Abstract
In aerobic respiration, the tricarboxylic acid cycle is pivotal to the complete oxidation of carbohydrates, proteins, and lipids to carbon dioxide and water. Plasmodium falciparum, the causative agent of human malaria, lacks a conventional tricarboxylic acid cycle and depends exclusively on glycolysis for ATP production. However, all of the constituent enzymes of the tricarboxylic acid cycle are annotated in the genome of P. falciparum, which implies that the pathway might have important, yet unidentified biosynthetic functions. Here we show that fumarate, a side product of the purine salvage pathway and a metabolic intermediate of the tricarboxylic acid cycle, is not a metabolic waste but is converted to aspartate through malate and oxaloacetate. P. falciparum-infected erythrocytes and free parasites incorporated [2,3-14C]fumarate into the nucleic acid and protein fractions. 13C NMR of parasites incubated with [2,3-13C]fumarate showed the formation of malate, pyruvate, lactate, and aspartate but not citrate or succinate. Further, treatment of free parasites with atovaquone inhibited the conversion of fumarate to aspartate, thereby indicating this pathway as an electron transport chain-dependent process. This study, therefore, provides a biosynthetic function for fumarate hydratase, malate quinone oxidoreductase, and aspartate aminotransferase of P. falciparum.
Keywords: Energy Metabolism, Mitochondrial Metabolism, Parasite Metabolism, Parasitology, Tricarboxylic Acid Cycle, Fumarate, Plasmodium falciparum, Purine Salvage Pathway
Introduction
Plasmodium falciparum is a parasitic protozoan, which causes the most severe form of malaria in humans. Because of the emergence in the parasite of widespread drug resistance to most of the first-line antimalarials, the development of new drugs that target the parasite metabolism is needed. Over several years, malarial parasites have been studied to understand various novel biochemical features, which not only gives opportunities for chemotherapeutic interventions but also stimulates broader scientific interests.
During the intraerythrocytic stages of P. falciparum, the tricarboxylic acid cycle does not seem to function like a conventional tricarboxylic acid cycle for the following reasons. (a) The bulk of the glucose is metabolized to lactate by anaerobic glycolysis, which is then secreted by the parasite as a metabolic waste (1) (2). As a result, unlike aerobic cells, in P. falciparum there is minimal carbon flow from the cytoplasm to the mitochondrial tricarboxylic acid cycle. (b) The multi-enzyme complex pyruvate dehydrogenase, which channels pyruvate into the tricarboxylic acid cycle through acetyl-CoA, has been found to localize on the apicoplast membrane in P. falciparum (3), unlike mammalian cells, where the enzyme is present on the inner mitochondrial membrane (4). (c) The enzyme isocitrate dehydrogenase generates NADPH rather than NADH (5), thereby implying that its main role is probably to act as a redox sensor rather than donating reducing equivalents to the electron transport chain. In addition, P. falciparum synthesizes ATP mainly by substrate level phosphorylation through glycolysis and not through oxidative phosphorylation (6), suggesting that the primary function of the tricarboxylic acid cycle in generating reducing equivalents for the electron transport chain might be dispensable for the parasite. However, genes for all of the enzymes of the tricarboxylic acid cycle are present and are expressed in P. falciparum (7, 8) during the intraerythrocytic stages, thereby implying that the pathway probably has important, yet unidentified biosynthetic functions.
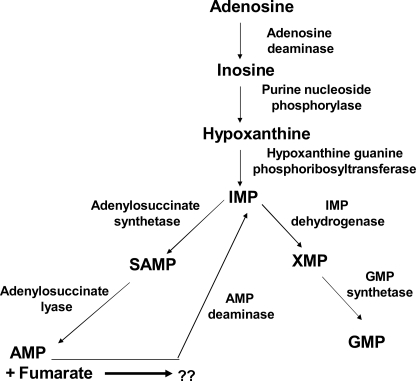
Unlike its human host, P. falciparum lacks the de novo purine biosynthetic pathway and is hence completely dependent on the purine salvage pathway to meet its purine nucleotide requirements (9). In the purine salvage pathway, hypoxanthine salvaged from the host is phosphoribosylated to IMP, which then branches out to form AMP and GMP (Fig. 1). IMP is converted to AMP in two steps, catalyzed by two distinct enzymes: (a) adenylosuccinate synthetase, which adds a molecule of aspartate to the 6-oxo position of IMP with a concomitant hydrolysis of GTP to GDP to produce succinyl-AMP (10); and (b) cleavage of succinyl-AMP to AMP and fumarate by adenylosuccinate lyase (11). Excess AMP is deaminated back to IMP by AMP deaminase. This cyclic conversion of IMP to AMP constitutes the purine nucleotide cycle (Fig. 1), which has been shown to play an important role under conditions (such as in muscle tissue) requiring excess ATP (12). The P. falciparum genome contains a homolog of AMP deaminase, suggesting the presence of the purine nucleotide cycle in the parasite. Apart from AMP, the other end product of this pathway is fumarate, which in most aerobic cells feeds into the tricarboxylic acid cycle (13). However, in view of a dysfunctional tricarboxylic acid cycle in P. falciparum, the metabolic fate of fumarate is not known.
FIGURE 1.
Purine salvage pathway in the intraerythrocytic P. falciparum. Adenosine and hypoxanthine salvaged from the host are converted to IMP, which then branches out to form AMP and GMP. Excess AMP is deaminated back to IMP to constitute purine nucleotide cycle. A molecule of fumarate is also generated for each molecule of AMP. The arrow and the question mark have been put in to emphasize that the reutilization pathway of fumarate has not been elucidated.
Our studies with metabolic tracers implicate fumarate generated from the purine salvage pathway as being incorporated into nucleic acids and proteins and not secreted out by the parasite. Fumarate gets converted to malate and then subsequently to aspartate through a metabolic pathway that involves fumarate hydratase, malate quinone oxidoreductase, and aspartate aminotransferase. This report shows functionality to a portion of the tricarboxylic acid cycle and also highlights possible metabolic cross-talk between purine salvage and the electron transport chain in P. falciparum.
EXPERIMENTAL PROCEDURES
Reagents
All reagents and chemicals, unless mentioned otherwise, were obtained from Sigma. [U-14C]Aspartate (200 mCi mmol−1) was obtained from the Board of Radiation and Isotope Technology (BRIT), India. [2,3-14C]Fumarate (10 mCi mmol−1) and 2,3-[13C]fumarate were obtained from Sigma. [8-3H]Hypoxanthine (19.2 Ci mmol−1) was obtained from GE Healthcare. Primers for gene cloning were custom synthesized at Sigma. Human O+ serum and erythrocytes for maintenance of the 3D7 strain of P. falciparum were obtained from healthy volunteers from a local hospital.
