Abstract
The structure of the 28 kDa complex of the first two RNA binding domains (RBDs) of nucleolin (RBD12) with an RNA stem–loop that includes the nucleolin recognition element UCCCGA in the loop was determined by NMR spectroscopy. The structure of nucleolin RBD12 with the nucleolin recognition element (NRE) reveals that the two RBDs bind on opposite sides of the RNA loop, forming a molecular clamp that brings the 5′ and 3′ ends of the recognition sequence close together and stabilizing the stem–loop. The specific interactions observed in the structure explain the sequence specificity for the NRE sequence. Binding studies of mutant proteins and analysis of conserved residues support the proposed interactions. The mode of interaction of the protein with the RNA and the location of the putative NRE sites suggest that nucleolin may function as an RNA chaperone to prevent improper folding of the nascent pre-rRNA.
Keywords: NMR/nucleolus/RBD/RNA binding protein/RNP
Introduction
Nucleolin, the most abundant nucleolar protein, is involved in several steps of ribosome biogenesis (Olson, 1990; Ginisty et al., 1999; Srivastava and Pollard, 1999). The specific and transient interaction of this multidomain protein with nascent pre-rRNA and ribosomal proteins is thought to be important for the proper folding of pre-rRNA and its packaging into pre-ribosomal particles (Ginisty et al., 1999). Nucleolin is highly conserved in vertebrates (Figure 1A), and structurally related proteins with the same multidomain organization are found in yeasts and plants (Ginisty et al., 1999; Srivastava and Pollard, 1999). The N-terminal acidic and basic region and the C-terminal domain rich in RGG repeats mediate protein–protein interactions with histone H1, U3 snoRNP and ribosomal proteins (Erard et al., 1988; Bouvet et al., 1998; Ginisty et al., 1998). The central region contains four RNA binding domains (RBD/RNP/RRM type) (Birney et al., 1993), which interact with pre-rRNA (Herrera and Olson, 1986; Ghisolfi-Nieto et al., 1996). The interaction of nucleolin with pre-rRNA is required for the first RNA processing step, which occurs in the 5′ ETS (Ginisty et al., 1998).
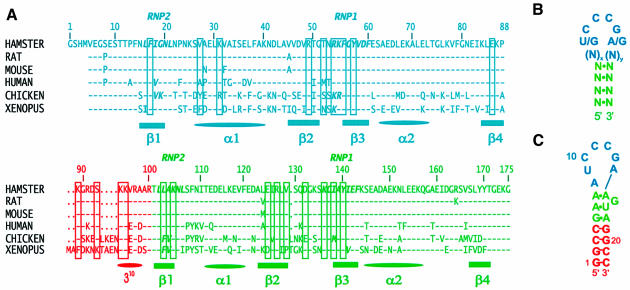
Fig. 1. (A) Sequence and numbering of the hamster nucleolin RBD12 used in this study and sequence alignment [BLAST (Altschul et al., 1997)] with the nucleolin RBD12 of rat, mouse, human, chicken and Xenopus. Residues from RBD1, RBD2 and the linker are shown in cyan, green and red, respectively. The secondary structure elements are indicated below the sequences. In the sequence alignment, a dash indicates that the residue is the same as in the hamster nucleolin and a dot indicates where there is a gap relative to Xenopus, which is six residues longer than hamster nucleolin. The residues involved in either protein–RNA interactions and/or interdomain interactions in the hamster nucleolin RBD12–sNRE complex are boxed. The conserved octapeptide RNP1 and hexapeptide RNP2 characteristic of the RBD/RNP/RRM motif (Birney et al., 1993) are indicated. (B) Schematic of the consensus NRE sequence and secondary structure. (C) Sequence and secondary structure of the sNRE used in these studies. Nucleotides 3–20 are the sequence identified by in vitro selection (Ghisolfi-Nieto et al., 1996).
In vitro and in vivo analysis identified stem–loop binding sites in the mouse and human pre-rRNA with a consensus (U/G)CCCG(A/G) in the loop: the nucleolin recognition element (NRE) (Ghisolfi-Nieto et al., 1996; Serin et al., 1996). The NRE has a minimum of 4 bp in the stem and a loop size of 7–14 nucleotides (Figure 1B). Putative NRE sites are found throughout the pre-rRNA (Serin et al., 1996). Thirty-three sites have been identified in human pre-rRNA, including 11 in the 5′ ETS (Serin et al., 1996). Truncation of the different RBDs of nucleolin showed that the RNA binding specificity for the NRE comes exclusively from the two most N-terminal RBDs (RBD12), which bind to the RNA stem–loops with the same 10–100 nM affinity as the full-length protein (Serin et al., 1997). This is one of only two proteins identified to date that binds an RNA stem–loop with two RBDs (Shi et al., 1997). Biochemical and genetic analysis of the interaction of nucleolin RBD12 with the NRE led to the proposal that each domain uses a different surface for the interaction with the RNA (Bouvet et al., 1997).
The RBD is one of the most common nucleic acid binding protein motifs. It has been found in hundreds of proteins, as a single domain or multiple domains (Birney et al., 1993). The RBD has a βαββαβ fold originally found for the RBD1 of U1A (Nagai et al., 1990). Proteins containing RBDs play a major role in the post-transcriptional regulation of gene expression (Burd and Dreyfuss, 1994; Varani and Nagai, 1998). They are involved in numerous pre-rRNA and mRNA processing events, including splicing, 3′-end processing, stability and transport. Most RBD-containing proteins also contain other domains that can mediate protein–protein interactions, suggesting that these proteins play central roles in the assembly of macromolecular RNP complexes (Varani and Nagai, 1998). Examples include the spliceosomal protein U1A, the family of SR proteins, as well as nucleolin. In spite of their biological importance, little is known about how the RBDs bind specifically to their RNA targets.
Only four RBD protein structures have been determined in complex with RNA, i.e. U1A (Oubridge et al., 1994; Allain et al., 1996), U2B″ (Price et al., 1998), sex-lethal (Handa et al., 1999) and poly(A) binding protein (PABP) (Deo et al., 1999), and one in complex with DNA, hnRNPA1 (Ding et al., 1999). U1A and U2B″ bind a stem–loop RNA using only one RBD. Sex-lethal and PABP both bind single-stranded RNA (UGU8 and A8, respectively) using two RBDs separated by a short 10–12 amino acid linker. hnRNPA1 uses two RBDs separated by a 17 amino acid linker and binds single-stranded DNA or RNA. However, in the complex, hnRNPA1 forms a dimer that binds two DNA molecules, each DNA strand binding RBD1 of one molecule and RBD2 of the other.
Here we present the first molecular insight into how a major nucleolar protein interacts with the pre-rRNA. The structure of the 28 kDa protein–RNA complex between nucleolin RBD12 and a 22 nucleotide RNA stem–loop that includes the 18 nucleotide sequence identified by in vitro selection (sNRE) (Figure 1C) was determined using multidimensional NMR spectroscopy. The RNA loop, which is largely unstructured in the free RNA, becomes ordered upon binding to the protein. The interaction of nucleolin RBD12 with the NRE stem–loop is unique among protein-complex structures determined to date. The two RBDs bind on opposite sides of the RNA loop, forming a molecular clamp which brings the 5′ and 3′ ends of the recognition sequence close together and stabilizing the stem–loop. The specific interactions observed in the structure explain the sequence specificity for the NRE sequence. Binding studies of mutant proteins and analysis of conserved residues provide additional support for the importance of the observed interactions with the RNA. The mode of interaction of the protein with the RNA and the location of the putative NRE sites suggest that nucleolin may function as an rRNA chaperone to prevent improper folding of the nascent pre-rRNA (Herschlag, 1995). This proposed chaperone activity of nucleolin may be analogous to the role of the hnRNP proteins in pre-mRNP assembly and pre-mRNA processing.
