. 16, 814–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carney J. P., Maser R. S., Olivares H., Davis E. M., Le Beau M., Yates J. R., 3rd, Hays L., Morgan W. F., Petrini J. H. (1998) Cell 93, 477–486 [DOI] [PubMed] [Google Scholar]
- 9. Stewart G. S., Maser R. S., Stankovic T., Bressan D. A., Kaplan M. I., Jaspers N. G., Raams A., Byrd P. J., Petrini J. H., Taylor A. M. (1999) Cell 99, 577–587 [DOI] [PubMed] [Google Scholar]
- 10. Waltes R., Kalb R., Gatei M., Kijas A. W., Stumm M., Sobeck A., Wieland B., Varon R., Lerenthal Y., Lavin M. F., Schindler D., Dörk T. (2009) Am. J. Hum. Genet. 84, 605–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Savitsky K., Bar-Shira A., Gilad S., Rotman G., Ziv Y., Vanagaite L., Tagle D. A., Smith S., Uziel T., Sfez S., Ashkenazi M., Pecker I., Frydman M., Harnik R., Patanjali S. R., Simmons A., Clines G. A., Sartiel A., Gatti R. A., Chessa L., Sanal O., Lavin M. F., Jaspers N. G., Taylor A. M., Arlett C. F., Miki T., Weissman S. M., Lovett M., Collins F. S., Shiloh Y. (1995) Science 268, 1749–1753 [DOI] [PubMed] [Google Scholar]
- 12. Lavin M. F., Shiloh Y. (1997) Annu. Rev. Immunol. 15, 177–202 [DOI] [PubMed] [Google Scholar]
- 13. Lavin M. F. (2008) Nat. Rev. Mol. Cell Biol. 9, 759–769 [DOI] [PubMed] [Google Scholar]
- 14. Bakkenist C. J., Kastan M. B. (2003) Nature 421, 499–506 [DOI] [PubMed] [Google Scholar]
- 15. Berkovich E., Monnat R. J., Jr., Kastan M. B. (2007) Nat. Cell Biol. 9, 683–690 [DOI] [PubMed] [Google Scholar]
- 16. You Z., Bailis J. M., Johnson S. A., Dilworth S. M., Hunter T. (2007) Nat. Cell Biol. 9, 1311–1318 [DOI] [PubMed] [Google Scholar]
- 17. Lee J. H., Paull T. T. (2005) Science 308, 551–554 [DOI] [PubMed] [Google Scholar]
- 18. Sun Y., Xu Y., Roy K., Price B. D. (2007) Mol. Cell. Biol. 27, 8502–8509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sun Y., Jiang X., Xu Y., Ayrapetov M. K., Moreau L. A., Whetstine J. R., Price B. D. (2009) Nat. Cell Biol. 11, 1376–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Uziel T., Lerenthal Y., Moyal L., Andegeko Y., Mittelman L., Shiloh Y. (2003) EMBO J. 22, 5612–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cerosaletti K., Concannon P. (2004) J. Biol. Chem. 279, 38813–38819 [DOI] [PubMed] [Google Scholar]
- 22. Lee J. H., Paull T. T. (2004) Science 304, 93–96 [DOI] [PubMed] [Google Scholar]
- 23. Dupré A., Boyer-Chatenet L., Gautier J. (2006) Nat. Struct. Mol. Biol. 13, 451–457 [DOI] [PubMed] [Google Scholar]
- 24. Kitagawa R., Bakkenist C. J., McKinnon P. J., Kastan M. B. (2004) Genes Dev. 18, 1423–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kitagawa R., Kastan M. B. (2005) Cold Spring Harb. Symp. Quant. Biol. 70, 99–109 [DOI] [PubMed] [Google Scholar]
- 26. van Attikum H., Gasser S. M. (2009) Trends Cell Biol. 19, 207–217 [DOI] [PubMed] [Google Scholar]
- 27. Cook P. J., Ju B. G., Telese F., Wang X., Glass C. K., Rosenfeld M. G. (2009) Nature 458, 591–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chapman J. R., Jackson S. P. (2008) EMBO Rep. 9, 795–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Melander F., Bekker-Jensen S., Falck J., Bartek J., Mailand N., Lukas J. (2008) J. Cell Biol. 181, 213–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huen M. S., Grant R., Manke I., Minn K., Yu X., Yaffe M. B., Chen J. (2007) Cell 131, 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kolas N. K., Chapman J. R., Nakada S., Ylanko J., Chahwan R., Sweeney F. D., Panier S., Mendez M., Wildenhain J., Thomson T. M., Pelletier L., Jackson S. P., Durocher D. (2007) Science 318, 1637–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J. (2007) Cell 131, 887–900 [DOI] [PubMed] [Google Scholar]