P. falciparum Culture Maintenance
The 3D7 strain of P. falciparum was cultured in vitro as described previously (14). Briefly, the parasites were maintained in human O+ erythrocytes isolated from O+ human blood collected from healthy volunteers at 5% hematocrit in RPMI 1640 (Sigma) buffered with HEPES containing 10% human serum and supplemented with 5% NaHCO3. The culture was grown at 37 °C in a candle jar. The medium was changed every 24 h, and parasitemia was estimated every 48 h by microscopic examination of Giemsa-stained smears. Free parasites were isolated by treating parasitized erythrocytes with 0.15% saponin. The parasitized culture was enriched for mature stages (trophozoites and schizonts) by centrifugation in a 70% Percoll (GE Healthcare) solution (15).
Metabolic Labeling
1 μCi of [U-14C]aspartate (25 μm) or [2,3-14C]fumarate (500 μm) was added to erythrocytes, parasitized erythrocytes (2% hematocrit and 6–8% parasitemia, 200 μl), or saponin-released free parasites (total protein: 2–3 mg ml−1). Although erythrocytes and parasitized erythrocytes were incubated for 24 or 48 h, saponin-released free parasites were incubated for 8 h at 37 °C in a 96-well flat-bottom plate placed in a candle jar. Parasites were in the trophozoite stage before labeling. After incubation, the spent medium of each cell type was collected and treated with 10% trichloroacetic acid for 15 min on ice followed by boiling for 10 min. The protein precipitate was removed by centrifugation for 30 min at 4 °C. The supernatant was collected, lyophilized, and reconstituted in 10–20 μl of distilled water. 2–3 μl of the concentrated medium was spotted on a 10 × 5 cm TLC silica gel 60 F254 plate (Merck) and air-dried. A mobile phase consisting of n-butanol, acetic acid, and water at a ratio of 4:1:1 was used to run the TLC, following which the TLC plate was dried and developed using a Fuji FLA-5000 phosphorimaging device.
For nucleic acid incorporation, the contents of each well were harvested onto glass fiber filters using a Combi-12 automated cell harvester (Molecular Devices, Sunnyvale, CA), washed extensively with distilled water, and dried. The incorporated radioactivity was measured as disintegrations per minute using a Wallac 1409 liquid scintillation counter (Wallac Oy, Turku, Finland).
To check the incorporation of radioactivity in proteins, the parasite culture was treated with 0.15% saponin solution for 2–3 min, which released the parasites from the erythrocytes. The parasite pellet was washed five or six times in PBS, resuspended in 1× SDS-gel loading buffer, boiled for 10 min, and subjected to 10% SDS-PAGE. The gel was dried and developed on a Fuji FLA-5000 phosphorimaging device. For 14C label distribution between spent medium and parasite lysate, saponin-released erythrocyte-free parasites were incubated with 1 μCi (0.5 mm) of [2,3-14C]fumarate for 8 h and processed as described under the supplemental “Methods.”
Metabolite Extraction, Separation, and Analysis
Free parasites, after incubation with the radioactive tracers [U-14C]aspartate, [2,3-14C]fumarate, and [8-3H]hypoxanthine, were washed thrice with 1× PBS to remove excess radioactive label, resuspended in distilled water, and lysed by rapid freeze-thawing. Soluble metabolites were extracted from the lysate by treatment with 0.5 m HClO4 for 30 min on ice using published protocols (16). Samples were then neutralized with 5 m KOH and centrifuged to remove the precipitate. The supernatant was concentrated by lyophilization and stored at −80 °C till further use. Purine and pyrimidine nucleotide monophosphates were separated using a reversed phase ion-pair HPLC (Genesis® C18, 4 μm, 150 × 4.6 mm, Grace Davison Discovery Sciences). The mobile phases were 8 mm tetrabutylammonium bisulfate (Spectrochem)/100 mm KH2PO4 (pH 6.0) (solution A) and 30% acetonitrile containing 8 mm tetrabutylammonium bisulfate and 100 mm KH2PO4 (pH 6.0) (solution B). The cell extracts were resuspended in a small volume of solution A and spiked with nucleotide monophosphate standards. After injection, the column was run isocratically with solution A for 2 column volumes followed by a gradient from 0 to 100% solution B for 60 min. The eluate was monitored at 254 nm with the flow rate set at 0.7 ml min−1. Peaks corresponding to the nucleotide fraction were collected, and the radioactive counts were measured as counts per minute using a Wallac 1409 (Wallac Oy) liquid scintillation counter.
Carbon-13 Nuclear Magnetic Resonance Experiments
Free parasites obtained from saponin treatment of parasitized erythrocytes were washed three times in 1× PBS and incubated with 10 mm [2,3-13C]fumarate for 4 h at 37 °C. Erythrocytes incubated with similar quantities of 13C-labeled tracers for the same amount of time served as controls. To monitor the effect of atovaquone, free parasites were treated with the drug (1 μm concentration) for 2 h followed by 4 h of incubation with [2,3-13C]fumarate. Following incubation, cell suspensions were frozen directly in liquid nitrogen and stored at −80 °C till further use. Before NMR spectra were acquired, the cell suspensions were disrupted by sonication and centrifuged, and the supernatants were collected for analysis.
After centrifugation for 30 min, the volume of the supernatant was made up to 450 μl with H2O, and 50 μl of D2O was added to obtain a final volume of 500 μl. 13C NMR spectra were collected on a 500-MHz Bruker NMR spectrometer equipped with a 5-mm broadband probe. Measurements were recorded at 25 °C under a bilevel broadband-gated proton decoupling with D2O lock.
Cloning, Expression, and Purification of P. falciparum Malate Dehydrogenase and P. falciparum Aspartate Aminotransferase
Details of cloning, expression, and purification of PfMDH4 and PfAAT are given under the supplemental “Methods.” PfAAT activity measurements were carried out as described by Winter and Dekker (17) with minor modifications.
RT-PCR
Total RNA from parasites was extracted using TRIzol reagent (Sigma), treated with DNase I (New England Biolabs), and extracted with phenol and chloroform. cDNA synthesis was carried out using MMLV reverse transcriptase (Amersham Biosciences) according to manufacturer's instructions. DNase-treated total RNA was used as a control in PCR reactions to rule out DNA contamination. Specific primers were used in separate PCR reactions for checking the expression of the genes. Primer sequences for RT-PCR are provided under the supplemental “Methods.”
Generation of Polyclonal Antibodies against P. falciparum MDH and AAT
The purified PfMDH (100 μg) was injected subcutaneously into a healthy New Zealand White rabbit after emulsification in complete Freund's adjuvant followed by two boosters in incomplete Freund's adjuvant, each separated by 14 days. Purified PfAAT (100 μg) was injected into mice (BALB/c), and subsequent injections were the same as in the case of PfMDH. Antiserum collected from the animals at the end of 14 days after the second booster was stored at 4 °C to allow complete coagulation of blood. The serum was separated by centrifugation. The supernatant was stored in aliquots in sterile tubes at −20 °C till further use.