Results and discussion
Structure determination and precision of the complex
Multidimensional NMR spectroscopy was used to determine the structure of the 28 kDa protein–RNA complex between nucleolin RBD12 and a 22 nucleotide RNA stem–loop. The sequences of the RNA and of hamster nucleolin RBD12 are shown in Figure 1. The sample preparation, NMR experiments, and spectral assignments of the protein and RNA components of the complex are described in Materials and methods. A total of 3246 structural constraints, including 3010 nuclear Overhauser effect (NOE) derived, were used to determine the structure of the complex (Table I). The most important among this large number of constraints are the 150 intermolecular NOE-derived constraints. Most of these were obtained from 2D and 3D 12C-13C filtered NOESY experiments (Otting and Wüthrich, 1990; Lee et al., 1994; Slijper et al., 1996; Zwahlen et al., 1997) where intermolecular and intramolecular NOEs could be observed separately. Intermolecular NOEs are observed for 9 nucleotides and 22 amino acid residues.
Table I. NMR and structure determination statistics for RBD12–sNRE complex.
| Input constraints | Protein | RNA | Total |
|---|---|---|---|
| No. of NOE derived distances | |||
| intramolecular | 2593 | 267 | 2860 |
| intra-residue | 451 | 110 | |
| sequential | 820 | 108 | |
| medium range (i + 2 to i + 4) | 491 | ||
| long range (>i + 4) | 831 | ||
| medium and long range | 49 | ||
| intermolecular | 150 | ||
| intra- and intermolecular | 3010 | ||
| Repulsive distance restraints | 7 | ||
| Hydrogen bond constraints (two per hydrogen bond) | 152 | 38 | 190 |
| intermolecular | 8 | ||
| Dihedral angle constraints | 22 | ||
| Base planarity constraints | 9 | ||
| Total distance, dihedral angle and base planarity constraints |
|
|
3246 |
| Final structure statistics | |||
| No. of refined structures | 19 | ||
| Average of XPLOR | |||
| total energy (kcal/mol) | 1131.9 ± 57.0 | ||
| NOE energy (kcal/mol) | 225.1 ± 20.3 | ||
| No. of violations >0.2 Å | 8–18 | ||
| R.m.s.d. to the average structure (Å) | Backbone | Heavy | |
| RNA (1–22) and protein (11–170) | 1.63 ± 0.47 | 1.84 ± 0.44 | |
| RNA only (1–22) | 1.29 ± 0.32 | 1.23 ± 0.26 | |
| RNA interface (5–18) | 0.99 ± 0.20 | 1.03 ± 0.18 | |
| protein only (11–170) | 1.43 ± 0.48 | 1.78 ± 0.45 | |
| RBD1 only (11–88) | 0.47 ± 0.10 | 0.90 ± 0.08 | |
| linker only (89–100) | 0.96 ± 0.35 | 1.85 ± 0.52 | |
| RBD2 only (101–170) | 0.64 ± 0.31 | 1.16 ± 0.27 | |
| Procheck analysis | |||
| residues in most favored regions | 64% | ||
| residues in additional allowed regions | 28% | ||
| residues in generously allowed regions | 6% | ||
| residues in disallowed regions | 2% | ||
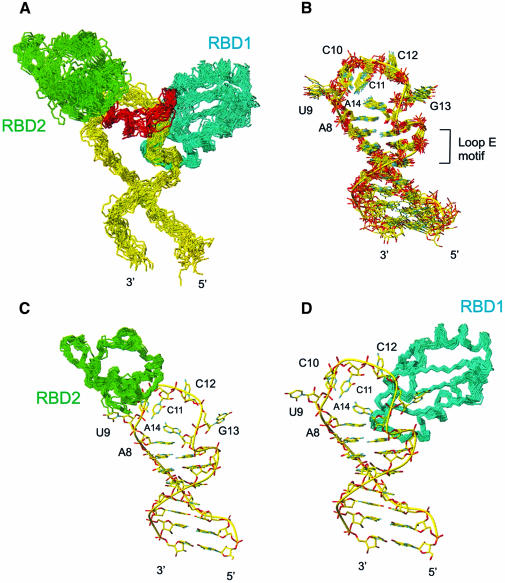
The large number of structural constraints, with an average of 17 NOE-derived constraints per amino acid and 19 per nucleotide, allowed the calculation of a precise structure of the protein–RNA complex (Figure 2; Table I). From 40 calculated structures, the 19 lowest energy structures form the ensemble of converged structures, superimposed in Figure 2, which were analyzed further. The r.m.s.d. to the mean for the backbone atoms of the whole complex (RNA and protein) is 1.63 Å (Figure 2A). Local superpositions of the RNA interface (G5–A18; Figure 2B), the protein backbone of RBD1 (Figure 2C) and the protein backbone of RBD2 (Figure 2D) have r.m.s.ds of 0.99, 0.47 and 0.64 Å, respectively. The precision of the structure is comparable to or better than that obtained on the previously determined protein–RNA complexes in solution (Allain et al., 1996, 1997; De Guzman et al., 1998; Mao et al., 1999; Stoldt et al., 1999; Ramos et al., 2000; Varani et al., 2000).
Fig. 2. Superpositions of the ensemble of the 19 lowest energy structures of the nucleolin RBD12–sNRE complex. (A) The complete nucleolin RBD12–RNA complex showing main chain atoms of RBD12 and backbone atoms of RNA, with backbone atoms of the protein and heavy atoms of the RNA superimposed. (B) The RNA alone, with the heavy atoms of the RNA interface (G5–A18) superimposed. (C) RBD2 structures superimposed on the backbone atoms, shown bound to the lowest energy structure of the RNA. (D) RBD1 structures superimposed on the backbone atoms, shown bound to the lowest energy structure of the RNA. The RNA, RBD1, RBD2 and the linker are shown in yellow, cyan, green and red, respectively.
The two RBDs have the same tertiary structure in the complex and free in solution
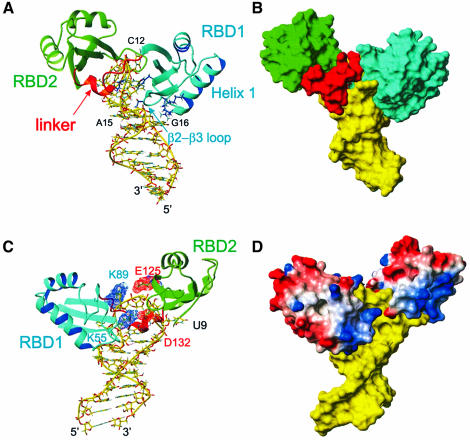
In the complex, RBD1 and RBD2 adopt the expected βαββαβ RBD fold (Nagai et al., 1990) (Figures 2 and 3) and essentially the same tertiary structure that they have in the free protein (Allain et al., 2000). The major difference between the bound and free structures of the individual RBDs is in the short β2–β3 loop of RBD1 (T52–K55), which is flexible in the structure of the free protein and becomes ordered in the complex because of its interaction with the RNA (Figure 3A). In the free protein, the two RBDs are connected by a flexible linker (Allain et al., 2000), which becomes completely ordered in the complex, as evident from 1H-15N NOE experiments (Kay et al., 1989). The structure of the complex shows that RBD1 binds primarily on the 3′ side of the RNA loop and the top of the stem (Figure 2D), RBD2 on the 5′ side of the loop (Figure 2C), and the linker in between, and that all three domains make extensive contacts with the RNA. The two RBDs and the linker showed no interdomain interactions in the free protein, but do interact in the complex (Figure 3), as discussed below.
Fig. 3. Overall description of the complex. The lowest energy structure is shown. (A) Stick (RNA) and ribbon (protein) representation of the complex showing how the RNA loop is ‘sandwiched’ between the two RBDs. RBD1 is located in the major groove side of the RNA and contacts C12, G13 and the loop E motif. RBD2 is located on the minor groove side and contacts U9 and C10. The linker is mostly located in the minor groove side on the RNA. The amino acid side chains from RBD1 V27, K31 (α-helix 1) and T52, R54 (β2–β3 loop), which contact the stem, as well as the inserting residues F56 and K94, are shown in blue. (B) Surface representation of the RNA and protein complex. The view is the same as in (A). (C) View of the complex showing that the two RBDs interact via two salt bridges (K89–E125 and K55–D132). Asp and Glu are shown in red and Lys and Arg in blue. The major groove face of the binding site is shown. (D) GRASP (Nicholls et al., 1991) representation of the complex with positively charged residues in blue and negatively charged residues in red. The color scheme is the same as Figure 2, except for the GRASP representation.