- 33. Doil C., Mailand N., Bekker-Jensen S., Menard P., Larsen D. H., Pepperkok R., Ellenberg J., Panier S., Durocher D., Bartek J., Lukas J., Lukas C. (2009) Cell 136, 435–446 [DOI] [PubMed] [Google Scholar]
- 34. Stewart G. S. (2009) Cell Cycle 8, 1532–1538 [DOI] [PubMed] [Google Scholar]
- 35. Guo Z., Kozlov S., Lavin M. F., Person M. D., Paull T. T. (2010) Science 330, 517–521 [DOI] [PubMed] [Google Scholar]
- 36. Kozlov S. V., Graham M. E., Peng C., Chen P., Robinson P. J., Lavin M. F. (2006) EMBO J. 25, 3504–3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pellegrini M., Celeste A., Difilippantonio S., Guo R., Wang W., Feigenbaum L., Nussenzweig A. (2006) Nature 443, 222–225 [DOI] [PubMed] [Google Scholar]
- 38. Daniel J. A., Pellegrini M., Lee J. H., Paull T. T., Feigenbaum L., Nussenzweig A. (2008) J. Cell Biol. 183, 777–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wood L. D., Halvorsen T. L., Dhar S., Baur J. A., Pandita R. K., Wright W. E., Hande M. P., Calaf G., Hei T. K., Levine F., Shay J. W., Wang J. J., Pandita T. K. (2001) Oncogene 20, 278–288 [DOI] [PubMed] [Google Scholar]
- 40. Heffernan T. P., Simpson D. A., Frank A. R., Heinloth A. N., Paules R. S., Cordeiro-Stone M., Kaufmann W. K. (2002) Mol. Cell. Biol. 22, 8552–8561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kozlov S., Gueven N., Keating K., Ramsay J., Lavin M. F. (2003) J. Biol. Chem. 278, 9309–9317 [DOI] [PubMed] [Google Scholar]
- 42. Chan L. S., Hansra G., Robinson P. J., Graham M. E. (2010) J. Proteome Res. 9, 4028–4037 [DOI] [PubMed] [Google Scholar]
- 43. Hsu J. L., Huang S. Y., Chow N. H., Chen S. H. (2003) Anal. Chem. 75, 6843–6852 [DOI] [PubMed] [Google Scholar]
- 44. Lemeer S., Jopling C., Gouw J., Mohammed S., Heck A. J., Slijper M., den Hertog J. (2008) Mol. Cell Proteomics 7, 2176–2187 [DOI] [PubMed] [Google Scholar]
- 45. Zhang N., Chen P., Khanna K. K., Scott S., Gatei M., Kozlov S., Watters D., Spring K., Yen T., Lavin M. F. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 8021–8026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jakob B., Scholz M., Taucher-Scholz G. (2000) Radiat. Res. 154, 398–405 [DOI] [PubMed] [Google Scholar]
- 47. Jakob B., Rudolph J. H., Gueven N., Lavin M. F., Taucher-Scholz G. (2005) Radiat. Res. 163, 681–690 [DOI] [PubMed] [Google Scholar]
- 48. Parra I., Windle B. (1993) Nat. Genet. 5, 17–21 [DOI] [PubMed] [Google Scholar]
- 49. Aten J. A., Bakker P. J., Stap J., Boschman G. A., Veenhof C. H. (1992) Histochem. J. 24, 251–259 [DOI] [PubMed] [Google Scholar]
- 50. Daub H., Olsen J. V., Bairlein M., Gnad F., Oppermann F. S., Körner R., Greff Z., Kéri G., Stemmann O., Mann M. (2008) Mol. Cell 31, 438–448 [DOI] [PubMed] [Google Scholar]
- 51. Matsuoka S., Ballif B. A., Smogorzewska A., McDonald E. R., 3rd, Hurov K. E., Luo J., Bakalarski C. E., Zhao Z., Solimini N., Lerenthal Y., Shiloh Y., Gygi S. P., Elledge S. J. (2007) Science 316, 1160–1166 [DOI] [PubMed] [Google Scholar]