Digitonin Permeabilization of Free Parasites and Western Blotting
Free parasites were treated with increasing concentrations of digitonin as described previously (18) with minor modifications. Total protein lysates were separated by SDS-PAGE (12%) and blotted onto a PVDF membrane using a Hoefer wet transfer apparatus (Hoefer®). Immunodetections were performed using the primary antibodies, mouse anti-PfAAT (1:2000), rabbit anti-PfMDH (1:2000), and rabbit anti-PfHSP60 (1:1000). As secondary antibody, anti-mouse or anti-rabbit IgG conjugated to horseradish peroxidase (Sigma) was used. Western blots were developed using the Pierce SuperSignal West Pico Chemiluminescent kit as described by the manufacturer.
Indirect Immunofluorescence
Indirect immunofluorescence was done as described previously (19) with minor modifications as described under the supplemental “Methods.”
RESULTS
Fumarate Is Incorporated into Nucleic Acid and Protein Fractions of P. falciparum
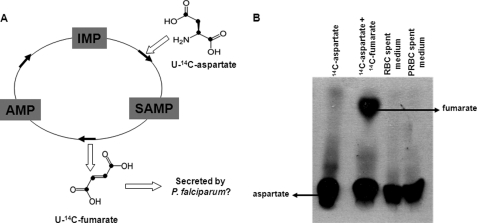
In the purine salvage pathway, the overall reaction of adenylosuccinate synthetase and adenylosuccinate lyase involves the transfer of an α-amino group from aspartate to the 6-oxo group of IMP to generate AMP, with the rest of the carbon skeleton of aspartate released as fumarate. To check whether P. falciparum secretes fumarate as a metabolic waste, PRBCs were incubated with 1 μCi of [U-14C]aspartate for 24 h, and spent medium was analyzed. RBCs were also incubated with the label as a control. [U-14C]Aspartate is expected to produce [U-14C]fumarate as a consequence of the purine salvage pathway (Fig. 2A), and if the latter is a metabolic waste, it should be secreted by PRBCs out into the medium. Upon examination by TLC of the spent medium of RBCs and PRBCs, no radioactive spot corresponding to the mobility of [14C]fumarate could be observed (Fig. 2B). To increase the sensitivity of the assay, the spent medium was concentrated before spotting, and the TLC plate was exposed for a longer time. Still, a spot corresponding to the mobility of fumarate could not be observed (data not shown).
FIGURE 2.
Fumarate is not secreted as a metabolic waste by P. falciparum. A, the carbon skeleton of [U-14C]aspartate (in which all of the carbon atoms are enriched in 14C (shown as bold circles)) is converted to [U-14C]fumarate in the parasite by the concerted actions of adenylosuccinate synthetase and adenylosuccinate lyase. Arrows indicate the direction of the reaction. B, radio-TLC analysis of the spent medium of erythrocytes (2% hematocrit) and parasitized erythrocytes (2% hematocrit and 8% parasitemia) incubated with 1 μCi of [U-14C]aspartate for 24 h. [U-14C]Aspartate and a mixture of [U-14C]aspartate and [2,3-14C]fumarate were used as TLC markers. A solvent mixture of n-butanol, acetic acid, and water in the ratio of 4:1:1 was used as the mobile phase for silica-TLC. The TLC plate was dried and developed using a Fuji FLA-5000 phosphorimaging device.
The failure to detect fumarate in the spent medium could also be because of limited uptake and metabolism of aspartate by PRBCs. However, PRBCs incubated with [U-14C]aspartate showed a significant amount of radioactivity in the nucleic acid and protein fractions (supplemental Fig. S1) indicating that [U-14C]aspartate was taken up and metabolized by the PRBCs. Saponin-released erythrocyte-free parasites also showed a dose-dependant incorporation of [U-14C]aspartate in to the nucleic acids (supplemental Fig. S1). Aspartate incorporation in the nucleic acids could be explained by the fact that P. falciparum synthesizes pyrimidines by the de novo pathway, in which the carbon skeleton of aspartate forms the backbone of the pyrimidine ring. These results imply that fumarate generated by the purine salvage pathway in P. falciparum is not secreted out in the medium and hence is not a metabolic waste.
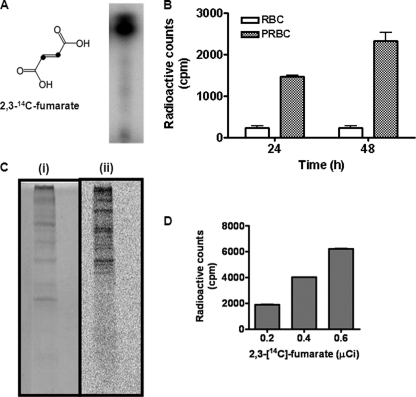
Therefore, to examine the metabolic fate of fumarate, RBCs and PRBCs were incubated with 1 μCi of [2,3-14C]fumarate for 24 and 48 h, and radioactivity was checked in nucleic acid fractions. The radiochemical purity of [2,3-14C]fumarate was established by TLC (Fig. 3A). PRBCs showed significant incorporation of radioactivity with counts in control erythrocytes being very low (Fig. 3B). Interestingly, radioactivity was also seen associated with the protein fraction of PRBCs, with all of the protein bands obtained from the cell lysate being radiolabeled (Fig. 3C). During the intraerythrocytic stages of P. falciparum life cycle, there exists an extensive metabolic cross-talk between the RBC and the parasite. Hence, metabolic labeling studies with [2,3-14C]fumarate were also done on saponin-released erythrocyte-free parasites. Here, the time of incubation of the parasites with [2,3-14C]fumarate was reduced to 8 h, and radioactivity in the nucleic acids was counted. A dose-dependent increase in radioactive incorporation in the nucleic acid fractions was obtained with [2,3-14C]fumarate, implicating the exclusive role of parasite metabolism through parasite-encoded enzymes (Fig. 3D).
FIGURE 3.
[2,3-14C]fumarate incorporation into nucleic acids and proteins by parasitized erythrocytes and free parasites. A, structure and radio-TLC analysis of [2,3-14C]fumarate (10 mCi mmol−1) in which the second and third carbon atoms are enriched in 14C. B, [2,3-14C]fumarate incorporation by parasitized erythrocytes after incubation with the radioactive label for 24 and 48 h. 1 μCi of [2,3-14C]fumarate was added to PRBCs and RBCs, and cells were harvested and counted for radioactivity as described under “Experimental Procedures.” C, parasitized erythrocytes incubated with 1 μCi of [2,3-14C]fumarate for 24 h were treated with 0.15% saponin solution to release free parasites from erythrocytes. The parasite pellet was washed, processed, and subjected to SDS-PAGE, which was stained by Coomassie Brilliant Blue (i) and developed using a Fuji FLA-5000 phosphorimaging device (ii). D, radioactive incorporation in the nucleic acid fractions of saponin released erythrocyte-free parasites incubated with varying amounts of [2,3-14C]fumarate.