The RNA structure has a loop E motif and an ordered loop
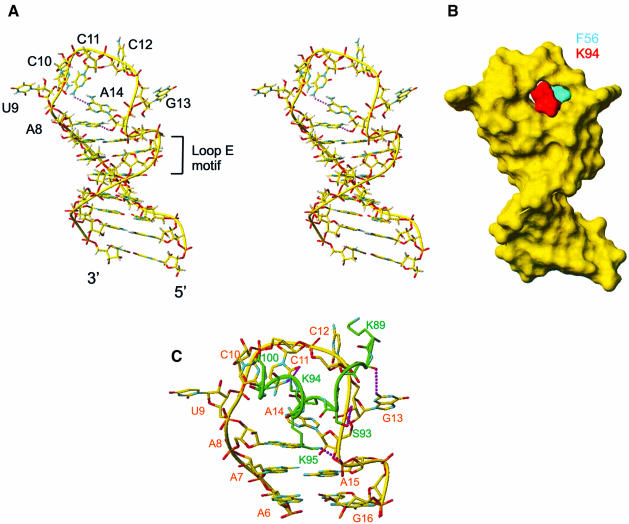
The RNA in the complex contains a structured seven nucleotide loop (A8–A14) and a stem with four Watson–Crick base pairs at the bottom and a loop E motif (Wimberly et al., 1993) (G5–A7, A15–A18) at the top (Figure 2). The loop is largely unstructured in the free RNA (P.Bouvet, F.H.-T.Allain, T.Dieckman, L.D.Finger and J.Feigon, unpublished results). In the RNA loop in the complex, A8 stacks on A7, and A14 stacks on A8. In addition to their stacking interaction, A8 and A14 are stabilized by a hydrogen bond between A8 N1 and A14 2′OH. The bases of C10 and C11 are stacked on each other inside the loop. C11 points down toward A14 in a position to form a hydrogen bond between A14 N1 and C11 4-amino group. The bases of U9, C12, and G13, which is in the syn conformation, stick out of the loop and are only stabilized by protein–RNA interactions. The 4 bp at the bottom of the stem, which do not participate in protein binding, form a regular A-form helix. At the top of the stem, the loop E motif, a commonly occurring motif (Leontis and Westhof, 1998) first structurally identified in loop E of eukaryotic 5S rRNA (Wimberly et al., 1993) and the sarcin–ricin loop of 28S rRNA (Szewczak et al., 1993), has the same structure as in the free sNRE RNA (P.Bouvet, F.H.-T.Allain, T.Dieckmann, L.D.Finger and J.Feigon, unpublished): a sheared G5–A18 base pair, an A6–U17– G16 base triple and a symmetric (trans-Hoogsteen) locally parallel A7–A15 base pair (Figure 4). The loop E motif is easily recognizable by the ‘S’ shape of the RNA backbone from A15 to U17.
Fig. 4. (A) Stereoview of the lowest energy structure of the RNA in the complex. A stick representation is shown. (B) Surface representation of the structure of the RNA with the two amino acids that insert into the loop shown in cyan (F56 from RBD1) and red (K94 from the linker). (C) Minor groove view of the RNA nucleotides that interact with the protein (A6–G16) and protein–RNA interactions from the linker (K89–R100). Only the heavy atoms are shown. Possible hydrogen bonds are shown in purple.
The RNA loop is sandwiched between RBD1 and RBD2 and the linker
In the complex, the two RBDs and the linker interact extensively with the RNA, RBD1 with six nucleotides, RBD2 with two nucleotides and the linker with five nucleotides. The RNA loop (A8–A14) is ‘sandwiched’ between the β-sheet surface of RBD1 on one side and the β-sheet surface of RBD2 and the linker on the other side (Figure 3A and B). RBD1 interacts with the ‘major groove side’ of the RNA and contacts A8, the 3′ end of the loop (C12, G13, A14), and A15, G16 at the top of the stem. The β2–β3 loop (T52, R54) and the C-terminus of α-helix 1 (V27, K31) contact the major groove of the loop E motif (G16, A15), A8 and A14 (Figure 3A). The β-sheet of RBD1 interacts with C12 and G13. RBD2 binds on the ‘minor groove side’ of the RNA and in contrast to RBD1 contacts only two nucleotides: U9 and C10 at the 5′ side of the loop. The protein–RNA interaction takes place mostly on the surface of its β-sheet. U9 is in contact with five residues from β1, β2, β3 and the β2–β3 loop (Figure 5A). C10 is sandwiched between C11 and Y140 (β3), and also interacts with R127 (β2) (Figure 5B). The 12 amino acid linker (K89–R100) makes numerous contacts to the RNA (Figure 4C). The aliphatic side chain of K89 interacts with the H5 and H6 of C12 (Figure 5C), the backbone of G90 and R91 have potential contacts with G13, amino acids S93 and K95 interact with the RNA backbone (Figure 4C), and K94 inserts in the RNA loop (Figure 4B).
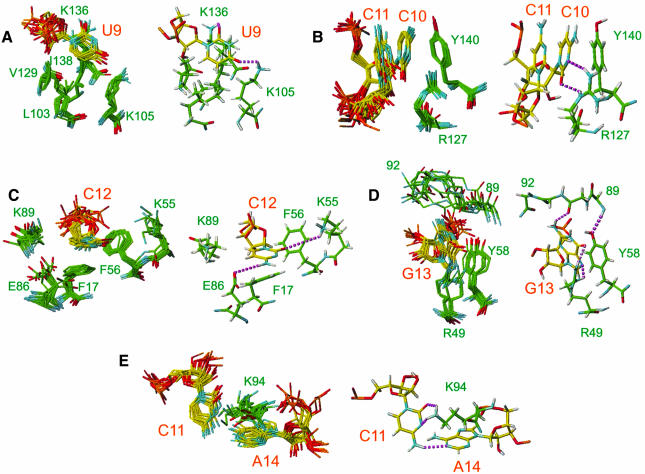
Fig. 5. Details of the protein–RNA interactions showing how binding specificity to the sequence 5′(UCCCGA)3′ is achieved. Recognition of (A) U9, (B) C10, (C) C12, (D) G13 and (E) C11 and A14 is illustrated. On the left side of each panel, the 10 lowest energy structures are superimposed. The heavy atoms of the relevant protein side chains and RNA bases are displayed. On the right side of each panel, a representative structure is shown. The protons are displayed in gray and proposed hydrogen bonds are shown by dashed lines in purple.
K94 and F56 insert into the RNA loop from opposite sides
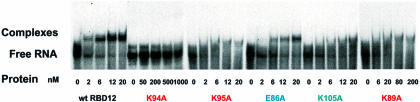
One of the key structural features of this protein–RNA complex is the insertion of the protein side chains of K94 and F56 into the RNA loop (Figures 3A and 4B) from opposite sides. K94 is in the linker region and F56 is at the N-terminus of the β3 strand of RBD1. F56 is stacked under the sugar ring of C12 (Figure 5C), which explains the unusual upfield chemical shift found for C12 ribose spin system (3.80 p.p.m. for H2′, 3.60 p.p.m. for H4′, and 3.09 and 2.41 p.p.m. for H5′, H5″). K94 is less well defined than F56 (Figure 5E). Nevertheless, in most of the structures the aliphatic part of K94 stacks on A14 and its amino group can potentially make two hydrogen bonds with C11 N3 and O2 (Figure 5E). The aliphatic protons of K94 are unusually upfield shifted (1.40 and 1.04 p.p.m. for Hβs, –0.16 p.p.m. for Hγ, 2.05 p.p.m. for Hε), consistent with stacking of this side chain on A14. The critical importance of this unusual interaction is confirmed by a gel shift assay, which showed that a K94A mutation abolishes detectable binding to the sNRE (Figure 6).
Fig. 6. Gel mobility shift assays on wild-type and mutated nucleolin RBD12 K89A, E86A, K95A, K105A and K94A proteins. The figure shows the results of several different gels, but controls with the wild-type protein were run for each experiment. There are no protein bands in the wells, which are not visible on the autoradiogram.