- 52. Beausoleil S. A., Jedrychowski M., Schwartz D., Elias J. E., Villén J., Li J., Cohn M. A., Cantley L. C., Gygi S. P. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12130–12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Maser R. S., Monsen K. J., Nelms B. E., Petrini J. H. (1997) Mol. Cell. Biol. 17, 6087–6096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Paull T. T., Rogakou E. P., Yamazaki V., Kirchgessner C. U., Gellert M., Bonner W. M. (2000) Curr. Biol. 10, 886–895 [DOI] [PubMed] [Google Scholar]
- 55. Lukas C., Bartek J., Lukas J. (2005) Chromosoma 114, 146–154 [DOI] [PubMed] [Google Scholar]
- 56. Jakob B., Scholz M., Taucher-Scholz G. (2003) Radiat Res. 159, 676–684 [DOI] [PubMed] [Google Scholar]
- 57. Taucher-Scholz G., Jakob B. (2007) Ion Irradiation as a Tool to Reveal the Spatiotemporal Dynamics of DNA Damage Response Processes in Genome Dynamics and Stability, Vol. 1 (Lankenau D.-H., ed) pp. 453–478, Springer-Verlag, Berlin [Google Scholar]
- 58. Lukas C., Melander F., Stucki M., Falck J., Bekker-Jensen S., Goldberg M., Lerenthal Y., Jackson S. P., Bartek J., Lukas J. (2004) EMBO J. 23, 2674–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Houldsworth J., Lavin M. F. (1983) Biochem. Int. 6, 349–356 [PubMed] [Google Scholar]
- 60. Cortez D., Wang Y., Qin J., Elledge S. J. (1999) Science 286, 1162–1166 [DOI] [PubMed] [Google Scholar]
- 61. Khanna K. K., Keating K. E., Kozlov S., Scott S., Gatei M., Hobson K., Taya Y., Gabrielli B., Chan D., Lees-Miller S. P., Lavin M. F. (1998) Nat. Genet. 20, 398–400 [DOI] [PubMed] [Google Scholar]
- 62. Mordes D. A., Cortez D. (2008) Cell Cycle 7, 2809–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lovejoy C. A., Cortez D. (2009) DNA Repair 8, 1004–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tian B., Yang Q., Mao Z. (2009) Nat. Cell Biol. 11, 211–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bensimon A., Schmidt A., Ziv Y., Elkon R., Wang S. Y., Chen D. J., Aebersold R., Shiloh Y. (2010) Sci. Signal. 3, rs3. [DOI] [PubMed] [Google Scholar]
- 66. So S., Davis A. J., Chen D. J. (2009) J. Cell Biol. 187, 977–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lukas C., Falck J., Bartkova J., Bartek J., Lukas J. (2003) Nat. Cell Biol. 5, 255–260 [DOI] [PubMed] [Google Scholar]
- 68. Soutoglou E., Misteli T. (2008) Science 320, 1507–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chen X., Zhao R., Glick G. G., Cortez D. (2007) Exp. Cell Res. 313, 1667–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fernandes N., Sun Y., Chen S., Paul P., Shaw R. J., Cantley L. C., Price B. D. (2005) J. Biol. Chem. 280, 15158–151864 [DOI] [PubMed] [Google Scholar]
- 71. Beamish H., Kedar P., Kaneko H., Chen P., Fukao T., Peng C., Beresten S., Gueven N., Purdie D., Lees-Miller S., Ellis N., Kondo N., Lavin M. F. (2002) J. Biol. Chem. 277, 30515–30523 [DOI] [PubMed] [Google Scholar]
- 72. Young D. B., Jonnalagadda J., Gatei M., Jans D. A., Meyn S., Khanna K. K. (2005) J. Biol. Chem. 280, 27587–27594 [DOI] [PubMed] [Google Scholar]
- 73. Falck J., Coates J., Jackson S. P. (2005) Nature 434, 605–611 [DOI] [PubMed] [Google Scholar]
- 74. Rasio D., Negrini M., Croce C. M. (1995) Cancer Res. 55, 6053–6057 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.