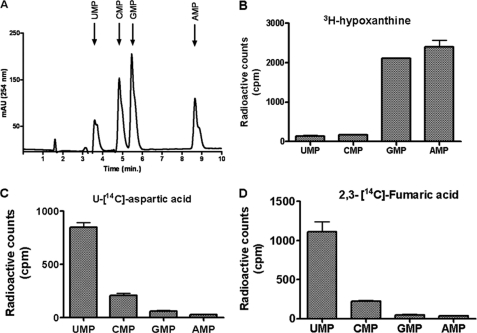
[2,3-14C]Fumarate Incorporation in Nucleic Acids Is through Pyrimidines and Not Purines
[2,3-14C]Fumarate incorporation in proteins indicates that fumarate is probably metabolized to an amino acid. Examination of the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (20) indicated that fumarate can be converted to three amino acids: aspartate, alanine, and glutamate. If fumarate is indeed converted to aspartate, then the nucleic acid incorporation could also be explained, as aspartate forms the backbone of pyrimidines. To explore this possibility, saponin-released free parasites were labeled with [2,3-14C]fumarate for 4 h. Following metabolite extraction, the samples were spiked with nucleotide monophosphate standards, which were then separated by ion-pair reversed phase HPLC (Fig. 4A). As a control, the nucleotide labeling pattern was assessed in parasites incubated with 1 μCi of [8-3H]hypoxanthine and [U-14C]aspartate. Free parasites incubated with [8-3H]hypoxanthine showed radioactive labeling of purines, GMP and AMP (Fig. 4B), whereas parasites treated with [U-14C]aspartate showed maximum radioactivity under UMP (Fig. 4C). Parasites incubated with [2,3-14C]fumarate showed maximum radioactive incorporation in UMP followed by CMP, with counts in GMP and AMP fractions being insignificant (Fig. 4D). These results indicate that fumarate incorporation into nucleic acids is not through the ribose moiety (as in that case, both purines and pyrimidines should be labeled) but specifically through the pyrimidine backbone. We therefore predict that fumarate is converted to aspartate and then incorporated into proteins and nucleic acids of P. falciparum.
FIGURE 4.
Purine and pyrimidine nucleotide labeling in free parasites by [8-3H]hypoxanthine, [U-14C]aspartate, and [2,3-14C]fumarate. A, separation of pyrimidine (UMP and CMP) and purine (GMP and AMP) mononucleotides on a C18 reversed phase HPLC. A mixture of nucleotide monophosphates was injected into the column and eluted with a linear gradient of solution B. B–D, soluble metabolites were extracted from free parasites that were incubated with 1 μCi of [3H]hypoxanthine (B), 1 μCi of [U-14C]aspartate (C), and 1 μCi of [2,3-14C]fumarate (D) and spiked with the nucleotide standards prior to HPLC. Peaks corresponding to the pyrimidine and purine nucleotide fractions were collected separately and concentrated, and incorporated radioactivity was measured as cpm using a Wallac 1409 liquid scintillation counter.
Intermediates in Fumarate Metabolism
Examination of the KEGG metabolic pathway database (20) highlights two possible routes for the formation of aspartate from fumarate. The first includes the metabolic intermediates malate and oxaloacetate, whereas the second is a direct conversion of fumarate to aspartate catalyzed by the enzyme aspartase (Fig. 5A). To distinguish between these two pathways, 0.5 mm fumarate, malate, and aspartate were added separately along with [2,3-14C]fumarate. If malate is not an intermediate in the metabolism of fumarate, and only a direct conversion operates in P. falciparum, then the presence of malate should not dilute the radioactive counts arising from [2,3-14C]fumarate incorporation. However, the radioactive counts in cultures incubated with both 1 μCi of [2,3-14C]fumarate and cold 0.5 mm fumarate, malate, or aspartate dropped to nearly 30–40% of the counts obtained from cultures in which only [2,3-14C]fumarate was added (supplemental Fig. S2). The drop in radioactive counts could be because of the existence of a common pathway that metabolizes both the radioactive tracer ([2,3-14C]fumarate) and its corresponding unlabeled metabolite as well as further downstream metabolic intermediates, or alternately, the drop in counts could be because of toxicity of these metabolic intermediates to the growth of P. falciparum. To check this, [8-3H]hypoxanthine labeling of nucleic acids was done in the presence or absence of these intermediates (supplemental Fig. S2). No difference in the percent incorporation was observed across untreated and treated parasite cultures, ruling out the toxicity of these intermediates to P. falciparum growth and thus supporting the existence of a common pathway for the metabolism of fumarate, malate, and oxaloacetate.
FIGURE 5.
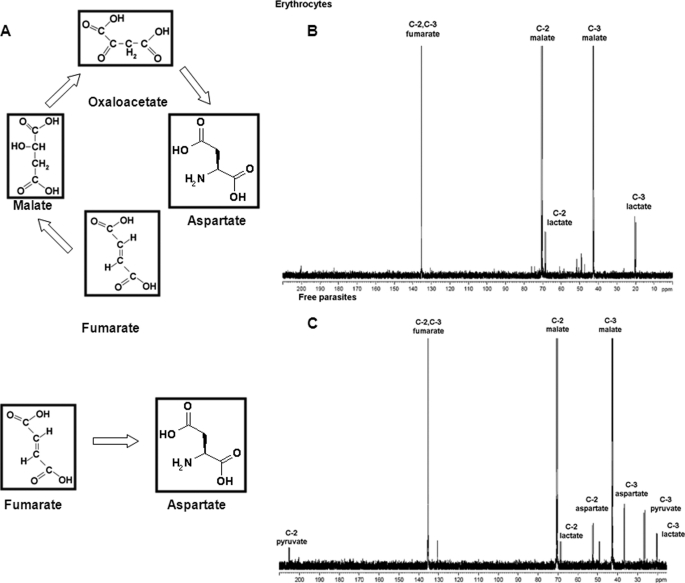
Metabolic intermediates in fumarate metabolism. A, pathways for fumarate metabolism. Fumarate can be converted to aspartate through the metabolic intermediates malate and oxaloacetate, or fumarate can be directly acted upon by aspartase to generate aspartate. B and C, RBCs (B) and PRBCs (C) incubated with 10 mm [2,3-13C]fumarate in 1× PBS at 37 °C for 4 h. Cell suspension was lysed and processed as described under “Experimental Procedures.” D2O was added to a final concentration of 10% as an internal lock to the cell lysate, and NMR spectra were recorded.
To further confirm the nature of the intermediary metabolites, free parasites were incubated with 10 mm [2,3-13C]fumarate for 4 h in 1× PBS and then processed for 13C NMR analysis. 13C proton-decoupled NMR of the parasites showed 13C enrichment in fumarate (chemical shift of C-2 and C-3 carbons in fumarate is at 136 ppm). The other dominant peaks were found at 70 and 42 ppm, which correspond, respectively, to the C-2 and C-3 of malate. In addition to these findings, 13C enrichment was also found in aspartate (C-2 δ, 52 ppm; C-3 δ, 36 ppm) and pyruvate (C-2 δ, 205 ppm; C-3 δ, 26 ppm) (Fig. 5C). Integration of the peak areas of fumarate, malate, aspartate pyruvate, and lactate in the 13C NMR spectrum shows the following relative enrichments: fumarate, 55.9 ± 16%; malate, 39.5 ± 17%; aspartate, 2.89 ± 0.62%; pyruvate, 2 ± 1.2%; and lactate, 1.2 ± 0.5%. On the other hand, RBCs incubated with [2,3-13C]fumarate showed enrichment in fumarate, malate, and lactate (Fig. 5B).