RBD1 and RBD2 interact via two salt bridges
The sandwiching of the RNA loop between the two RBDs brings these domains and the linker into contact with one another (Figure 3). Two salt bridges are formed on opposite sides of the loop, between K55 and D132 and between K89 and E125 (Figure 3C and D). There is no direct spectroscopic evidence for the formation of these salt bridges, but they are found in most of the converged structures. These two pairs of interacting residues are conserved or replaced by residues that can form an equivalent salt bridge in all of the nucleolins (Figure 1A). These domain interactions, along with the linker–RBD1 interaction discussed above, help clamp the RNA loop between the two RBDs.
The structure effectively explains the requirement for both RBDs and the linker to achieve RNA binding specificity (Serin et al., 1997) since all three parts of the protein interact with the RNA and with each other.
Sequence-specific recognition of the NRE sequence UCCCGA
The protein–RNA and RNA–RNA interactions found in the nucleolin RBD12–sNRE complex reveal the molecular basis for the sequence specificity for the NRE loop consensus (U/G)CCCG(A/G) (Ghisolfi-Nieto et al., 1996; Bouvet et al., 1997). Each nucleotide is recognized by specific hydrogen bond and stacking interactions (Figure 5). The first nucleotide in the consensus sequence, U9, is recognized by K105 and K136. In the consensus sequence, G is tolerated as well, and modeling of a G at this position shows that a G would also be recognized via intermolecular hydrogen bonds to the G N3 and O6 instead of the U O2 and O4, respectively. C10 is stacked between C11 and Y140. The base functional groups of C10 O2 and N3 are hydrogen bonded to R127. This protein–RNA interaction confirms a genetic screen that predicted the interaction between R127 and C10 (Bouvet et al., 1997). The C12 base is stacked on F17 and its sugar ring is stacked on F56. The H5–H6 edge of C12 is in van der Waals contact with the aliphatic side chain of K89, and its 4-amino and O2 are in contact with E86 and K55, respectively. Note that K55 and K89 contribute to both a protein–RNA interaction and an interdomain interaction (Figure 3). G13 is in the syn conformation, stacks on Y58, and is recognized via three intermolecular hydrogen bonds, two with R49 and one with the main chain carbonyl of R91. The last two nucleotides of the consensus sequence, C11 and A14, interact with each other via a hydrogen bond between C11 4-amino and A14 N1 and with K94. A G would be tolerated in place of A14, with C11 4-amino hydrogen bonding with G O6 instead of A N1.
All of the protein residues that interact with the RNA consensus nucleotides are either conserved or replaced by an amino acid that would conserve the interaction in human, mouse, rat and chicken nucleolin (Figure 1A), consistent with the fact that all these proteins specifically recognize the NRE sequence. The nucleolin of Xenopus does not bind the NRE sequence (P.Bouvet, unpublished) and this can partially be explained by the presence of a phenylalanine at the critical K89 position. Mutation to alanine of some of the residues that interact with the RNA leads to weaker (E86A, K89A, K105A) or total loss of binding (K94A) in gel shift assays (Figure 6), further confirming their role in NRE binding. Replacement of any nucleotide in the (U/G)CCCG(A/G) consensus sequence would lead to steric clash and/or the loss of one or more hydrogen bonds in the structure.
The specific protein contacts to the top of the loop E motif will be discussed in detail elsewhere (manuscript in preparation), since they are likely to be specific to this stem sequence with hamster nucleolin. There are a series of hydrogen bond and van der Waals contacts from six amino acid side chains [T52, R54, V27, K31 (Figure 3A), S93 and K95 (Figure 4C)] to A8, A14, A15 and G16, which primarily recognize the S-shape of the loop E motif backbone. While the specific recognition of the loop consensus should be universal, the NRE does not have a sequence requirement in the stem and those specific interactions may be a result of the in vitro selection. Consistent with this, V27 and T52 are not conserved among the different nucleolins. The specific contacts to A8 and the top of the loop E motif probably account for the higher binding affinity of nucleolin for the sNRE (2–5 nM) compared with other RNA targets with different stem sequences (10–100 nM) (Ghisolfi-Nieto et al., 1996).
Comparison between nucleolin RBD12 and RBD1 of spliceosomal U1A and U2B″
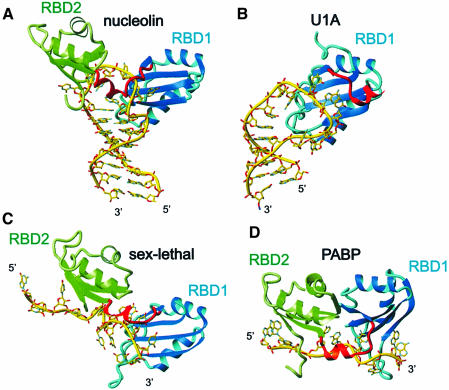
Nucleolin and the spliceosomal proteins U1A (Oubridge et al., 1994) and U2B″ (Price et al., 1998) are the only RBD proteins that specifically bind RNA stem–loops for which protein–RNA complex structures have been determined. It is interesting to compare the mode of recognition by one RBD versus two. The RBD1 of U1A and U2B″ contacts a much larger loop from the major groove side only. In contrast, nucleolin RBD12 binds on both the minor and major groove sides of the RNA loop (Figure 3). The structure of RBD1 and its position on the major groove side of the RNA is very similar in all three complexes. In contrast, the linker position (in U1A and U2B″ the 14 residues C-terminal to the RBD) is very different (Figure 7A and B). The linker of nucleolin RBD12 is positioned primarily in the minor groove of the RNA loop, whereas in U1A and U2B″ the linker is on the major groove side of the RNA loop. In all of these complexes, sequence-specific recognition involves insertion of amino acid side chains into the loop, but for U1A and U2B″ seven amino acids from the β2–β3 loop insert, while in nucleolin only two residues insert, F56 of RBD1 β-sheet and K94 from the linker (Figure 2D).
Fig. 7. Comparison between nucleolin RBD12–sNRE complex and the other RBD–RNA complexes. (A) Nucleolin RBD12–sNRE complex. (B) U1A RBD1 bound to U1 snRNA stem–loop II (Oubridge et al., 1994). (C) Sex-lethal RBD12–UGU8 complex (Handa et al., 1999). (D) PABP RBD12–A8 complex (Deo et al., 1999). Note that the location of the amino acids on the surface of the β-sheet varies among the different RBDs. In all panels, RBD1 is shown as a ribbon in cyan and RBD2 is in green. The RNA is in yellow, represented as sticks.
All three complexes also have contacts to the top of the stem, but they are mediated differently. In the U1A complex (Oubridge et al., 1994), the C⋅G base pair is recognized by a side chain from the β2–β3 loop (R52), and in the U2B″–U2A′ complex the U⋅U base pair is recognized by K20 from the α1–β2 loop. In the nucleolin complex, two base pairs of the stem are contacted by six amino acids. Note that the equivalent residues of V27 and K31 (Figure 3A), K23 and K27, respectively, in U2B″, recognize the last U of the RNA loop in the U2B″–U2A′–RNA complex (Price et al., 1998).
Comparison with other RBD12–RNA complexes
Structures of only two other RBD12–RNA complexes have been reported: sex-lethal (Handa et al., 1999) and PABP (Deo et al., 1999), both of which bind mRNA (Figure 7). These three complexes have several features in common. They all adopt a common topology for the protein, the βαββαβ RBD fold, and the two RBDs are separated by a linker, which forms a short helix at its C-terminus adjacent to RBD2. Like nucleolin RDB12, the linkers are ordered and have important interactions with the RNA, and the two RBDs interact with each other in the complex. This interaction is fairly weak in sex-lethal and in nucleolin, where the two domains interact via only two salt bridges. It is much stronger in the PABP complex, where 550 Å2 of solvent-accessible surface are buried between the two domains (Deo et al., 1999) (Figure 7D).
