Abstract
Control of polyene macrolide production in Streptomyces natalensis is mediated by the transcriptional activator PimM. This regulator, which combines an N-terminal PAS domain with a C-terminal helix-turn-helix motif, is highly conserved among polyene biosynthetic gene clusters. PimM, truncated forms of the protein without the PAS domain (PimMΔPAS), and forms containing just the DNA-binding domain (DBD) (PimMDBD) were overexpressed in Escherichia coli as GST-fused proteins. GST-PimM binds directly to eight promoters of the pimaricin cluster, as demonstrated by electrophoretic mobility shift assays. Assays with truncated forms of the protein revealed that the PAS domain does not mediate specificity or the distinct recognition of target genes, which rely on the DBD domain, but significantly reduces binding affinity up to 500-fold. Transcription start points were identified by 5′-rapid amplification of cDNA ends, and the binding regions of PimMDBD were investigated by DNase I protection studies. In all cases, binding took place covering the −35 hexamer box of each promoter, suggesting an interaction of PimM and RNA polymerase to cause transcription activation. Information content analysis of the 16 sequences protected in target promoters was used to deduce the structure of the PimM-binding site. This site displays dyad symmetry, spans 14 nucleotides, and adjusts to the consensus TVGGGAWWTCCCBA. Experimental validation of this binding site was performed by using synthetic DNA duplexes. Binding of PimM to the promoter region of one of the polyketide synthase genes from the Streptomyces nodosus amphotericin cluster containing the consensus binding site was also observed, thus proving the applicability of the findings reported here to other antifungal polyketides.
Keywords: Antibiotics, DNA Operators, Gene Regulation, Transcription Promoter, Transcription Regulation, LuxR, PAS, Streptomyces, Pimaricin, Polyene
Introduction
Streptomycetes are filamentous soil bacteria that have a complex life cycle that involves differentiation and sporulation. These bacteria are well known for their ability to produce a great variety of secondary metabolites, including therapeutic molecules like antibiotics, immunosuppressants, or anticancer agents. Production of these compounds is regulated in response to nutritional status alteration and a variety of environmental conditions, and hence occurs in a growth-phase-dependent manner, at the transition between the rapid growth phase and the stationary growth phase and is usually accompanied by morphological differentiation (1). The control of secondary metabolite production is a complex process involving multiple levels of intertwined regulation. Whereas the higher levels are composed by regulatory genes that exert a pleiotropic control over various aspects of secondary metabolism, the lowest is composed by regulatory genes that only affect a single antibiotic biosynthetic pathway. The latter genes are usually found within the respective antibiotic biosynthesis gene cluster, a feature that has greatly facilitated their study.
Pimaricin is a tetraene macrolide antifungal antibiotic produced by S. natalensis (2). As a polyene, its antifungal activity lies in its interaction with membrane ergosterol, but unlike in other polyenes, this action is not exerted via permeabilization of the membrane (3). Like other macrocyclic polyketides, pimaricin is synthesized by the action of so-called type I modular polyketide synthases (4), and its biosynthetic gene cluster has been characterized (5–8). The gene cluster contains 19 open reading frames, including two pathway-specific regulatory genes, pimR and pimM (see Ref. 9 for a review). PimR is the archetype of a new class of regulators that combines an N-terminal domain corresponding to the Streptomyces antibiotic regulatory protein (SARP)2 family of transcriptional activators with a C-terminal half homologous to guanylate cyclases and large ATP-binding regulators of the LuxR family. Gene disruption of pimR totally blocked pimaricin production (10), thus confirming its role as transcriptional activator. PimM constitutes a second transcriptional activator of pimaricin biosynthesis. It is a regulator that combines an N-terminal PAS sensory domain (11, 12) with a C-terminal HTH motif of the LuxR type for DNA binding. PAS domains were first found in eukaryotes and were named after homology to the Drosophila period protein (Per), the aryl hydrocarbon receptor nuclear translocator protein (ARNT), and the Drosophila single-minded protein (Sim). Unlike the majority of prokaryotic PAS domain-containing regulators, which function as sensor kinases of two-component systems (11), PimM does not belong to a two-component system. Inactivation of pimM from the S. natalensis chromosome resulted in complete loss of pimaricin production, suggesting that PimM is a second positive regulator of pimaricin biosynthesis (13). The pimM regulatory model is an attractive paradigm because PimM homologous regulatory proteins have been found to be encoded in all known biosynthetic gene clusters of antifungal polyketides, such as the amphotericin (AmphRIV) (14), candicidin (FscRI) (15), nystatin (NysRIV) (16), or filipin (PteF) (17).
Gene expression analyses by reverse transcription-polymerase chain reaction (RT-PCR) of the pimaricin gene cluster revealed the targets for the PimM regulatory protein. According to these analyses, the genes responsible for initiation (pimS0) and the first cycles of polyketide chain extension (pimS1), were among the major targets for regulation, although other pim genes were also differentially affected, thus accounting for the lack of pimaricin production (13). We now report the direct binding of PimM to upstream sequences of eight promoters of the pimaricin gene cluster in Streptomyces natalensis and provide evidence that binding specificity relies on the DNA-binding domain, whereas the PAS domain significantly reduces the affinity of binding to target promoters. Footprinting analysis has allowed the identification of so far unknown boxes in the promoters of these genes. This study constitutes the first molecular characterization of the mode of action of a polyene macrolide regulator and makes PimM the first pathway-specific regulator of antibiotic biosynthesis, not belonging to the SARP family, whose binding site has been determined.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Cultivation
S. natalensis ATCC 27448 was routinely grown in YEME medium (18) without sucrose. Sporulation was achieved in TBO medium (19) at 30 °C. Escherichia coli strain DH5α was used as a host for DNA manipulation. E. coli BL21 (DE3) was used for expression studies.
Plasmids and DNA Manipulation
pUC19 (New England Biolabs) was used as the routine cloning vector, and pGEX-2T (GE Healthcare) was the vector used to construct PimM expression plasmids. Plasmid and genomic DNA preparation, DNA digestion, fragment isolation, and transformation of E. coli were performed by standard procedures (20). PCRs were carried out using Phusion DNA polymerase as described by the enzyme supplier (Finnzymes). DNA sequencing was accomplished by the dideoxynucleotide chain termination method using the PerkinElmer Amplitaq Gold Big Dye-terminator sequencing system with an Applied Biosystems ABI 3130 DNA genetic analyzer (Foster City, CA).
Construction of Expression Plasmids
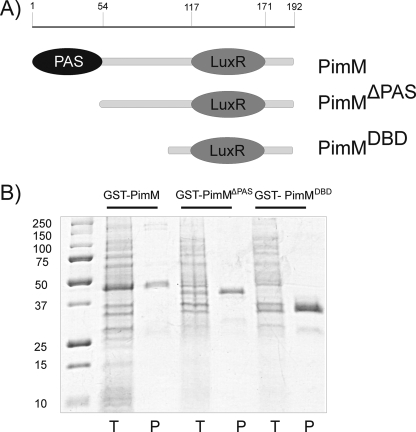
The pimM gene was amplified for insertion into the GST expression vector pGEX-2T using PCR. The forward primer used (5′-TACAGGATCCATGGCGAGCCTTGATAGAACATTGACCATCCAGCAGG-3′) introduced a unique BamHI site at the 5′ end of the gene, whereas the reverse primer (5′-GGAATTCGCCTGTGCCCGCTCACTTCACG-3′) carries an EcoRI site 12 nucleotides downstream from the TGA translational stop codon. The amplified DNA fragment was digested with BamHI and EcoRI and cloned into the same sites of pGEX-2T to generate pJM. The amplified DNA fragment was sequenced from the expression vector in order to discard any mistakes introduced by the DNA polymerase. Similarly, a truncated version of pimM lacking the N-terminal PAS domain (first 150 nucleotides) was amplified using the forward primer (5′-TACAGGATCCAATTTACTGGAAGGTAAGCACCAGCG-3′) and the same reverse primer. Cloning of the amplified and digested DNA fragment into pGEX-2T yielded pJMΔPAS. The LuxR DNA-binding domain of pimM (PimMDBD) was amplified using the forward primer (5′-TACAGGATCCGCCGGGGACGCCGAGGGG-3′) and the same reverse primer. This generates a GST-PimMDBD fusion protein, which includes the last 93 residues of PimM (Fig. 2A).