Free parasite culture incubated with [14C]fumarate was used to compare the radioactive counts metabolized and retained within the parasite with that secreted. The spent medium had 48 ± 7% of the input counts, whereas 28 ± 5% of the counts were associated with the cellular fraction. Upon separation of the spent medium on TLC, all the counts were found to be mainly in the lower spot corresponding to malate. It should be noted here that the spot corresponding to the mobility of malate might well include other polar metabolites such as aspartate, pyruvate, and lactate. Taken together with the NMR data, it would appear that this spot should largely contain malate.
Enzymes Involved in the Fumarate to Aspartate Conversion Pathway
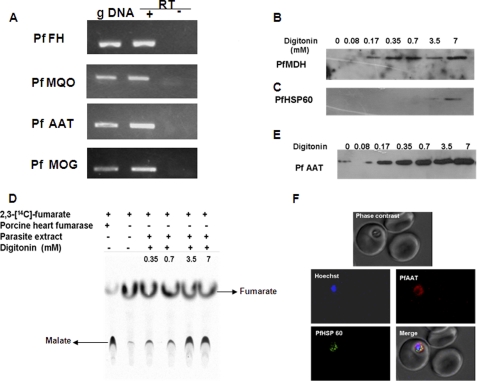
Enzymes catalyzing the conversion of fumarate to aspartate are fumarate hydratase (FH), MDH, or malate quinone oxidoreductase (MQO) and aspartate aminotransferase (AAT). The P. falciparum genome contains homologs of all of these enzymes. Previously, it was shown that malate dehydrogenase is cytosolic (21, 22), but the subcellular localization of other enzymes is unknown. RT-PCR analysis showed the expression of FH, MQO, and AAT (Fig. 6A).
FIGURE 6.
Expression and subcellular localization of enzymes involved in fumarate to aspartate conversion pathway. A, RT-PCR amplification of fragments of PfFH, PfMQO, PfAAT, and malate oxoglutarate (PfMOG) translocator. gDNA indicates reactions in which genomic DNA was used as a template for PCR. + and − RT indicate reactions with and without reverse transcriptase, respectively. −RT was included as a control to rule out genomic DNA contamination. B and C, Western blotting of the different subcellular fractions of P. falciparum parasites obtained with increasing concentrations of digitonin with anti-PfMDH and anti-PfHSP60 antisera. D, radio-TLC analysis for fumarate hydratase activity in different parasite cellular fractions obtained by digitonin treatment. Activity was assessed by the formation of [14C]malate from [2,3-14C]fumarate. The mobile phase consisted of a 4:1:1 mixture of n-butanol, acetic acid, and water. The silica-TLC plate was dried and developed using a Fuji FLA-5000 phosphorimaging device. E, Western blotting of subcellular fractions of P. falciparum by anti-PfAAT antisera. Low levels of PfAAT seen in the absence of digitonin are due to background lysis of cells. F, indirect immunofluorescence analysis of parasitized erythrocytes with anti-PfAAT and anti-PfHSP60 antisera. Anti-mouse and anti-rabbit IgG antibodies conjugated with Alexa Fluor dyes were used as secondary antibodies. Hoechst was used as the nuclear stain.
Fumarate hydratase is an enzyme that catalyzes the reversible conversion of fumarate to malate. Free parasites were treated with increasing concentrations of digitonin to prepare distinct cytosolic and organellar protein fractions. Digitonin is a non-ionic detergent used to solubilize cellular and organellar membranes. At lower concentrations of digitonin, cell membrane is permeabilized and cytosolic proteins are released, whereas at subsequent higher concentrations, the organellar and nuclear membranes are solubilized and organellar extracts are obtained. As PfMDH is cytosolic, it was used as a cytosolic marker in the experiments reported here. The digitonin concentration required to fully release cytoplasmic proteins from free parasites was obtained by using antisera against the cytoplasmic marker protein, malate dehydrogenase. On the other hand, antisera against P. falciparum heat shock protein 60 (PfHSP60) (a kind gift from Prof. G. Padmanabhan, Indian Institute of Science) were used as mitochondrial marker. From the Western blot analysis, these values were obtained as 0.17 and 3.5 mm, respectively (Fig. 6, B and C).
The digitonin extracts were probed for P. falciparum fumarate hydratase through its enzymatic activity for the formation of [14C]malate from [14C]fumarate. Porcine heart fumarase (Sigma) was used as a positive control. P. falciparum fumarate hydratase (PfFH) activity was found more in the cell extracts obtained by permeabilization with higher digitonin concentrations of 3.5 and 7.0 mm (Fig. 6D). This indicates that PfFH is not cytosolic but probably mitochondrial. In support of this, PfFH shows a very high probability (0.89) of being targeted to mitochondria as predicted by the MitoProt II 1.0a4 program (23).
Antisera raised against recombinant P. falciparum aspartate aminotransferase (PfAAT) specifically recognized a protein of 50 kDa, the expected molecular mass of the enzyme. Earlier studies on the cloning, expression, and substrate specificity of recombinant PfAAT revealed that the enzyme catalyzes the transamination of α-ketomethylthiobutyrate to generate methionine (24). These authors speculated that this enzyme has an important role in methionine recycling (24); however, the subcellular localization of this enzyme was not known. Our Western blotting analysis on digitonin extracts showed that PfAAT was present in the cytosolic extracts (Fig. 6E). This was further confirmed by confocal microscopy (Fig. 6F) where no colocalization of PfAAT (red) was found with the mitochondrial marker, PfHSP60 (green). The Km and kcat values for the conversion of α-ketoglutarate to glutamate by PfAAT are reported by Berger et al. (24). The Km for aspartate was 6 mm when α-ketoglutarate was used at a concentration of 10 mm. However, it was not known whether the enzyme could catalyze the reverse reaction of oxaloacetate to aspartate. Hence, recombinant purified PfAAT was characterized for its ability to catalyze the conversion of oxaloacetate to aspartate. The enzyme was found to be active in the direction of the formation of aspartate with a Km value for glutamate at 8 ± 1 mm and oxaloacetate at 5 ± 0.5 mm. Hence, the enzyme has a similar propensity to catalyze the reaction in both the forward and reverse reactions.
Atovaquone Inhibits the Conversion of Fumarate to Aspartate
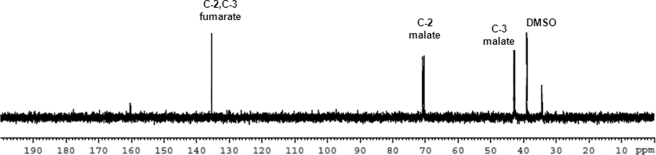
The conversion of malate to oxaloacetate can occur through malate dehydrogenase (MDH) or MQO. The latter generates reduced coenzyme Q (CoQH2), from coenzyme Q (CoQ) along with the oxidation of malate to oxaloacetate. CoQH2 is then oxidized directly by complex III, and the reducing equivalents are transferred to cytochrome c. Atovaquone is a specific and potent inhibitor of the enzymatic activity of P. falciparum complex III (25), and thus, treatment of P. falciparum parasites should lead to a depletion of CoQ. We examined the effect of this depletion of CoQ on the conversion of fumarate to aspartate. Although 13C enrichment in malate was unaffected in atovaquone treated parasites, which was expected as fumarate hydratase is unaltered by atovaquone, these parasites did not show 13C enrichment in aspartate (Fig. 7). This indicates that malate to oxaloacetate conversion is mediated chiefly through the putative P. falciparum malate quinone oxidoreductase and also establishes a metabolic link between the purine salvage pathway and the electron transport chain.