An interesting common feature of the RBD12–RNA complexes is that in all three structures RBD2 binds the 5′ end and RBD1 binds the 3′ end of the target RNA sequence. However, the nucleolin RBD12–RNA complex is unique in that nucleolin RBD12 brings the 5′ and 3′ ends of the NRE in close proximity to each other, so that the NRE consensus nucleotides (U9–A14) form an upside-down ‘U’ (Figures 4 and 7A). In contrast, sex-lethal (Handa et al., 1999) and PABP (Deo et al., 1999), which bind UG(U)8 and A8, respectively, interact along the length of the RNA and seem to stretch it out, resulting in a large separation between the 3′ and 5′ ends of the RNA (Figure 7C and D). It is remarkable that the same scaffold (RBD1–linker–RBD2) can shape RNA so differently. PABP stretches poly(A) via extensive interdomain interactions, while nucleolin brings together the ends of the RNA by inserting two amino acid side chains (K94 and F56) into the RNA loop and stabilizing the C11–A14 interaction with K94. In the sex-lethal and PABP complexes, there are no intramolecular hydrogen bonds in the RNA, while in the nucleolin complex two hydrogen bonds are formed, induced by protein binding.
The linker also plays a distinctly different role among the RBD–RNA complexes. The extended region of the linker (K89–D92 in nucleolin) in all the complexes contributes to the RNA binding specificity via interactions with its main chain, but in U1A it interacts with a C, in PABP with an A, in sex-lethal with a U, and in nucleolin and U2B″ with a G (G13). The position of the helix of the linker relative to the RNA backbone also differs among the three complexes (Figure 7). Furthermore, in the recently solved trimolecular complex between two U1A N-terminal regions (1–102) bound to symmetrical sites on their mRNA regulatory element, the helical part of the linker functions as a dimerization domain to form an RNA-binding-dependent interdomain interaction (Varani et al., 2000).
Diverse recognition of RNA by RBD proteins
With several RBD–RNA and RBD12–RNA complexes determined, it is interesting to compare the sequence-specific recognition by these diverse domains. The number of nucleotides that are contacted by a single RBD varies from 0 for RBD2 of U1A (Lu and Hall, 1995) to 12 for RBD1 of U2B″ (Price et al., 1998). The RBD1 and RBD2 of nucleolin interact with six and two nucleotides, respectively, whereas for sex-lethal (Handa et al., 1999) it is six and three, and for PABP (Deo et al., 1999) three and four nucleotides that contact RBD1 and RBD2, respectively. Although all of the RBDs interact with the RNA primarily with their β-sheet, the location of the amino acids on the surface of the β-sheet varies among the different RBDs (Figure 7). In all of the RBD–RNA complexes, RNA bases stack on the aromatic or hydrophobic side chains of the conserved residues of the RNP2(β1) and RNP1(β3) sequence, but it is difficult to predict which base will be involved in stacking. In the RBD–RNA complexes solved to date, there are four examples of a Y or F in β3, next to an R in β2. In all cases, the RNA base stacks on the aromatic side chain, and the arginine side chain interacts with the base, but it is a G in nucleolin RBD1 (G13), a C in nucleolin RBD2 (C10) (Figure 5), a U in sex-lethal RBD1 and an A in PABP RBD1. Therefore, elucidating a code for RNA recognition by an RBD may be difficult. Recognition of RNA by RBD proteins is not simply determined by RBD–RNA interactions, but also by RBD–interdomain interactions, linker–RBD interactions, RNA intramolecular interactions, and often protein–protein interactions, as in the trimolecular U2B″–U2A′–RNA (Price et al., 1998) and U1A–RNA–U1A (Varani et al., 2000) complexes and hnRNPA1 dimers (Ding et al., 1999).
Implications in understanding the role of nucleolin in ribosome biogenesis
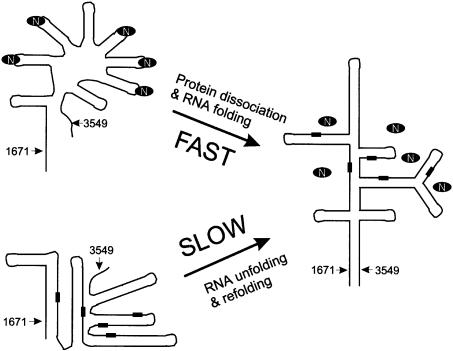
The structure of the nucleolin RBD12–RNA complex and the locations of the putative NRE binding sites suggest a role for nucleolin as an RNA chaperone (Herschlag, 1995). The 5′ ETS RNA and 28S RNA divergent domains in mouse and human, where two-thirds of the putative nucleolin binding sites were found (Serin et al., 1996), are highly rich in G and C (40% each) (Renalier et al., 1989). Based on phylogeny (Renalier et al., 1989) and electron microscopy (Wellauer et al., 1974; Schibler et al., 1975) studies, these regions are predicted to form a secondary structure composed of very long helices of mostly G⋅C base pairs in the mature pre-rRNA, e.g. region 1671–3549 in the human 5′ ETS (Renalier et al., 1989) (Figure 8). Correct folding of these helices, which is required for pre-rRNA processing, probably requires an RNA chaperone. We propose nucleolin as a candidate for such chaperone activity. Nucleolin is the most abundant nucleolar protein, binds RNA as soon as it is transcribed (Ghisolfi-Nieto et al., 1996), and as shown here specifically recognizes a GC-rich sequence. The majority of the putative NRE sequences are found within regions which are thought to be double stranded in the mature pre-rRNA, in agreement with the fact that nucleolin is not associated with the mature ribosomes (Serin et al., 1996). We suggest that nucleolin binds the (U/G)CCCG(G/A) sequences on the pre-rRNA as they are transcribed, inducing the formation of and/or stabilizing RNA stem–loops by bringing the 5′ and 3′ ends of the consensus sequence close to each other. Furthermore, the protein–RNA complex sequesters the bases of the NRE sequence, making them inaccessible for base pairing. Binding of nucleolin will therefore prevent stable alternative RNA structures from forming prior to completion of transcription. Subsequently, the ribonucleoprotein complex formed will unfold and the long stable RNA helices will form, since dissociation of nucleolin from the RNA (Kd = 10–100 nM) is expected to be faster than the unfolding of a stable alternative G–C-rich RNA structure free of protein (Herschlag, 1995).
Fig. 8. Proposed model of the RNA chaperone activity of nucleolin for proper folding of the 5′ ETS region between nucleotides 1671 and 3549 of human 47S pre-rRNA. A schematic representation of the predicted secondary structure of this region in the mature pre-rRNA based on phylogeny (Renalier et al., 1989) and electron microscopy (Wellauer et al., 1974; Schibler et al., 1975) studies is shown on the right. The putative NRE binding sites in this sequence are indicated by black rectangles. They are all found in double-stranded regions of the mature pre-rRNA, so nucleolin (indicated by the black oval ring) is not expected to be bound. On the left side of the figure are shown schematically two alternate structures that the RNA can adopt with (top) or without (bottom) nucleolin. Without nucleolin, the RNA can be kinetically trapped in alternative stable structures, which have to unfold to form the mature pre-rRNA, with the result that formation of the mature pre-rRNA will be slow. The bound nucleolin promotes and/or stabilizes stem–loops at the NRE consensus sites, preventing the formation of alternative stable helices, and then dissociates to allow the final structure to form.
This model for a chaperone activity for nucleolin is consistent with earlier reports based on CD measurements on pre-rRNA (Sipos and Olson, 1991) and gel mobility experiments on DNA (Sapp et al., 1986), which showed that nucleolin promotes the reassociation of complementary nucleic acid strands. Nucleolin may play a role analogous to that of hnRNP proteins in pre-mRNP assembly and pre-mRNA processing (Krecic and Swanson, 1999). RNA chaperone activity has been demonstrated for hnRNPA1 (Portman and Dreyfuss, 1994). Based on the crystal structure of hnRNPA1 with DNA, it has been proposed that this protein will also fold the RNA into stem–loops (Ding et al., 1999). Interestingly, HIV-1 nucleocapsid protein also binds sequence specifically to RNA stem–loops (De Guzman et al., 1998) and is thought to have an RNA chaperone activity (Rein et al., 1998). Thus, this model for the interaction of nucleolin with pre-rRNA may be common to these other RNA chaperones.