FIGURE 2.
Purification of GST fusion proteins in E. coli BL21. A, scheme of PimM and its truncated versions. Numbers indicate amino acid residues from the N terminus. B, purification of GST-PimM, GST-PimMΔPAS, and GST-PimMDBD proteins by affinity chromatography on glutathione-Sepharose. Lanes T, total E. coli cell extract; lanes P, purified proteins after affinity chromatography. Left lane, molecular size markers (in kDa).
Expression and Purification of GST Fusion Proteins
E. coli BL21(DE3) cells were grown at 18 °C in 600 ml of LB medium containing 100 μg/ml ampicillin until an A600 of 0.7 was reached and then induced by adding isopropyl 1-thio-β-d-galactopyranoside to a final concentration of 0.1 mm and grown for an additional 14 h at 18 °C. Cells were harvested, resuspended in 50 mm Tris-HCl, pH 8.0, and lysed by sonication using an ultrasonic processor XL apparatus (Misonix Inc.). The insoluble material was separated by centrifugation, and the soluble fraction was applied to a glutathione-Sepharose 4B column (Amersham Biosciences). Protein was eluted with 10 mm reduced glutathione in 50 mm Tris-HCl, pH 8.0, and conserved in 20% glycerol at −80 °C before use. Protein elution was monitored at 280 nm, and the presence of the fusion protein was assessed by SDS-PAGE. Enzyme concentration and yield were determined by the Bradford method (21) using bovine serum albumin as a standard.
Isolation of Total RNA
For RNA extraction, 300 μl of culture (48 h of growth in YEME medium without sucrose; stationary phase of growth) was added to 600 μl of RNA Protect Bacteria Reagent (Qiagen), mixed, and maintained for 5 min at room temperature. Then cells were harvested by centrifugation and frozen directly in liquid nitrogen. Cell pellets were resuspended in 900 μl of lysis solution (400 μl of acid phenol, 100 μl of chloroform/isoamyl alcohol (24:1), 400 μl of RLT buffer (Qiagen)) and disrupted with a FastPrepTM FP120 (BIO 101) apparatus by using the lysing matrix B. Then 400 μl of lysate were mixed with 400 μl of chloroform/isoamyl alcohol to remove phenol, and after centrifugation, the upper phase was mixed with 300 μl of RLT buffer (Qiagen) and 250 μl of ethanol. RNeasy Mini Spin columns were then used for RNA isolation according to the manufacturer's instructions. DNA was removed by a double treatment with RNase-free DNase (Qiagen) in the column plus an additional treatment with Turbo DNA-Free (Ambion) in solution. Total RNA concentration was determined with a NanoDrop ND-1000 spectrophotometer (Thermo Scientific), and quality and integrity were checked in a bioanalyzer 2100 apparatus (Agilent Technologies).
RT-PCR Experiments
Transcription was studied by using the SuperScriptTM One-Step RT-PCR system with Platinum® TaqDNA polymerase (Invitrogen), using 100 ng of total RNA as template. Conditions were as follows: first strand cDNA synthesis, 45 °C for 40 min followed by heating at 94 °C for 2 min; amplification, 30 cycles of 98 °C for 15 s, 59–71 °C (depending of the set of primers used) for 30 s, and 72 °C for 1 min. Primers (17–22-mers; supplemental Table S1) were designed to cover intergenic regions, generating PCR products of ∼400–600 bp. Negative controls were carried out with each set of primers and Platinum® TaqDNA polymerase in order to confirm the absence of contaminating DNA in the RNA preparations. The identity of each amplified product was corroborated by direct sequencing of the PCR product.
Rapid Amplification of cDNA Ends (RACE)
The 5′-ends of certain transcripts were identified by using a 5′-RACE system (Invitrogen), following the manufacturer's instructions (version 2.0). Briefly, first strand cDNA synthesis was carried out using 3.7 μg of RNA, reverse transcriptase, and the gene-specific primer (numbers 1 in supplemental Table S2). The cDNA was purified using the SNAP columns provided in the kit, and poly(dC) tails were added to the 3′-ends using terminal deoxynucleotidyl transferase. PCR amplification of the tailed cDNA was carried out using the 5′-RACE abridged anchor primer with the first nested primer (numbers 2 in supplemental Table S2). A dilution of the PCR mixture then was subjected to reamplification using the abridged universal amplification primer with the second nested primer (numbers 3 in supplemental Table S2). The PCR products were gel-purified and sequenced. When cDNA tailing with poly(dC) did not permit the identification of the transcription start point, poly(dA) tails were added to the 3′-ends of cDNA. In these cases, second strand cDNA synthesis was necessary prior to nested amplifications and was carried out using the 3′-RACE adapter primer (Invitrogen). PCR amplification of the cDNA was then carried out using the abridged universal amplification primer with the first nested primer (numbers 2 in supplemental Table S2). Final nested amplification was carried out as before.
DNA-Protein Binding Assays
DNA binding tests were performed by EMSA. The DNA fragments used for EMSA were amplified by PCR using the primers listed in supplemental Table S3 and labeled at both ends with digoxigenin with the DIG Oligonucleotide 3′-End Labeling Kit, 2nd Generation (Roche Applied Science). A standard binding reaction contained 2 ng of labeled DNA probe, 40 mm Tris-HCl, pH 8.0, 0.4 mm MgCl2, 5 mm KCl, 0.1 mm DTT, 7.8 mm glutathione, 0.005% Nonidet P-40, 40 mg/ml poly(dI-dC), 20.6% glycerol in a 25-μl final volume. The reaction was performed as described (22). The samples were loaded onto a 5% polyacrylamide (29:1) native gel in 0.5× TBE buffer. After electrophoresis (4 h, 70 V, 4 °C), DNA was electroblotted onto a nylon membrane (HyBond-N, Amersham Biosciences) in 0.5× TBE buffer (30 min, 200 mA). The DNA was fixed by UV cross-linking, detected with anti-digoxigenin antibodies, and developed by chemiluminiscence with the CDP-StarTM reagent (Roche Applied Science). When required, the intensity of bands was determined by using a scanner (Hewlett-Packard) with a Gel-Pro analyzer 3.1 program (Media Cybernetics).
To obtain DNA duplexes for the validation of the binding site, one of the following oligonucleotide pairs (P1, GGCACTGTCTAGCGAGACTAGGGAATTCCCTAGAACCGACGCTTTCACAC and GTGTGAAAGCGTCGGTTCTAGGGAATTCCCTAGTCTCGCTAGACAGTGCC; P2, GGCACTGTCTAGCGAGACTAGGGCCCTAGAACCGACGCTTTCACAC and GTGTGAAAGCGTCGGTTCTAGGGCCCTAGTCTCGCTAGACAGTGCC; P3, GGCACTGTCTAGCGAGACTAGGGGCCGCCCTAGAACCGACGCTTTCACAC and GTGTGAAAGCGTCGGTTCTAGGGCGGCCCCTAGTCTCGCTAGACAGTGCC) was annealed by heating at 95 °C for 10 min and slowly cooled to room temperature in a solution of 100 mm NaCl, followed by PAGE purification and 3′-end labeling.