FIGURE 7.
Atovaquone treatment inhibits the 13C enrichment of aspartate in parasites incubated with [2,3-13C]fumarate. Free parasites treated with 1 μm atovaquone for 2 h were then pulsed with 10 mm [2,3-13C]fumarate for 2 h. Following incubation, the parasites were processed as described under “Experimental Procedures.” DMSO, dimethyl sulfoxide.
DISCUSSION
It has been shown that P. falciparum does not incorporate labeled glucose (1, 26) significantly into the tricarboxylic acid cycle intermediates. However, metabolite analysis of P. falciparum extracts in the intraerythrocytic stages by NMR (27) and ESI-MS/MS (28) indicate the presence of fumarate, and its intracellular concentration is estimated at about 0.2 mm (27). The most convincing evidence for the presence of an intracellular pool of fumarate comes from the study by Painter et al. (29). That study involves the generation of transgenic P. falciparum parasites carrying Saccharomyces cerevisiae dihydroorotate dehydrogenase (ScDHODH) along with the endogenous PfDHODH. Both of the enzymes oxidize dihydroorotate to orotate, but unlike PfDHODH, which transfers the reducing equivalents to ubiquinone (CoQ), ScDHODH uses fumarate as the electron acceptor. The other difference between the two enzymes is their subcellular localization. Whereas PfDHODH is localized to the mitochondrial inner membrane (30), ScDHODH-GFP fusion protein is localized to the cytoplasm (29). Although Painter et al. (29) have not speculated on the source of fumarate, these results imply that there is a sufficient intracellular pool of cytosolic fumarate to sustain the activity of ScDHODH in the transgenic parasites. The source of this cytosolic fumarate must be the purine salvage pathway, which generates a molecule of fumarate along with the synthesis of a molecule of AMP. In the absence of a functional tricarboxylic acid cycle during the intraerythrocytic stages of P. falciparum, the cytosolic fumarate generated from the purine salvage pathway as shown by radio-TLC is not secreted out and therefore must be metabolized in the cell.
Our results show that fumarate becomes incorporated into proteins and nucleic acids and also specifically labels pyrimidines and not purines. Previously, P. falciparum (31), Plasmodium knowlesi (32), and Plasmodium lophurae (33) have been shown to fix radiolabeled CO2 into amino acids glutamate, aspartate and alanine, and it was proposed to be mediated through the concerted enzymatic action of carboxykinases followed by transaminases. However, we ruled out such a pathway to explain our results because we used [2,3-14C]fumarate and not [1,4-14C]fumarate (in which the carboxylic acid group carbons are labeled). The latter, but not the former, can give rise to [14C]CO2.
Earlier, Fry and Beesley (34) observed that isolated mitochondria of Plasmodium yoelli and P. falciparum could reduce cytochrome c in the presence of NADH, α-glycerophosphate, and succinate, which are substrates for NADH dehydrogenase, α-glycerophosphate dehydrogenase, and succinate dehydrogenase, respectively. The authors (34) found that fumarate inhibits the reduction of cytochrome c, implicating the presence of a fumarate reductase activity in P. falciparum. On the other hand, Uyemura et al. (35) found that fumarate increases the mitochondrial membrane potential in isolated P. yoelli parasites. These two results are completely contradictory with respect to the direction of fumarate metabolism. Fry and Beesley's (34) results implicate a fumarate reductase activity in which fumarate is reduced to succinate with concomitant oxidation of reduced ubiquinone (CoQH2) to CoQ, whereas the results obtained by Uyemura et al. (35) imply that fumarate (by the action of fumarate hydratase) is converted to malate, which is a substrate for malate quinone oxidoreductase and hence would be oxidized to oxaloacetate. The oxidation of malate to oxaloacetate generates CoQH2, which then feeds the electron transport chain at complex III.
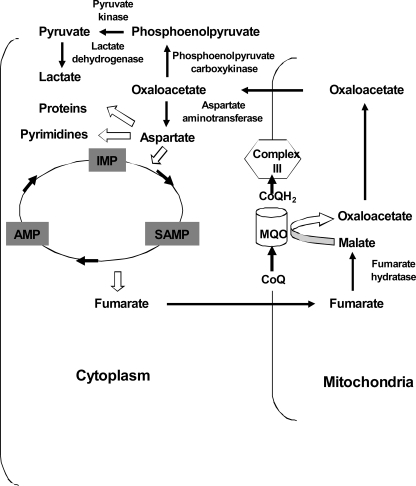
Our 13C NMR experiments showed peaks corresponding to malate, aspartate, and pyruvate. This excludes a direct route of conversion of fumarate to aspartate, which is supported by the absence of a homolog of aspartase in the P. falciparum genome (7). The observation of [13C]pyruvate can be explained by the action of phosphoenolpyruvate carboxykinase on oxaloacetate to generate phophoenol pyruvate, which then forms pyruvate by the action of pyruvate kinase (Fig. 8). Both phosphoenolpyruvate carboxykinase and pyruvate kinase have been shown to be expressed in the intraerythrocytic stages of P. falciparum (8). We did not see any 13C enrichment for succinate in the 13C NMR spectrum of free parasites incubated with [2,3-13C]fumarate, indicating the absence of a fumarate reductase activity and also suggesting that succinate dehydrogenase is not bidirectional under the conditions of this study. This is unlike in Trypanosoma cruzi, a pathogenic protozoan responsible for Chagas disease. T. cruzi intact cells upon incubation with [1-13C]glucose show 13C enrichment at the C-2 position of succinate (36). A functional fumarate reductase has been identified, with knockdown of this enzyme leading to the accumulation of fumarate in the spent medium of T. cruzi cells (37).
FIGURE 8.
Model for fumarate metabolism in P. falciparum. Cytosolic fumarate generated from the purine salvage pathway is metabolized to malate by mitochondrial fumarate hydratase, which then is converted to oxaloacetate by malate quinone oxidoreductase. In the latter step, CoQ is reduced to CoQH2, which is subsequently oxidized back to CoQ by complex III. Finally, oxaloacetate is converted to aspartate by a cytosolic aspartate aminotransferase.