Materials and methods
Construction of wild-type and mutant nucleolin RBD12 and protein purification
The wild-type RBD12 subdomain of hamster nucleolin was constructed by direct cloning of the PCR product using oligonucleotide primers 5′R12 (5′-cgtatccatatggtggaaggttcagaatcaactaacacctttc-3′) and 3′R12 (5′-cgcggatcctatcatcccttctcccc-agtatagtaaag-3′). The PCR product was introduced into pet-15b vector. Mutated nucleolin RBD12 genes were produced using a PCR strategy with mutated oligonucleotide primers and PCR products were also introduced into pet-15b vector. Unlabeled, 15N- and 15N,13C-labeled proteins were expressed in BL21(DE3)pLysS cells, and grown and purified as described (Serin et al., 1997; Allain et al., 2000). Final sample conditions were 1–2 mM protein in 50 mM potassium phosphate buffer pH 6.2, 100 mM KCl.
Gel shift assays
Assays were performed as described (Serin et al., 1997). Labeled RNA (10 fmol) was incubated in 10 µl of TMKC buffer (20 mM Tris–HCl pH 7.4, 4 mM MgCl2, 200 mM KCl, 20% glycerol, 1 mM dithiothreitol, 0.5 mg/ml tRNA, 4 µg/ml bovine serum albumin) with the indicated amount of protein for 15 min at room temperature. The reactions were then directly loaded onto an 8% polyacrylamide gel (acrylamide:bisacrylamide 60:1) with 5% glycerol in 0.5× TBE and run at room temperature. The gels were dried and subjected to autoradiography. Gel shifts with mutated protein were each performed twice with two independent protein and RNA preparations.
Preparation of the isotopically labeled sNRE RNA and protein–RNA complexes.
RNA samples with the sequence 5′-GGCCGAAAUCCCGAAGUAGGCC-3′ (Figure 1C) were prepared by enzymatic synthesis as previously described (Dieckmann and Feigon, 1997). The 18 underlined nucleotides are the SELEX consensus sequence and the additional nucleotides were added to stabilize the stem and increase the transcription yields. Samples were synthesized as unlabeled, uniformly 13C,15N labeled, 13C,15N-G labeled, 13C,15N-A labeled, and 13C,15N-U and 13C,15N-C labeled.
Complexes were prepared by addition of aliquots of lyophilized RNA (usually 3–4 times) until a 1:1 stoichiometry was reached. The titration was monitored by observing the appearance of resolved resonances from the complex and disappearance of resonances from the free protein in 2D 1H-15N HMQC (Bax et al., 1983) spectra. Samples were prepared with 13C,15N-labeled or 15N-labeled protein complexed with unlabeled RNA (two samples) and labeled RNA complexed with 15N-labeled protein (four samples). Final sample conditions were 1–1.5 mM protein–RNA complex in a 1:1 ratio, in 50 mM potassium phosphate, 100 mM KCl pH 6.2 in D2O or 90%H2O/10%D2O. Samples were exchanged between H2O and D2O as needed by lyophilization. The protein–RNA complex is stable at temperatures ranging from 275 to 328 K.
NMR spectroscopy and assignment methodology
All the NMR spectra were acquired at 500 or 600 MHz on Bruker DRX spectrometers. Spectra were processed with Bruker Xwinnmr and analyzed by Felix97 (MSI, Inc.) and XEASY (Bartels et al., 1995). All the spectra listed below of nucleolin RBD12 in complex with RNA were recorded at 293 and 303 K. Additional NOESY spectra were recorded at 318 and 323 K.
A series of 3D 15N NOESY-HSQC, 15N TOCSY-HSQC (Cavanagh et al., 1996), 13C HSQC-NOESY (Majumdar and Zuiderweg, 1993) and 13C NOESY-HMQC (Marion et al., 1989) (in D2O) (τm = 150 ms for all 3D NOESY spectra) were acquired on the nucleolin RBD12–sNRE complex. A 2D homonuclear TOCSY (50 ms, in D2O) was acquired on the 15N-only labeled protein in complex to assign the aromatic side chains. A 1H-15N TROSY (Czisch and Boelens, 1998; Pervushin et al., 1998) was used to resolve overlapping amide resonances. These spectra and comparison to the spectra of the free protein (Allain et al., 2000) were sufficient to unambiguously assign nucleolin RBD12 in complex with the RNA. Complete assignments for the backbone and side chain resonances from V5 to G175 were obtained.
2D homonuclear NOESY in H2O (1-1 echo at 278 K) and D2O, TOCSY (50 ms) and DQF-COSY acquired on the complex with unlabeled RNA, and two 3D 13C NOESY-HMQC and HCCH-TOCSY (Bax et al., 1990) (both in D2O) acquired on the complex with the fully labeled RNA were sufficient to obtain complete non-exchangeable resonance assignments of the RNA in the complex, except for a few H5′ and H5″. 1H-13C HSQC were acquired on the protein–RNA complexes with the G-only, A-only and C,U-only 13C,15N-labeled RNA to confirm the assignments obtained using the fully labeled RNA in complex (Dieckmann and Feigon, 1997). U9 and G13 imino protons were not observable, consistent with the absence of hydrogen bonds for them in the structure. All other imino but no amino proton resonances in the loop were assigned. The acquisition of spectra at the higher temperatures was essential for resolving critical overlapping resonances, in particular the aromatic resonances of C10 and C11 and for detecting the Hδ and Hε of F17, which were only observable at 323 K due to a slower ring flipping rate than usual due to its stacking on C12. A similar behavior was observed in the U1A–RNA complex (Howe et al., 1998).
In order to observe only intramolecular NOEs or intermolecular NOEs, a set of 2D and 3D spectra with 12C and/or 13C double X-filtered experiments was acquired on the samples where either only the RNA or the protein was isotopically labeled. A set of four 2D 12C-13C filtered 1H-1H NOESY spectra (Otting and Wüthrich, 1990; Slijper et al., 1996) and a 3D 1H-13C double half-filtered HMQC-NOESY (Lee et al., 1994; Zwahlen et al., 1997) were obtained on the samples with the fully labeled protein complexed with unlabeled RNA and with the unlabeled protein complexed with the fully labeled RNA.
Structure calculation of the protein–RNA complex
Interproton distance constraints were obtained from 2D homonuclear NOESY spectra in D2O with presaturation of the residual HDO signal (τm = 30, 100, 150 and 200 ms) at 303, 318 and 328 K, a 1-1 echo NOESY (τm = 300 ms) in 90% H2O/10% D2O at 278 K, and several 3D 15N and 13C separated NOESY spectra (τm = 150 ms) recorded in either 90% H2O/10% D2O or 99.9% D2O at 303 and 318 K. The volumes of the NOE cross-peaks assigned in the 2D homonuclear (in D2O) and 3D 15N-separated NOESY were integrated by SPSCAN and converted into distance constraints using the CALIBA subroutine of XEASY. Distance restraints derived from the assigned NOEs were given qualitative upper limits of 3.5, 5 or 7 Å plus a pseudo atom correction based on the intensity of the cross-peak, and those from the 2D 1-1 echo NOESY were given qualitative upper limits of 5 and 7 Å. In total, 3010 NOE-derived distance restraints were used, including 2593 protein, 267 RNA and 150 intermolecular. In the early rounds of calculations, C11 was found to interact with the surface of the β-sheet of RBD2 in some of the structures. Since C11 H6 and H5 did not show any intermolecular NOEs to protein side chains and their linewidths were not broadened by conformational exchange, we included seven repulsive restraints (lower bound of 4 Å and an upper bound of 20 Å). Seventy-six hydrogen bond constraints within the protein were added based on the observation of slowly exchanging amide protons when the protein–RNA complex was freshly transferred from H2O to D2O. Nineteen hydrogen bond constraints within the RNA were used: 12 in the four G⋅C base pairs of the stem, four in the two non-Watson–Crick base pairs and three in the base triple of the loop E motif. An additional four intermolecular hydrogen bond restraints (two between R49 and G13 and two between R127 and C10) were included in the last run of structure calculations. Those were based on the unusual downfield chemical shifts of R49 Hε (9.30 p.p.m.) and R127 Hε (9.70 p.p.m.) (Jiang et al., 1999), and on the previous round of structure calculations, which showed that such hydrogen bonds were likely. The 22 δ dihedral angles of the RNA were constrained to C2′ endo (145 ± 30°) or C3′ endo (90 ± 10°) based on the size of the 3JH1′-H2′ coupling and the intensity of the H1′-H2′ TOCSY cross-peaks (Varani et al., 1996). A8, U9, C12, G13, A14, A15 and G16 are C2′ endo range and all other residues are C3′ endo range. Nine weak base-planarity constraints in the RNA stem and the loop E of the RNA (one per base pair and three for the base triple) were used.