Footprinting Assays
DNase I footprinting assays were performed by the fluorescent labeling procedure (23), using the GST-PimMDBD protein form that has the highest affinity for DNA. The DNA fragments used were the same as those used for EMSA experiments, cloned into pUC19, and amplified by PCR using the universal and reverse primers, one of them labeled with 6-carboxyfluorescein. In each case, the same labeled oligonucleotide served to prime the sequencing reaction used as the molecular size marker. The PCR products were purified after agarose-gel electrophoresis, and DNA concentrations were determined with a NanoDrop ND-1000 spectrophotometer (Thermo Scientific).
The reaction components were the same as described above for the EMSA reaction. Labeled DNA fragment (0.28 pmol) and GST- PimMDBD protein were added to a final volume of 56 μl and incubated at 30 °C for 10 min. Lyophilized bovine pancreas DNase I (Roche Applied Science grade I) was reconstituted in 20 mm Tris-HCl, pH 7.0, 50 mm NaCl, 100 μg/ml BSA, 1 mm DTT, 10% glycerol to a final concentration of 2.5 × 10−3 units/μl. Nuclease digestions were carried out with 0.01 units (4 μl) at 30 °C for 1 min and stopped with 120 μl of 40 mm EDTA in 9 mm Tris-HCl, pH 8.0. After phenol/chloroform purification and ethanol precipitation, samples were loaded in an Applied Biosystems ABI 3130 DNA genetic analyzer (Foster City, CA). Results were analyzed with the PEAK SCANNER program (Applied Biosystems).
Bioinformatic Analysis
To calculate the information content (Ri value) of individual sequences (24) and to obtain the logo of the binding site of the regulator PimM, we used the BiPad server (25). The candidate sequences to contain promoters were analyzed using the Patser algorithm (26), implemented in the Web resource Regulatory Sequence Analysis Tools (27). The pseudocount value was set to 10, and the alphabet parameter was adjusted to the GC content of the Streptomyces genome: AT, 0.15; CG, 0.35. The matrices used to search for regions −35 and −10 were those derived from the alignments of class C and class A promoters of Bourn and Babb (28). To search for a combination of “class C-n nt of separation-class A,” we included n columns of null values in the combined matrix.
RESULTS
Organization of pim Cluster Transcriptional Units
Organization of the pimaricin gene cluster and transcription of the genes were previously largely deduced by analyzing gene chromosomal arrangement and by the different expression profiles of certain genes in RT-PCR experiments (10, 13); however, a more accurate identification of operons was needed in order to define an overall picture of the transcriptional arrangement of the pim genes.
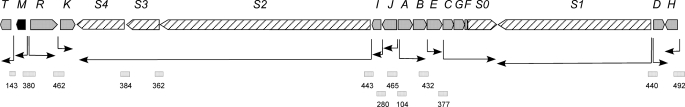
Of the 19 genes belonging to the pimaricin cluster, three of them (pimS1, pimD, and pimH) are presumed to be transcribed as monocistronic units, as can be deduced from their chromosomal arrangement (Fig. 1). Previous work had identified another five genes (pimT, pimM, pimR, pimK, and pimE) that are also transcribed as monocistrons (10, 13, 29, 30). The remaining genes of the cluster could be transcribed as polycistronic units. In fact, pimA and pimB, which encode a putative heterodimer ABC transporter involved in pimaricin secretion (31), show overlapping coding sequences (the pimB start codon is located 23 bp upstream from the pimA stop codon); they are thought to be translationally coupled and have been demonstrated to be transcribed as a bicistronic operon (13). However, evidence for the remaining putative polycistronic units was lacking.
FIGURE 1.
Organization of the pimaricin gene cluster. The pointed boxes indicate the direction of transcription. The pimM gene is indicated in black, and the polyketide synthase genes are shown as striped pointed boxes. The arrows indicate deduced transcriptional units. Square boxes, DNA fragments used in mobility shift experiments (sizes (bp) are indicated below).
We thus decided to analyze the possible co-transcription of neighboring genes by RT-PCR experiments. Total RNA was prepared from S. natalensis after growth for 48 h (when pimaricin is actively produced (5)). Primers were designed to obtain cDNAs corresponding to unabated transcription between two genes (supplemental Table S1). Transcripts were analyzed after 30 PCR cycles. A primer pair designed to amplify a cDNA of the lysA gene (encoding diaminopimelate decarboxylase) was used as an internal control (10). These analyses were carried out at least three times for each primer pair. Following this strategy, we corroborated the co-transcription of pimS2, pimS3, and pimS4, which was previously proposed based on their identical expression profile in RT-PCR experiments using total RNA from S. natalensis ΔpimM mutants and the absence of apparent transcriptional terminators in the short intergenic regions between them (13). Similarly, pimC, pimG, pimF, and pimS0 could also be co-transcribed because unabated transcription was observed between the upstream and the downstream gene. No transcripts were detected linking pimI and pimS2 or linking pimJ and pimI, thus suggesting that both pimI and pimS2 should have their own promoters (see below). Fig. 1 shows the deduced organization of transcriptional units.
Heterologous Expression of PimM and of Its Truncated Versions
The involvement of the product of the gene pimM in the regulation of the biosynthesis of the pimaricin molecule has been suggested on the basis of gene inactivation experiments (13) but has not been proven in vitro. Heterologous expression of PimM was carried out as a GST fusion protein following cloning into the pGEX-2T vector and transformation into E. coli BL21(DE3). A significant proportion of GST-PimM fusion protein was found in the soluble fraction and was purified by glutathione affinity chromatography (Fig. 2B). The purified GST-PimM fraction contained a detectable band on a Coomassie Blue-stained SDS-polyacrylamide gel consonant with the expected 47-kDa fusion protein. The identity of the fusion protein was verified by MALDI-TOF MS. GST-PimM fusion protein was fairly unstable, lasting only for 2 weeks when stored in 10 mm reduced glutathione, 50 mm Tris-HCl, pH 8.0, 20% glycerol at −80 °C.
PimM could not be separated from GST by using thrombin because, regardless of lacking canonical proteolytic sites in its sequence, it was completely degraded upon digestion. However, given that GST-tagged proteins have been successfully used in EMSAs (32), the fusion protein GST-PimM was used for in vitro experiments.
Similarly, two truncated versions of PimM (i) lacking the first 49 amino acid residues, which correspond to the PAS domain, and (ii) containing just the N-terminal LuxR DNA-binding domain (DBD; last 93 residues) were also expressed as GST fusion proteins (see “Experimental Procedures”) (Fig. 2A). The identity of the fusion proteins was also verified by MALDI-TOF MS. Interestingly, GST-PimMDBD turned out to be far more stable than GST-PimM, lasting up to 3 months at −80 °C.