PfFH, which catalyzes the first step of fumarate to malate conversion, seems to be localized in the mitochondria, given the increased activity of PfFH in mitochondrial extracts. However, as the purine salvage pathway in P. falciparum is cytosolic, the generated fumarate must enter the mitochondria. Several mitochondrial carrier proteins have been found to be present in P. falciparum that could mediate the reversible transport of metabolites into the mitochondria (38). One of them is the malate-oxoglutarate translocator, which transports malate into mitochondria in exchange for oxoglutarate (38). RT-PCR analysis shows that this translocator is expressed in the parasite (Fig. 6A). However, this transporter has not been characterized for its substrate specificity, and it is possible that this translocator might be involved in the transport of fumarate from the cytosol to the mitochondria.
P. falciparum obtains most of its amino acids from host hemoglobin degradation (39), and thus, the parasite may not completely rely on this metabolic pathway for its aspartate requirements. However, the conversion of fumarate to aspartate, as shown in Fig. 8, might serve a key role in replenishing the electron transport chain, thereby contributing to the maintenance of the mitochondrial membrane potential through the conversion of malate to oxaloacetate. The mitochondrial membrane potential has been speculated to play an important role in the transport of proteins, solutes and ions across the mitochondria. Additionally, the conversion of fumarate to aspartate might be a possible regulatory mechanism by which purine salvage pathway modulates the de novo pyrimidine biosynthetic pathway in P. falciparum. To what extent it does is yet to be confirmed. Further, metabolism of fumarate ensures a mechanism to prevent its feedback inhibition of the purine salvage pathway.
Our 13C NMR observations seem to indicate that the bulk of fumarate is converted to malate, with a small proportion further converted to aspartate. However, this relative enrichment of 13C represents a steady state condition. Aspartate feeds into synthesized proteins, and its carbon skeleton through pyrimidine nucleotides feeds into nucleic acids; with both of these processes being highly active in the trophozoite stage of the parasite, the steady state concentration of free 13C-enriched aspartate would be expected to be low. Also, it should be noted that apart from labeling proteins and nucleic acids through pyrimidines, [13C]aspartate is also utilized for AMP synthesis from IMP. This would regenerate [13C]fumarate, which would add to the pool of unutilized [13C]fumarate and reenter the pathway. Further, comparison of the relative levels of secreted malate with that metabolized and retained intracellularly using [14C]fumarate (500 μm) indicated a partitioning of 48 and 28%, respectively. A recent report by Olszewski et al. (40) highlights the fact that glutamine feeds the tricarboxylic acid cycle intermediates through 2-oxoglutarate in P. falciparum. The authors (40) have shown that malate is secreted out and also fumarate, but the latter to a much lesser extent. Indeed, our observations agree with the findings of Olszewski et al. (40). However, a moderate flux of malate to oxaloacetate does seem to be operating in the parasite, which might be of physiological relevance. In a recent report by Storm and Müller (41), it was found that phosphoenolpyruvate carboxylase (an enzyme that converts phosphoenolpyruvate to oxaloacetate) knockout parasites could be generated only in the presence of excess malate. Further, growth of these parasites could be recovered with malate or aspartate. These observations show that malate to oxaloacetate conversion seems to take place physiologically and that oxaloacetate is at a critical node in parasite metabolism. Conversion of malate to oxaloacetate cannot occur through PfMDH, as this enzyme has been shown to catalyze only the reduction reaction. We also examined different conditions that are known to activate MDH from other organisms to catalyze malate to oxaloacetate conversion, namely the presence of citrate and aspartate aminotransferase. But these failed to alter the direction of conversion by recombinant PfMDH (data not shown). MQO is a CoQ-dependent dehydrogenase, and the absence of 13C enrichment in aspartate in parasites incubated with atovaquone and [2,3-13C]fumarate indicates that this enzyme from the parasite catalyzes the oxidation of malate to oxaloacetate. Indeed, PfMQO exhibits high homology to MQO from Helicobacter pylori (42), which has been shown to convert malate to oxaloacetate and not the reverse. A genetic approach that obliterates fumarate production would throw more light on the importance of this pathway.
Overall, our findings suggest roles for the enzymes fumarate hydratase, malate quinone oxidoreductase, and aspartate aminotransferase. Fumarate hydratase in P. falciparum belongs to the type I class, which is unlike type II fumarate hydratase found in the human host (38). A counterpart of malate quinone oxidoreductase is absent in the human host. A recent study that appeared while this manuscript was under revision reports that PfAAT inhibition is lethal to the parasite (43). These enzymes could therefore be targeted for the development of antimalarial chemotherapeutics. Our findings also underscore the key role of mitochondrial dicarboxylate transporters in P. falciparum. Fumarate generated in the cytosol has to be translocated into the mitochondria, and our results suggest the need for transport of oxaloacetate from mitochondria to cytosol. Animal mitochondria under normal physiological conditions do not transport oxaloacetate significantly, and only a minor amount is transported through the dicarboxylate or oxoglutarate-malate translocator. On the other hand, plant mitochondria transport oxaloacetate at rapid rates across the inner mitochondrial membrane (44). A BLAST search highlighted several translocators with unknown functions in P. falciparum (38), which will require further characterization with regard to their cellular localization and specificity for different dicarboxylic acids.
Supplementary Material
Acknowledgments
We thank GlaxoSmithKline for atovaquone. We also thank Drs. Govindaraju, Mahesh, and Debu (New Chemistry Unit, Jawaharlal Nehru Centre for Advanced Scientific Research) for help with the 13C NMR experiments and Prof. G. Padmanabhan and Dr. Vathsala (Indian Institute of Science) for providing PfHSP60 antibodies.
This work was supported by the Department of Science and Technology, India.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Methods” and Figs. S1 and S2.
- Pf
- Plasmodium falciparum
- MDH
- malate dehydrogenase
- AAT
- aspartate aminotransferase
- FH
- fumarate hydratase
- CoQ
- coenzyme Q
- CoQH2
- reduced coenzyme Q
- Sc
- Saccharomyces cerevisiae
- DHODH
- dihydroorotate dehydrogenase
- MQO
- malate quinone oxidoreductase
- MDH
- malate dehydrogenase
- RBC
- red blood cell
- PRBC
- parasitized red blood cell.