Structure calculations were carried out using the simulated annealing protocol implemented in X-PLOR 3.8 (Brünger, 1992) as described (Howe et al., 1998). Forty structures were calculated, starting from randomized RNA and protein chains. The 19 lowest energy structures from the ensemble of calculated structures were further analyzed and described (Table I). The hydrogen bonds described in the paper are present in the majority of the 19 lowest energy structures (distance between heavy atoms of <4 Å and acceptor proton–donor angle >120° when applicable). Figures of the structures were generated with MOLMOL (Koradi et al., 1996).
Coordinates
The 19 conformers of nucleolin RBD12–sNRE complex have been deposited in the RCSB Protein Data Bank (accession code 1FJE).
Acknowledgments
Acknowledgements
The authors thank Dr R.D.Peterson and Mr J.E.Masse for help in NMR data acquisition, and Mr E.Feinstein for figure preparation. This work is supported by NIH grant GM37254 to J.F., Association pour la Recherche contre le Cancer and the CNRS to P.B., and HFSPO and UCLA JCCC postdoctoral fellowships to F.H.-T.A.
References
- Allain F.H.-T., Gubser,C.C., Howe,P.W.A., Nagai,K. Neuhaus,D. and Varani,G. (1996) Specificity of ribonucleoprotein interaction determined by RNA folding during complex formulation. Nature, 380, 646–650. [DOI] [PubMed] [Google Scholar]
- Allain F.H.-T., Howe,P.W.A., Neuhaus,D. and Varani,G. (1997) Structural basis of the RNA-binding specificity of human U1A protein. EMBO J., 16, 5764–5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allain F.H.-T., Gilbert,D.E., Bouvet,P. and Feigon,J. (2000) Solution structure of the two N-terminal RNA-binding domains of Nucleolin and NMR study of the interaction with its RNA target. J. Mol. Biol., 303, 227–241. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J.H., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels C., Xia,T.H., Billeter,M., Guntert,P. and Wuthrich,K. (1995) The program Xeasy for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR, 6, 1–10. [DOI] [PubMed] [Google Scholar]
- Bax A., Griffey,R.H. and Hawkins,B.L. (1983) Correlation of proton and nitrogen-15 chemical shifts by multiple quantum NMR. J. Magn. Reson., 55, 301–315. [Google Scholar]
- Bax A., Clore,G.M. and Gronenborn,A.M. (1990) 1H-1H correlation via isotropic mixing of 13C magnetization, a new three-dimensional approach for assigning 1H and 13C spectra of 13C-enriched proteins. J. Magn. Reson., 88, 425–431. [Google Scholar]
- Birney E., Kumar,S. and Krainer,A.R. (1993) Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res., 21, 5803–5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet P., Jain,C., Belasco,J.G., Amalric,F. and Erard,M. (1997) RNA recognition by the joint action of two nucleolin RNA-binding domains: genetic analysis and structural modeling. EMBO J., 16, 5235–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet P., Diaz,J.-J., Kindbeiter,K., Madjarn,J.-J. and Amalric,F. (1998) Nucleolin interacts with several ribosomal proteins through its RGG domain. J. Biol. Chem., 273, 19025–19029. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. (1992) X-PLOR (Version 3.1) Manual. Yale University Press, New Haven, CT. [Google Scholar]
- Burd C.G. and Dreyfuss,G. (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science, 265, 615–621. [DOI] [PubMed] [Google Scholar]
- Cavanagh J., Fairbrother,W.J., Palmer,A.G.,III and Skelton,N.J. (1996) Protein NMR Spectroscopy: Principles and Practice. Academic Press, San Diego, CA. [Google Scholar]
- Czisch M. and Boelens,R. (1998) Sensitivity enhancement in the TROSY experiment. J. Magn. Reson., 134, 158–160. [DOI] [PubMed] [Google Scholar]
- De Guzman R.N., Wu,Z.R., Stalling,C.C., Pappalardo,L., Boret,P.N. and Summers,M.F. (1998) Structure of the HIV-1 nucleocapsid protein bound to the SL3 ψ-RNA recognition element. Science, 279, 384–388. [DOI] [PubMed] [Google Scholar]
- Deo R.C., Bonanno,J.B., Sonenberg,N. and Burley,S.K. (1999) Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell, 98, 835–845. [DOI] [PubMed] [Google Scholar]
- Dieckmann T. and Feigon,J. (1997) Assignment methodology for larger oligonucleotides: application to an ATP-binding RNA aptamer. J. Biomol. NMR, 9, 259–272. [DOI] [PubMed] [Google Scholar]
- Ding J., Hayashi,M.K., Zhang,Y., Manche,L., Krainer,A.R. and Xu,R.-M. (1999) Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev., 13, 1102–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erard M.S., Belenguer,P., Caizergues-Ferrer,M., Pantaloni,A. and Amalric,F. (1988) A major nucleolar protein, nucleolin, induces chromatin decondensation by binding to histone H1. Eur. J. Biochem., 175, 525–530. [DOI] [PubMed] [Google Scholar]
- Ghisolfi-Nieto L., Joseph,G., Puvion-Dutilleul,F., Amaric,F. and Bouvet,P. (1996) Nucleolin is a sequence specific RNA-binding protein—characterization of targets on pre-ribosomal RNA. J. Mol. Biol., 260, 34–53. [DOI] [PubMed] [Google Scholar]
- Ginisty H., Amalric,F. and Bouvet,P. (1998) Nucleolin functions in the first step of ribosomal RNA processing. EMBO J., 17, 1476–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginisty H., Sicard,H., Roger,B. and Bouvet,P. (1999) Structure and functions of nucleolin. J. Cell Sci., 112, 761–772. [DOI] [PubMed] [Google Scholar]
- Handa N., Nureki,O., Kurimoto,K., Kim,I., Sakamoto,H., Shimura,Y., Muto,Y. and Yokoyama,S. (1999) Structural basis for recognition of the tra mRNA precursor by the sex-lethal protein. Nature, 398, 579–585. [DOI] [PubMed] [Google Scholar]
- Herrera A.H. and Olson,M.O. (1986) Association of protein C23 with rapidly labeled nucleolar RNA. Biochemistry, 25, 6258–6264. [DOI] [PubMed] [Google Scholar]
- Herschlag D. (1995) RNA chaperones and the RNA folding problem. J. Biol. Chem., 270, 20871–20874. [DOI] [PubMed] [Google Scholar]
- Howe P.W.A., Allain,F.H.-T., Varani,G. and Neuhaus,D. (1998) Determination of the NMR structure of the complex between U1A protein and its RNA polyadenylation inhibition element. J. Biomol. NMR, 11, 59–84. [DOI] [PubMed] [Google Scholar]
- Jiang F., Gorin,A., Hu,W.D., Majumdar,A., Baskerville,S., Xu,W.J., Ellington,A. and Patel,D.J. (1999) Anchoring an extended HTLV-1 Rex peptide within an RNA major groove containing junctional base triples. Structure Fold. Des., 7, 1461–1472. [DOI] [PubMed] [Google Scholar]
- Kay L.E., Torchia,D.A. and Bax,A. (1989) Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry, 28, 8972–8979. [DOI] [PubMed] [Google Scholar]
- Koradi R., Billeter,M. and Wüthrich,K. (1996) MOLMOL—a program for display and analysis of macromolecular structures. J. Mol. Graph., 14, 51–55. [DOI] [PubMed] [Google Scholar]
- Krecic A.M. and Swanson,M.S. (1999) hnRNP complexes: composition, structure and function. Curr. Opin. Cell Biol., 11, 363–371. [DOI] [PubMed] [Google Scholar]