GST-PimM Binds to Several Promoters of the Pimaricin Gene Cluster
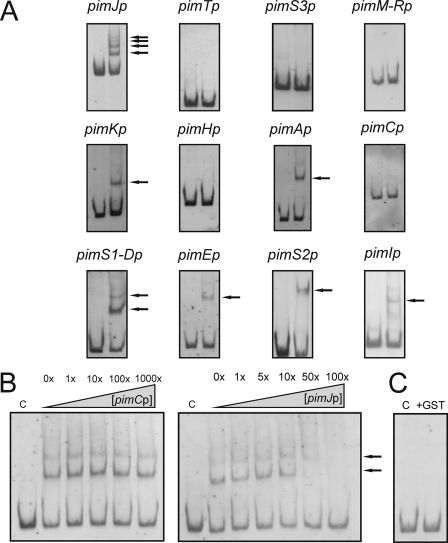
Incubation of GST-PimM with each labeled DNA fragment from the putative promoter regions selected was assessed (Fig. 3) using an EMSA (see “Experimental Procedures”). For each experiment, two negative control reactions were performed: absence of protein and use of GST (isolated separately). The appearance of a retarded band(s) was only observed upon incubation of GST-PimM with seven of the promoter regions analyzed. In these cases, the intensity of the retarded band(s) was diminished by the addition of the same unlabeled DNA (Fig. 3B). These regions were the pimK promoter (one retardation band), the pimS2 promoter (one retardation band), the pimI promoter (one retardation band), the pimJ promoter (four retardation bands), the pimA promoter (one shifted band), the pimE promoter (one retardation band), and the pimS1-D promoter (two retardation bands) (Fig. 3A). Promoter regions, such as pimTp, pimM-Rp, pimCp, or pimHp, were not retarded, indicating that PimM does not interact directly with them. In all cases, control reactions made with pure GST protein were negative, excluding a possible binding of this protein to the promoters (Fig. 3C). The specificity of binding of GST-PimM to target promoters was tested by competition with promoters that do not interact with PimM. Fig. 3B shows a competition experiment between pimJp and pimCp. The addition of 1–1000-fold higher concentrations of pimCp competitor DNA failed to diminish the intensities of the pimJp retardation bands.
FIGURE 3.
GST-PimM DNA binding assay results. A, EMSA of GST-PimM binding to different putative promoter regions. Left lane, control without protein; right lane, 60 μm GST-PimM protein. The pimJ promoter region was used as a positive control. The arrows indicate the DNA-protein complexes. Promoter names are indicated above the pictures. All experiments were carried out with 2 ng of labeled DNA probe. B, left panel, competition experiment between pimJp and pimCp. Note that 1000-fold higher concentrations of unlabeled pimCp competitor DNA failed to decrease the intensities of the pimJp retardation bands. Right panel, competition experiment between labeled pimJp and unlabeled pimJp. Both experiments were performed with 19 μm GST-PimM protein. C, control reaction with the pimJ promoter region and 60 μm pure GST protein. In all cases, lane C indicates control without protein.
The appearance of several shifted bands in some EMSAs indicates that several DNA-protein complexes were formed due to binding of increasing amounts of PimM. This can be explained by the presence of various binding sites for the protein. Once one binding site is occupied, further protein can bind other binding sites, thus accounting for the DNA-protein complexes of lower electrophoretic mobility.
Binding Ability Relies on the DNA-binding Domain
To test the effect of the deletion of the PAS domain from the PimM N-terminal region on the ability of the protein to bind to its cognate promoters, two truncated versions of PimM were created and fused to GST. These fusion proteins, GST-PimMΔPAS and GST-PimMDBD, contain the last 143 and 93 amino acids of PimM, starting at the residues Ser50 and Ser100, respectively (Fig. 2A).
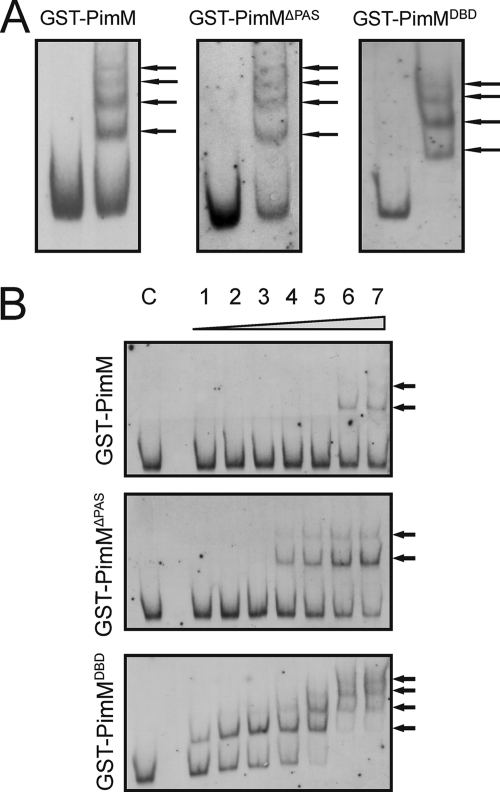
Binding of the different forms of the protein to different promoters was also studied by EMSA. Fig. 4 shows the results with the pimJ promoter region. Interestingly, the absence of the PAS domain did not seem to affect PimM binding ability because no significant differences between GST-PimM and its truncated forms were found regarding the number of shifted bands when the assay was carried out with 60 μm protein, probably indicating that at such protein concentration, all binding sites are occupied (Fig. 4A). Moreover, the pattern of regions retarded and not retarded and the maximum number of shifted bands in each case did not vary upon removal of the PAS domain (not shown), thus suggesting that the distinct recognition of target promoters is also independent of the PAS domain.
FIGURE 4.
GST-PimMDBD binds promoters with highest affinity. Analysis by EMSA of the binding of the different GST fusion proteins to the pimJ promoter. A, binding of the different GST fusion proteins in a protein-saturated assay (60 μm). B, lane C, control without protein; lane 1, 19 nm protein; lane 2, 95 nm protein; lane 3, 190 nm protein; lane 4, 950 nm protein; lane 5, 1.9 μm protein; lane 6, 9.5 μm protein; lane 7, 19 μm protein. The arrows indicate the DNA-protein complexes. Top, GST-PimM; middle, GST-PimMΔPAS; bottom, GST-PimMDBD.
PAS Domain Reduces Binding Affinity
In order to investigate the binding affinity of the different forms of the protein, we used a gradient of protein concentration and the same amount of labeled probe (2 ng) to perform EMSAs. Quantification of the integral optical density of retarded and unretarded bands at the lowest concentration of protein able to produce a detectable shift in the labeled probe (Fig. 4B) was used to get an estimate of protein affinities. Quantification was performed as indicated under “Experimental Procedures.”
Although 9.5 μm GST-PimM was required to produce the first shifted band in EMSA, 0.95 μm GST-PimMΔPAS and 19 nm GST-PimMDBD were sufficient to produce the same effect (Fig. 4B). Values of integral optical density for the first shifted band were 938, 866, and 901 for GST-PimM, GST-PimMΔPAS, and GST-PimMDBD, respectively, whereas integral optical density values for the unretarded band were 1724, 1968, and 1672. Values for the control reactions (with no protein) were 2589, 2711, and 2741, thus indicating that the film was not saturated. These results indicate that the three fusion proteins bind a similar proportion of labeled probe at very different concentrations, hence revealing that truncated forms of the protein have significantly higher affinity. Thus, GST-PimMDBD shows a 50-fold higher affinity than GST-PimMΔPAS and a 500-fold higher affinity than GST-PimM. Similar results were obtained when we studied other promoters (not shown).
DNase I Protection Studies Reveal Binding Sites in pim Promoters
To determine the PimM binding sequences, the promoter regions shown above to be retarded in EMSA were studied by DNase I protection analysis. GST-PimMDBD protein (25.5 μm) was tested using 5′-end fluorescein-labeled DNA fragments (23). All analyses were carried out in triplicate.