REFERENCES
- 1. Bryant C., Voller A., Smith M. J. (1964) Am. J. Trop. Med. Hyg. 13, 515–519 [DOI] [PubMed] [Google Scholar]
- 2. Kirk K., Horner H. A., Kirk J. (1996) Mol. Biochem. Parasitol. 82, 195–205 [DOI] [PubMed] [Google Scholar]
- 3. Foth B. J., Stimmler L. M., Handman E., Crabb B. S., Hodder A. N., McFadden G. I. (2005) Mol. Microbiol. 55, 39–53 [DOI] [PubMed] [Google Scholar]
- 4. Chen J., Jones M. (1976) Arch. Biochem. Biophys. 176, 82–90 [DOI] [PubMed] [Google Scholar]
- 5. Wrenger C., Muller S. (2003) Eur. J. Biochem. 270, 1775–1783 [DOI] [PubMed] [Google Scholar]
- 6. Vaidya A. B., Mather M. W. (2009) Annu. Rev. Microbiol. 63, 249–267 [DOI] [PubMed] [Google Scholar]
- 7. Gardner M. J., Hall N., Fung E., White O., Berriman M., Hyman R. W., Carlton J. M., Pain A., Nelson K. E., Bowman S., Paulsen I. T., James K., Eisen J. A., Rutherford K., Salzberg S. L., Craig A., Kyes S., Chan M. S., Nene V., Shallom S. J., Suh B., Peterson J., Angiuoli S., Pertea M., Allen J., Selengut J., Haft D., Mather M. W., Vaidya A. B., Martin D. M., Fairlamb A. H., Fraunholz M. J., Roos D. S., Ralph S. A., McFadden G. I., Cummings L. M., Subramanian G. M., Mungall C., Venter J. C., Carucci D. J., Hoffman S. L., Newbold C., Davis R. W., Fraser C. M., Barrell B. (2002) Nature 419, 512–51912368865 [Google Scholar]
- 8. Bozdech Z., Llinás M., Pulliam B. L., Wong E. D., Zhu J., DeRisi J. L. (2003) PLoS Biol. 1, E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Downie M. J., Kirk K., Mamoun C. B. (2008) Eukaryot. Cell 7, 1231–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Raman J., Mehrotra S., Anand R. P., Balaram H. (2004) Mol. Biochem. Parasitol. 138, 1–8 [DOI] [PubMed] [Google Scholar]
- 11. Bulusu V., Srinivasan B., Bopanna M. P., Balaram H. (2009) Biochim. Biophys. Acta 1794, 642–654 [DOI] [PubMed] [Google Scholar]
- 12. Aragon J. J., Lowenstein J. M. (1980) Eur. J. Biochem. 110, 371–377 [DOI] [PubMed] [Google Scholar]
- 13. Aragón J. J., Tornheim K., Goodman M. N., Lowenstein J. M. (1981) Curr. Top. Cell. Regul. 18, 131–149 [DOI] [PubMed] [Google Scholar]
- 14. Trager W., Jensen J. B. (1976) Science 193, 673–675 [DOI] [PubMed] [Google Scholar]
- 15. Rivadeneira E. M., Wasserman M., Espinal C. T. (1983) J. Protozool. 30, 367–370 [DOI] [PubMed] [Google Scholar]
- 16. Cassera M. B., Hazleton K. Z., Riegelhaupt P. M., Merino E. F., Luo M., Akabas M. H., Schramm V. L. (2008) J. Biol. Chem. 283, 32889–32899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Winter H. C., Dekker E. E. (1989) Plant Physiol. 89, 1122–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hodges M., Yikilmaz E., Patterson G., Kasvosve I., Rouault T. A., Gordeuk V. R., Loyevsky M. (2005) Mol. Biochem. Parasitol. 143, 29–38 [DOI] [PubMed] [Google Scholar]
- 19. Tonkin C. J., van Dooren G. G., Spurck T. P., Struck N. S., Good R. T., Handman E., Cowman A. F., McFadden G. I. (2004) Mol. Biochem. Parasitol. 137, 13–21 [DOI] [PubMed] [Google Scholar]
- 20. Kanehisa M., Goto S. (2000) Nucleic Acids Res. 28, 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lang-Unnasch N. (1992) Mol. Biochem. Parasitol. 50, 17–25 [DOI] [PubMed] [Google Scholar]
- 22. Pradhan A., Mukherjee P., Tripathi A. K., Avery M. A., Walker L. A., Tekwani B. L. (2009) Mol. Cell. Biochem. 325, 141–8 [DOI] [PubMed] [Google Scholar]
- 23. Claros M. G., Vincens P. (1996) Eur. J. Biochem. 241, 779–786 [DOI] [PubMed] [Google Scholar]
- 24. Berger L. C., Wilson J., Wood P., Berger B. J. (2001) J. Bacteriol. 183, 4421–4434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mather M. W., Darrouzet E., Valkova-Valchanova M., Cooley J. W., McIntosh M. T., Daldal F., Vaidya A. B. (2005) J. Biol. Chem. 280, 27458–27465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lian L. Y., Al-Helal M., Roslaini A. M., Fisher N., Bray P. G., Ward S. A., Biagini G. A. (2009) Malar. J. 8, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Teng R., Junankar P. R., Bubb W. A., Rae C., Mercier P., Kirk K. (2009) NMR Biomed. 22, 292–302 [DOI] [PubMed] [Google Scholar]
- 28. Olszewski K. L., Morrisey J. M., Wilinski D., Burns J. M., Vaidya A. B., Rabinowitz J. D., Llinás M. (2009) Cell Host Microbe 5, 191–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Painter H. J., Morrisey J. M., Mather M. W., Vaidya A. B. (2007) Nature 446, 88–91 [DOI] [PubMed] [Google Scholar]
- 30. Krungkrai J. (1995) Biochim. Biophys. Acta 1243, 351–360 [DOI] [PubMed] [Google Scholar]
- 31. Blum J. J., Ginsberg H. (1984) J. Protozool. 31, 167–169 [DOI] [PubMed] [Google Scholar]
- 32. Sherman I. W., Ting I. P. (1968) Comp. Biochem. Physiol. 24, 639–642 [DOI] [PubMed] [Google Scholar]
- 33. Sherman I. W., Ting I. P. (1966) Nature 212, 1387–1388 [DOI] [PubMed] [Google Scholar]
- 34. Fry M., Beesley J. E. (1991) Parasitology 102, 17–26 [DOI] [PubMed] [Google Scholar]
- 35. Uyemura S. A., Luo S., Vieira M., Moreno S. N., Docampo R. (2004) J. Biol. Chem. 279, 385–393 [DOI] [PubMed] [Google Scholar]
- 36. Frydman B., de los Santos C., Cannata J. J., Cazzulo J. J. (1990) Eur. J. Biochem. 192, 363–368 [DOI] [PubMed] [Google Scholar]
- 37. Coustou V., Biran M., Besteiro S., Rivière L., Baltz T., Franconi J. M., Bringaud F. (2006) J. Biol. Chem. 281, 26832–26846 [DOI] [PubMed] [Google Scholar]
- 38. van Dooren G. G., Stimmler L. M., McFadden G. I. (2006) FEMS Microbiol. Rev. 30, 596–630 [DOI] [PubMed] [Google Scholar]
- 39. Sherman I. W. (1979) Microbiol. Rev. 43, 453–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Olszewski K. L., Mather M. W., Morrisey J. M., Garcia B. A., Vaidya A. B., Rabinowitz J. D., Llinás M. (2010) Nature 466, 774–778 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41. Storm J., Müller S. (2010) Malar. J. 9, Suppl. 2, P49 [Google Scholar]
- 42. Kather B., Stingl K., van der Rest M. E., Altendorf K., Molenaar D. (2000) J. Bacteriol. 182, 3204–3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wrenger C., Müller I. B., Schifferdecker A. J., Jain R., Jordanova R., Groves M. R. (2011) J. Mol. Biol. 405, 956–971 [DOI] [PubMed] [Google Scholar]
- 44. Laloi M. (1999) Cell. Mol. Life Sci. 56, 918–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.