- Lee W., Revington,M.J., Arrowsmith,C. and Kay,L.E. (1994) A pulsed field gradient isotope-filtered 3D C-13 HMQC-NOESY experiment for extracting intermolecular NOE contacts in molecular complexes. FEBS Lett., 350, 87–90. [DOI] [PubMed] [Google Scholar]
- Leontis N.B. and Westhof,E. (1998) A common motif organizes the structure of multi-helix loops in 16 S and 23 S ribosomal RNAs. J. Mol. Biol., 283, 571–583. [DOI] [PubMed] [Google Scholar]
- Lu J.R. and Hall,K.B. (1995) An RBD that does not bind RNA: NMR secondary structure determination and biochemical properties of the C-terminal RNA binding domain from the human U1A protein. J. Mol. Biol., 247, 739–752. [DOI] [PubMed] [Google Scholar]
- Majumdar A. and Zuiderweg,E.R.P. (1993) Improved C-13-resolved HSQC-NOESY spectra in H2O, using pulsed field gradients. J. Magn. Reson. Ser. B, 102, 242–244. [Google Scholar]
- Mao H., White,S.A. and Williamson,J.R. (1999) A novel loop–loop recognition motif in the yeast ribosomal protein L30 autoregulatory RNA complex. Nature Struct. Biol., 6, 1139–1147. [DOI] [PubMed] [Google Scholar]
- Marion D., Kay,L.E., Sparks,S.W., Torchia,D.A. and Bax,A. (1989) Three-dimensional heteronuclear NMR of 15N-labeled proteins. J. Am. Chem. Soc., 111, 1515–1517. [Google Scholar]
- Nagai K., Oubridge,C., Jessen,T.H., Li,J. and Evans,P.R. (1990) Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein-A. Nature, 348, 515–520. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–96. [DOI] [PubMed] [Google Scholar]
- Olson M.O.J. (1990) The role of proteins in nucleolar structure and function. In Strauss,P.R. and Wilson,S.H. (eds), The Eukaryotic Nucleus–Molecular Biochemistry and Macromolecular Assemblies. The Telford Press, Caldwell, NJ, Vol. 2, pp. 519–559. [Google Scholar]
- Otting G. and Wüthrich,K. (1990) Heteronuclear filters in two-dimensional [1H,1H]-NMR spectroscopy: combined use with isotope labelling for studies of macromolecular conformation and intermolecular interactions. Q. Rev. Biophys., 23, 39–96. [DOI] [PubMed] [Google Scholar]
- Oubridge C., Ito,N., Evans,P.R., Teo,C.-H. and Nagai,K. (1994) Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature, 372, 432–438. [DOI] [PubMed] [Google Scholar]
- Pervushin K., Reik,R., Wider,G. and Wuthrich,K. (1998) Transverse relaxation-optimized spectroscopy (TROSY) for NMR studies of aromatic spin systems in 13C-labeled proteins. J. Am. Chem. Soc., 120, 6394–6400. [Google Scholar]
- Portman D.S. and Dreyfuss,G. (1994) RNA annealing activities in HeLa nuclei. EMBO J., 13, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price S.R., Evens,P.R. and Nagai,K. (1998) Crystal structure of the spliceosomal U2B′′–U2A′ protein complex bound to a fragment of U2 small nuclear RNA. Nature, 394, 645–650. [DOI] [PubMed] [Google Scholar]
- Ramos A., Grunert,S., Adams,J., Micklem,D.R., Proctor,M.R., Freund,S., Bycroft,M., St Johnston,D. and Varani,G. (2000) RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J., 19, 997–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein A., Henderson,L.E. and Levin,J.G. (1998) Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci., 23, 297–301. [DOI] [PubMed] [Google Scholar]
- Renalier M.H., Mazan,S., Joseph,N., Michot,B. and Bachellerie,J.P. (1989) Structure of the 5′-external transcribed spacer of the human ribosomal RNA gene. FEBS Lett., 249, 279–284. [DOI] [PubMed] [Google Scholar]
- Sapp M., Knippers,R. and Richter,A. (1986) DNA binding properties of a 110kDa nucleolar protein. Nucleic Acids Res., 14, 6803–6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U., Wyler,T. and Hagenbuchle,O. (1975) Changes in size and secondary structure of the ribosomal transcription unit during vertebrate evolution. J. Mol. Biol., 94, 503–517. [DOI] [PubMed] [Google Scholar]
- Serin G., Joseph,G., Faucher,C., Ghisolfi,L., Bouche,G., Amalric,F. and Bouvet,P. (1996) Localization of nucleolin binding sites on human and mouse pre-ribosomal RNA. Biochimie, 78, 530–538. [DOI] [PubMed] [Google Scholar]
- Serin G., Joseph,G., Ghisolfi,L., Bauzan,M., Erard,M., Amalric,F. and Bouvet,P. (1997) Two RNA-binding domains determine the RNA-binding specificity of nucleolin. J. Biol. Chem., 272, 13109–13116. [DOI] [PubMed] [Google Scholar]
- Shi H., Hoffman,B.E. and Lis,J.T. (1997) A specific RNA hairpin loop structure binds the RNA recognition motifs of the Drosophila SR protein B52. Mol. Cell. Biol., 17, 2649–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipos K. and Olson,M.O.J. (1991) Nucleolin promotes secondary structure in ribosomal RNA. Biochem. Biophys. Res. Commun., 177, 673–678. [DOI] [PubMed] [Google Scholar]
- Slijper M., Kaptein,R. and Boelens,R. (1996) Simultaneous C-13 and N-15 isotope editing of biomolecular complexes: application to a mutant Lac repressor headpiece DNA complex. J. Magn. Reson. B, 111, 199–203. [DOI] [PubMed] [Google Scholar]
- Srivastava M. and Pollard,H.B. (1999) Molecular dissection of nucleolin’s role in growth and cell proliferation: new insights. FASEB J., 13, 1911–1922. [PubMed] [Google Scholar]
- Stoldt M., Wohnert,J., Ohlenschlager,O., Gorlach,M. and Brown,L.R. (1999) The NMR structure of the 5S rRNA E-domain–protein L25 complex shows preformed and induced recognition. EMBO J., 18, 6508–6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczak A.A., Moore,P.B., Chan,Y.-L. and Wool,I.G. (1993) The conformation of the sarcin/ricin loop from 28S ribosomal RNA. Proc. Natl Acad. Sci. USA, 90, 9581–9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani G. and Nagai,K. (1998) RNA recognition by RNP proteins during RNA processing. Annu. Rev. Biophys. Biomol. Struct., 27, 407–445. [DOI] [PubMed] [Google Scholar]
- Varani G., Aboul-ela,F. and Allain,F.H.-T. (1996) NMR investigation of RNA structure. Prog. NMR Spectrosc., 29, 51–127. [Google Scholar]
- Varani L., Gunderson,S.I., Mattaj,I.W., Kay,L.E., Neuhaus,D. and Varani,G. (2000) The NMR structure of the 38 kDa U1A protein–PIE RNA complex reveals the basis of cooperativity in regulation of polyadenylation by human U1A protein. Nature Struct. Biol., 7, 329–335. [DOI] [PubMed] [Google Scholar]
- Wellauer P.K., Dawid,I.B., Kelley,D.E. and Perry,R.P. (1974) Secondary structure maps of ribosomal RNA. II. Processing of mouse L-cell ribosomal RNA and variations in the processing pathway. J. Mol. Biol., 89, 397–407. [DOI] [PubMed] [Google Scholar]
- Wimberly B., Varani,G. and Tinoco,I.,Jr (1993) The conformation of loop E of eukaryotic 5S ribosomal RNA. Biochemistry, 32, 1078–1087. [DOI] [PubMed] [Google Scholar]
- Zwahlen C., Legault,P., Vincent,S.J.F., Greenblatt,J., Konrat,R. and Kay,L.E. (1997) Methods for measurement of intermolecular NOEs by multinuclear NMR spectroscopy: application to a bacteriophage lambda N-peptide/boxB RNA complex. J. Am. Chem. Soc., 119, 6711–6721. [Google Scholar]