Results of the analysis of the pimKp promoter region showed a protected stretch extending for 24 bp of the coding strand. This protected region is located at nucleotide positions −114 to −91 with respect to the pimK translational ATG start site. The protection of the reverse strand of pimKp was 5 nucleotides larger than that of the coding strand (positions −113 to −85), and both regions are displaced 1–6 nucleotides (supplemental Fig. S1).
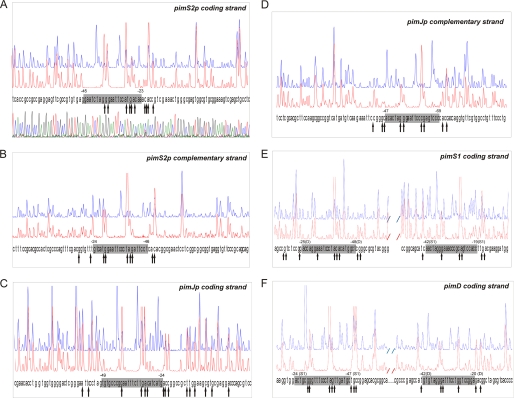
Footprinting assays of the pimS2p region revealed a 23-nucleotide protection in the coding strand (positions −189 to −167 with respect to the pimS2 translation start site). In the bottom strand, the protected sequence was also 23 bp long, spanning from position −191 to −169 (Fig. 5, A and B). In this case, both protected regions were displaced by 2 nucleotides.
FIGURE 5.
Identification of binding sites. DNase I footprints of the GST-PimMDBD protein bound to the promoter regions of pimS2 (A and B), pimJ (C and D), and pimS1-D (E and F). In each panel, the upper electropherogram (blue line) shows the control reaction. The protected nucleotide sequence is boxed; hypersensitive sites (arrows) are also indicated. Sequencing reactions are not included except in A. Coordinates are from the transcriptional start point. In the case of the bidirectional promoter pimS1-Dp, the gene to which coordinates refer is indicated in parentheses.
Results of the analysis of the pimIp region showed a protected region extending for 23 bp of the coding strand (positions −121 to −99 with respect to the pimI translational start site). The length of the protection of the reverse strand of the pimI promoter was the same (positions −120 to −98). Both protected regions were displaced just one position (supplemental Fig. S1).
In the case of the pimJ promoter, a protected region of 26 nucleotides was observed in the coding strand of pimJ (positions −205 to −180 from the pimJ translation start codon). In the bottom strand, the protected sequence was 23 bp long, at positions −225 to −203 (Fig. 5, C and D). These protected regions were almost completely displaced (i.e. they overlap by only 3 nucleotides and are thought to belong to two different operators). It is interesting to note that in this case, and contrary to what has been observed with other pim promoters, protection in the coding strand is not accompanied by an equivalent/similar protection in the opposite strand. Further experimental analyses will be required to establish the precise reason for this result. Both protected regions could constitute two independent operators whose overlapping arrangement would preclude a canonical protection (see “Discussion”).
Results of the analysis of the pimAp region showed a protected region extending for 22 bp of the coding strand (positions −41 to −20 with respect to the pimA translational start site). The protection of the reverse strand of the pimA promoter was 1 nucleotide larger than that of the coding strand (positions −45 to −23). In this case, both protected regions were displaced by 2–3 nucleotides (supplemental Fig. S1).
Footprinting assays of the pimEp region revealed a 24-nucleotide protection in the coding strand (positions −46 to −23 with respect to the pimE translation start site). In the bottom strand, the protected sequence was also 24 bp long, spanning from position −43 to −20 (supplemental Fig. S1). In this case, both protected regions were displaced by 3 nucleotides.
It is noteworthy that when we carried out the footprinting analysis with the bidirectional pimS1-Dp promoter, two protected areas were observed in each strand, in agreement with the appearance of two retardation bands in EMSA experiments. The two areas are 69 nucleotides apart in the S. natalensis chromosome. The first protected area, extending for 24 bp of the pimS1 coding strand, is located at nucleotide positions −112 to −89 with respect to the pimS1 ATG translational start site. The protection of the reverse strand was also 24 bp long (positions −117 to −94), both regions being displaced 5 nucleotides. The second protected area extended for 23 bp of the coding strand of pimD, at positions −79 to −57 from the pimD translational start site. In the bottom strand, the protected sequence was also 23 bp long, spanning from position −84 to −62 (Fig. 5, E and F). In this case, both regions were also displaced by 5 nucleotides.
In almost all cases, DNase I-hypersensitive positions flanked the protected sequence, indicating altered DNA topology after incubation with GST-PimMDBD. Some hypersensitive positions were also found inside the target sequences, typically two areas (Fig. 5), thus suggesting that PimM bends DNA, making those positions accessible to DNase I digestion.
Characterization of Promoters
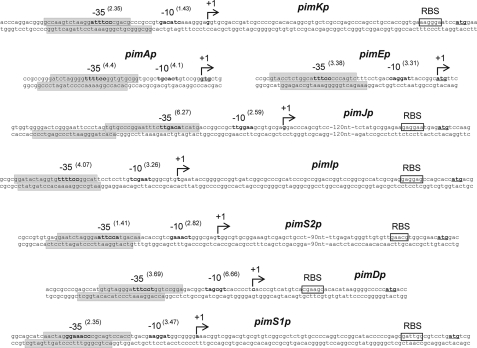
To determine the transcriptional start sites of target promoters and to corroborate the monocistronic nature of some genes, such as pimJ and pimI, 5′-RACE experiments were carried out. Once the +1 sites were known, the corresponding −10 and −35 boxes of each promoter were established by comparison with the matrices reported by Bourn and Babb (28) for Streptomyces that take into account the nucleotides occurring in 13-nucleotide stretches, including the −10 or −35 consensus hexamers (see “Experimental Procedures”). Results are summarized in Fig. 6.
FIGURE 6.
Transcriptional start site of promoters regulated by PimM and protected regions. The position of the transcriptional start site was determined by 5′-RACE. The putative −10 and −35 hexanucleotides are in boldface type. Scores resulting from the comparison with the matrices reported by Bourn and Babb (28) for Streptomyces are indicated in parentheses. The TSP is indicated by a bent arrow and boldface type. Nucleotides showing homology with the 16 S RNA, which could form a ribosome-binding site, are framed with a box labeled RBS. The start codon is shown in boldface type and underlined. Protected nucleotide sequences are indicated with shaded boxes. Note that pimA and pimE are transcribed as leaderless mRNAs, thus lacking RBS.
The pimK transcription start point (TSP) is located at a guanine 69 bp upstream from the ATG codon. Analysis of the region upstream of the TSP revealed that the −10 box with the highest score to the consensus Streptomyces was GAC-ATC, centered at 10 nucleotides from the start site. A search using combined class C-class A matrices (28) revealed a −35 box ATTTCC separated by 13 nucleotides, with a score of 2.35 for the 35-nt promoter sequence. It is noteworthy that the protected region observed in the footprinting assays is only 22 nt away from the TSP site, covering the −35 hexamer box (Fig. 6).
A single RACE product of ∼490 bp was observed for pimS2, thus supporting the conclusion that this gene has its own promoter, and consequently pimI is transcribed as monocistronic unit. The TSP of pimS2 is located at a thymine 145 bp upstream from the ATG codon. Analysis of the region upstream of the TSP revealed the presence of a −10 box GAAACT (score 2.82), centered at 10 nucleotides from the start site, and a −35 box ATTCCA separated by 14 nucleotides. As in the former case, the protected region in the coding strand lies 22 nt upstream from the TSP site, covering the −35 hexamer box of the promoter.
For pimI, a single RACE product of 410 bp was observed, thus supporting the conclusion that this gene has its own promoter. The pimI transcription start point corresponds to a thymine located 76 bp upstream from the ATG codon (Fig. 6). The sequence TCGAAT (score 3.26), centered at position −10, constitutes the −10 consensus, and a −35 box TTTTCC (score 4.07) was identified at a 14-nt distance. Interestingly, the protected region in the pimI sense strand is 23 nucleotides away from the TSP site, and again both protected regions cover the −35 box of pimIp.
The pimJ TSP is located at a guanine 156 bp upstream from the ATG codon. Analysis of the region upstream of the TSP revealed that the −10 box with the highest score was TTGGAA, centered at 11 nucleotides from the start site, and the −35 box TTGACA (score 6.27) at a 14-nt distance. The protected region in the pimJ sense strand is only 24 nucleotides away from the TSP site and covers the −35 hexamer box of the promoter (Fig. 6).
For pimA, the TSP was identified at the first guanine of the GTG start codon, thus indicating that this gene is transcribed as a leaderless mRNA and that the starts of transcription and translation coincide in pimA. The Patser analysis of the upstream sequence revealed TGCACT and TTTTCC as the −10 and −35 boxes (scores 4.1 and 4.4, respectively). Both boxes are separated by 14 nt, with the −10 hexamer centered at 9 nt from the TSP. Both protected regions, in the sense and antisense strands, covered the −35 box of the promoter.
The TSP of pimE was located at the adenine of the ATG codon, thus indicating that this gene is also transcribed as a leaderless mRNA. The analysis of the upstream sequence using the matrices of Bourn and Babb revealed a clear promoter, with the −10 box CAGGAT located 8 nt upstream from the observed TSP and the −35 box TTTCCC (score 3.38) separated by 15 nt. As in former cases, the protected regions covered the −35 hexamer box of the promoter.
The pimS1 TSP is located at an adenine 70 bp upstream from the ATG codon, the −10 box AAGGAT is centered at 10 nt from the TSP, and the −35 box GAAACC is at a 16-nt distance (Fig. 6). Again protected regions covered the −35 hexamer box of the promoter.
In the case of pimD, the TSP was located at a cytosine situated 37 bp upstream from the ATG codon. The −10 and −35 boxes (TAGCGT and TTTCCT, respectively) were centered at positions −10 and −30 from the TSP and are separated by 14 nt (Fig. 6). The protected region in the pimD sense strand is 20 nucleotides away from the TSP site. As in former cases, both protected regions cover the −35 box of the promoter.
Information Content Analysis of the Nucleotide Sequences
An information-based model of the binding site was constructed, taking into account the 16 protected regions observed in the footprinting assays. A sequence logo (33) that depicts the binding site is shown in Fig. 7. This site spans 14 nucleotides and adjusts to the consensus TVGGGAWWTCCCBA (where V represents A, C, or G; W is A or T; and B is C, G, or T). It is noteworthy that the binding site displays dyad symmetry.
FIGURE 7.
Sequence logo of PimM binding site. Sequence logo of the nucleotides sequences that constitute the PimM binding site. The logo was constructed with the 16 sequences shown to be protected by PimM binding in footprinting experiments. The height of each letter is proportional to the frequency of the base, and the height of the letter stack shows the conservation in bits at that position (33). Note the high conservation of the eight central positions. The total information (Rsequence) for the binding site is 11.42 bits (0.71 bits/base). An alignment of the 16 binding sites is indicated below. Information contents (Ri) of each binding site are shown.
The individual information contents of the binding sites, or Ri (24), allowed us to analyze the requirements of PimDBD for DNA recognition. All operators showed high Ri values, ranging from 6 bits in the case of the binding site at the pimE promoter to 18 bits in one of the operators of the pimJ promoter (Fig. 7). The total information (Rsequence) for the binding site showed a mean value of 11.42 (0.71 bits/base).
Validation of the Consensus Binding Site
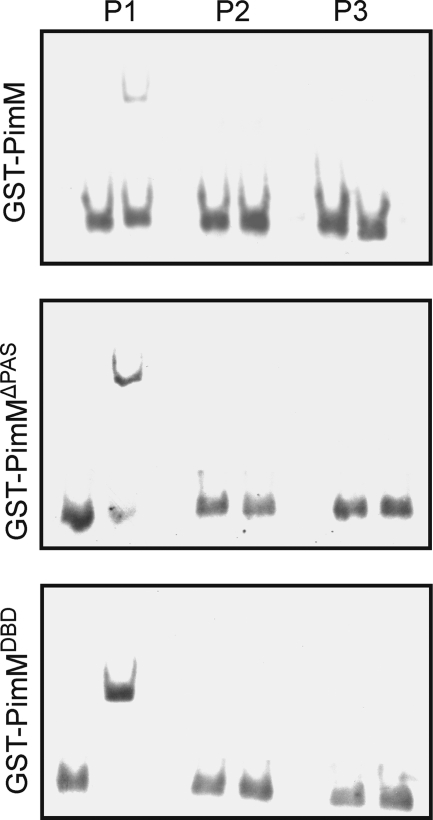
In order to validate the consensus binding site, three DNA duplexes were constructed: one containing the canonical binding site TAGGGAATTCCCTA (P1), a second one with the four central positions deleted (TAGGGCCCTA) (P2), and a third one where those central positions were replaced by GCCG (TAGGGGCCGCCCTA) (P3). The binding of the different forms of the protein to the duplexes was then studied by EMSA. Interestingly, only the P1 duplex was able to form a complex with the proteins, and it did it with the three of them (Fig. 8). This result validates the proposed binding site and demonstrates that the three forms of the protein bind the same site. Furthermore, the fact that the absence of the PAS domain did not affect protein binding demonstrates that binding specificity is independent of the PAS domain.
FIGURE 8.
Validation of PimM binding site. Shown is analysis by EMSA of the binding of the different GST fusion proteins to synthetic duplexes. Base composition of the duplexes is indicated under “Experimental Procedures.” Left lane, control without protein; right lane, 9 μm protein. Top, GST-PimM; middle, GST-PimMΔPAS; bottom, GST-PimMDBD. Note that only the P1 duplex, with the consensus binding site, forms a complex with the proteins.
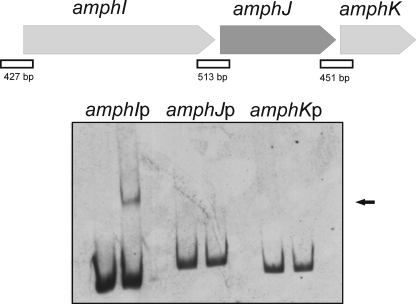
GST-PimM Binds the Promoter Region of amphI
Given the high degree of conservation of PimM homologous regulators from other polyene producers, and in order to check the general applicability of the consensus binding site, we searched for the presence of the consensus in the amphotericin cluster from S. nodosus. We thus found the sequence ACTAGGGATTTCCTGCCG in the putative promoter region of the gene amphI, which encodes one of the six modular polyketide synthase proteins involved in amphotericin biosynthesis. This sequence fits well to the consensus (Ri of 10.49), which prompted us to use this region to perform EMSA assays with GST-PimM. As shown in Fig. 9, GST-PimM binds strongly to the S. nodosus amphI promoter region, whereas it does not to the upstream regions of the genes amphJ and amphK, which do not contain sequences that fit the consensus. This result proves the applicability of the findings reported here to other antifungal polyketides.
FIGURE 9.
GST-PimM binds the amphI promoter. Shown is analysis by EMSA of GST-PimM binding to different putative promoter regions of some amphotericin genes. Left lane, control without protein; right lane, 60 μm GST-PimM protein. The arrow indicates the DNA-protein complex. Putative promoter names are indicated above the pictures. All experiments were carried out with 2 ng of labeled DNA probe. The organization of the studied genes in the S. nodosus chromosome is indicated above. Square boxes indicate DNA fragments used in mobility shift experiments.
DISCUSSION
On the left-hand side of the pimaricin gene cluster there is a gene, pimM, whose product is a regulator that combines an N-terminal PAS sensor domain (11, 12) with a C-terminal HTH motif of the LuxR type for DNA binding. The presence of a PAS-like domain within PimM suggests that this protein could respond to the energy levels in the cell (34), whereas the HTH motif suggested the ability of PimM to bind DNA (35) and thus regulate the expression of pimaricin genes. The majority of prokaryotic PAS domains function as sensor modules of sensor kinases of two-component systems (11), but this is not the case of PimM. The absence of pimaricin production upon disruption of the gene by removal of the HTH domain clearly indicated that PimM behaves as an activator of pimaricin biosynthesis (13). Highly similar regulators are encoded by all characterized polyene macrolide biosynthetic gene clusters, including amphotericin (AmphRIV) (14), candicidin (FscRI) (15), nystatin (NysRIV) (16), or filipin (PteF) (17). Among them, only NysRIV and PimM have been characterized to some extent. Preliminary results indicated that both PimM and NysRIV regulators might follow a similar regulatory pattern for the expression of their respective polyenes (13), thus suggesting that this pattern could be shared by the homologous regulatory genes found in other polyene biosynthetic gene clusters.
Electrophoretic mobility shift assays have been used here to prove the direct binding of PimM to certain promoters of the pim genes. Given that GST-tagged proteins have been successfully used in EMSAs (22, 32), GST-PimM fusion proteins were used for in vitro experiments. Both the complete PimM protein and truncated forms of the protein lacking the PAS domain (PimMΔPAS) and containing just the DNA-binding domain (PimMDBD) were found to bind with high affinity to the promoter regions of the same pim genes and to produce the same pattern of shifted bands in each case, thus suggesting that binding specificity is independent of the PAS domain. Moreover, the presence of the PAS domain severely reduced the affinity of binding to target promoters, thus suggesting that the role of the PAS domain could be to limit affinity of PimM toward its targets. Similar results have been observed with response regulators of two-component systems, such as PhoB from E. coli or PhoP from Streptomyces coelicolor, where the presence of the N-terminal domain reduces the binding efficiency of the C-terminal DBD domain (22, 36).
One shift band was observed upon incubation of PimM with the pimKp, pimS2p, pimIp, pimAp, or pimEp promoter demonstrating that this regulator binds directly to these regions. Two shift bands were obtained with the pimS1-D bidirectional promoter region at increasing protein concentrations (not shown). The formation of two complexes of different molecular weight indicates that more than one binding site is present in this region, a result that was confirmed by footprinting studies where two distinct protection areas were observed. In the case of the pimJ promoter, four retardation bands were observed in EMSAs, whereas only two protected areas were observed in footprinting experiments. This result, bearing in mind that weak DNA-protein interactions are frequently stabilized once in the polyacrylamide gel, could be explained considering that two of the retardation bands are due to unstable interactions, although the possibility that they are derived from protein-protein interactions cannot be excluded.
EMSA results explain previous gene expression analyses in S. natalensis wild type and ΔpimM mutant by RT-PCR (13) and demonstrate that the control exerted by PimM on the expression of the genes pimK, pimS2S3S4, pimI, pimJ, pimAB, pimE, pimS1, and pimD takes place through direct binding to the promoters of these genes. The lack of binding to the promoter region of pimC, which is thought to drive the expression of the multicistronic operon pimCGFS0, suggests that the lack of expression of these genes upon gene disruption of pimM (13) is mediated by the action of another hierarchical regulator that would be activated by PimM. Further experimental analyses will be required to test this hypothesis.
The large number of promoters directly controlled by PimM is particularly interesting. This regulator binds to eight of the 12 promoters present in the pimaricin cluster, in some cases to promoters of consecutive genes (e.g. pimI, pimJ, and pimS2S3S4). To our knowledge, this is unprecedented for an antibiotic pathway-specific regulator because all antibiotic pathway-specific regulators characterized to date, which are mostly SARPs, have been proved to bind at most two or three promoters.
Footprinting analyses revealed protected sequences of 22–29 nucleotides in target promoters. Typically, the protected region in the sense strand of the activated gene is accompanied by a protection in the complementary strand, both protected regions being slightly displaced (see Fig. 6). This could be explained by the binding of one monomer of the protein to each strand. The only exception is the protection achieved when we used the promoter region pimJp. In this case, protection in the sense strand is not accompanied by a protection in the opposite strand, thus providing evidence that protein monomers bind DNA from one face of the DNA. A possible explanation for this result came from the determination of the binding site sequence, which revealed that, in fact, this DNA region contains two independent operators with an overlapping arrangement. It is plausible that this arrangement precludes a canonical protection, maybe by steric hindrance. Future experimental approaches, now under way, will hopefully determine the reason for this result.
All protected sequences were flanked by DNase I-hypersensitive positions, suggesting an altered DNA topology after incubation with GST-PimMDBD. Some hypersensitive positions were also found inside the target sequences, typically two areas, thus suggesting that PimM bends DNA, making those positions accessible to DNase I digestion (37).
Analysis of the protected sequences in the DNase I protection assays together with the identification of transcriptional start points in the target promoters revealed that there was a consistent overlap of the regulator binding site with the putative −35 region of each promoter. This interaction is presumed to enable protein-protein contacts between RNA polymerase and PimM as an important functional aspect in transcriptional activation. This corresponds to a Class II activation mechanism where PimM would contact domain 4 of the RNA polymerase σ subunit, resulting in recruitment of RNA polymerase to the promoter (38).
A comparison of the 16 protected sequences shown in this article permitted the development of an information-based model of the binding site. A sequence logo (33) that depicts the binding site is shown in Fig. 7. This site spans 14 nucleotides and adjusts to the consensus TVGGGAWWTCCCBA. The conservation of the eight central positions is very clear and emphasizes the importance of these bases to establish specific protein-DNA contacts. Most of the positions of the core display values near to or higher than 1 bit; thus, given that proteins usually bind DNA from one face, it is expected that these binding site positions are located at the major groove of a B-form of DNA (39). It is noteworthy that the binding site displayed dyad symmetry. This is in agreement with the binding sites of regulators of the LuxR type, such as LuxR (40), CepR (41), or TraR (42). Dyad symmetry is necessary for binding to a protein that has a 2-fold rotational symmetry in its DNA-binding domain; thus, it is very likely that the DNA-binding domain of PimM will have a 2-fold rotational symmetry.
Confirmation of the binding site was obtained by comparing EMSAs using the different forms of the protein and (46–50-bp) DNA duplexes possessing or lacking the consensus. Either the modification or the deletion of the four central nucleotides of the consensus abrogated DNA binding, thus validating the proposed binding site. Furthermore, the absence of the PAS domain did not prevent protein binding, thus corroborating that specificity does not rely on the PAS domain. This is in clear contrast with previous studies carried out with other transcription factors, such as Drosophila bHLH/PAS transcription factors, where the PAS domain mediates all of the features conferring specificity and the distinct recognition of target genes (43).
To prove the general applicability of the consensus binding site, we searched for the presence of the consensus in the amphotericin cluster from S. nodosus. Amphotericin has been the leading antifungal drug for many years and is still considered the “gold standard” in antifungal chemotherapy. Analysis of the regions upstream of the biosynthetic genes allowed us to identify a matching sequence in the putative promoter region of amphI, one of the polyketide synthase genes involved in macrolide construction. EMSA assays with this region showed that PimM can bind sequences that fit the consensus in other polyene biosynthetic gene clusters, thus proving the general applicability of the binding site reported and suggesting that the orthologous regulators of polyene biosynthesis share the same regulatory pattern.
PimM constitutes, to our knowledge, the first transcriptional regulator of antibiotic biosynthesis, not belonging to the SARP family, whose target binding site has been determined, and this study represents the first molecular characterization of the mode of action of a polyene macrolide regulator. Polyenes represent a major class of antifungal agents that are also active against parasites, enveloped viruses, and prion diseases (4), but despite their general interest, very little is known about the regulation of their biosynthesis. This report now provides important clues toward understanding the regulation of polyene biosynthesis and will prove valuable for maximizing yields of these important compounds.
Supplementary Material
Acknowledgments
We thank Dr. A. Rodríguez-García for advice and critical contributions to this work and J. Merino, B. Martín, M. A. Vidales, and A. Casenave for excellent technical assistance.
This work was supported by Spanish Ministerio de Ciencia e Innovación Grants BIO2007-67585 and BIO2010-19911 (to J. F. A.), Junta de Castilla y León Grupo de Excelencia GR117, Ministerio de Ciencia e Innovación F.P.U. Fellowships AP2005-3644 (to J. S.-A.) and AP2007-02055 (to T. D. P.), a fellowship from the University of León (to S. M. G.), and Junta de Castilla y León Contract E-24-2009-0053914 (to C. M. V.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Fig. S1.
- SARP
- Streptomyces antibiotic regulatory protein
- RACE
- rapid amplification of cDNA ends
- DBD
- DNA-binding domain
- TSP
- transcription start point
- nt
- nucleotide(s).
REFERENCES
- 1. Bibb M. (1996) Microbiology 142, 1335–1344 [DOI] [PubMed] [Google Scholar]
- 2. Aparicio J. F., Mendes M. V., Antón N., Recio E., Martín J. F. (2004) Curr. Med. Chem. 11, 1645–1656 [DOI] [PubMed] [Google Scholar]
- 3. te Welscher Y. M., ten Napel H. H., Balagué M. M., Souza C. M., Riezman H., de Kruijff B., Breukink E. (2008) J. Biol. Chem. 283, 6393–6401 [DOI] [PubMed] [Google Scholar]
- 4. Caffrey P., Aparicio J. F., Malpartida F., Zotchev S. B. (2008) Curr. Top. Med. Chem. 8, 639–653 [DOI] [PubMed] [Google Scholar]
- 5. Mendes M. V., Recio E., Fouces R., Luiten R., Martín J. F., Aparicio J. F. (2001) Chem. Biol. 8, 635–644 [DOI] [PubMed] [Google Scholar]
- 6. Mendes M. V., Antón N., Martín J. F., Aparicio J. F. (2005) Biochem. J. 386, 57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aparicio J. F., Martín J. F. (2008) Mol. Biosyst. 4, 804–809 [DOI] [PubMed] [Google Scholar]
- 8. Kells P. M., Ouellet H., Santos-Aberturas J., Aparicio J. F., Podust L. M. (2010) Chem. Biol. 17, 841–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Martín J. F., Aparicio J. F. (2009) Methods Enzymol. 459, 215–242 [DOI] [PubMed] [Google Scholar]
- 10. Antón N., Mendes M. V., Martín J. F., Aparicio J. F. (2004) J. Bacteriol. 186, 2567–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Taylor B. L., Zhulin I. B. (1999) Microbiol. Mol. Biol. Rev. 63, 479–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hefti M. H., Françoijs K. J., de Vries S. C., Dixon R., Vervoort J. (2004) Eur. J. Biochem. 271, 1198–1208 [DOI] [PubMed] [Google Scholar]
- 13. Antón N., Santos-Aberturas J., Mendes M. V., Guerra S. M., Martín J. F., Aparicio J. F. (2007) Microbiology 153, 3174–3183 [DOI] [PubMed] [Google Scholar]
- 14. Carmody M., Byrne B., Murphy B., Breen C., Lynch S., Flood E., Finnan S., Caffrey P. (2004) Gene 343, 107–115 [DOI] [PubMed] [Google Scholar]
- 15. Chen S., Huang X., Zhou X., Bai L., He J., Jeong K. J., Lee S. Y., Deng Z. (2003) Chem. Biol. 10, 1065–1076 [DOI] [PubMed] [Google Scholar]
- 16. Sekurova O. N., Brautaset T., Sletta H., Borgos S. E., Jakobsen Ø. M., Ellingsen T. E., Strøm A. R., Valla S., Zotchev S. B. (2004) J. Bacteriol. 186, 1345–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Omura S., Ikeda H., Ishikawa J., Hanamoto A., Takahashi C., Shinose M., Takahashi Y., Horikawa H., Nakazawa H., Osonoe T., Kikuchi H., Shiba T., Sakaki Y., Hattori M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12215–12220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kieser T., Bibb M. J., Buttner M. J., Chater K. F., Hopwood D. A. (2000) Practical Streptomyces Genetics, John Innes Foundation, Norwich, UK [Google Scholar]
- 19. Aparicio J. F., Colina A. J., Ceballos E., Martín J. F. (1999) J. Biol. Chem. 274, 10133–10139 [DOI] [PubMed] [Google Scholar]
- 20. Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory Press, NY [Google Scholar]
- 21. Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 22. Sola-Landa A., Rodríguez-García A., Franco-Domínguez E., Martín J. F. (2005) Mol. Microbiol. 56, 1373–1385 [DOI] [PubMed] [Google Scholar]
- 23. Rodríguez-García A., Ludovice M., Martín J. F., Liras P. (1997) Mol. Microbiol. 25, 219–228 [DOI] [PubMed] [Google Scholar]
- 24. Schneider T. D. (1997) J. Theor. Biol. 189, 427–441 [DOI] [PubMed] [Google Scholar]
- 25. Bi C., Rogan P. K. (2006) BMC Bioinformatics 7, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hertz G. Z., Stormo G. D. (1999) Bioinformatics 15, 563–577 [DOI] [PubMed] [Google Scholar]
- 27. van Helden J. (2003) Nucleic Acids Res. 31, 3593–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bourn W. R., Babb B. (1995) Nucleic Acids Res. 23, 3696–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mendes M. V., Recio E., Antón N., Guerra S. M., Santos-Aberturas J., Martín J. F., Aparicio J. F. (2007) Chem. Biol. 14, 279–290 [DOI] [PubMed] [Google Scholar]
- 30. Vicente C. M., Santos-Aberturas J., Guerra S. M., Payero T. D., Martín J. F., Aparicio J. F. (2009) Microb. Cell Fact. 8, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Aparicio J. F., Caffrey P., Gil J. A., Zotchev S. B. (2003) Appl. Microbiol. Biotechnol. 61, 179–188 [DOI] [PubMed] [Google Scholar]
- 32. Mendes M. V., Tunca S., Antón N., Recio E., Sola-Landa A., Aparicio J. F., Martín J. F. (2007) Metab. Eng. 9, 217–227 [DOI] [PubMed] [Google Scholar]
- 33. Schneider T. D., Stephens R. M. (1990) Nucleic Acids Res. 18, 6097–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ponting C. P., Aravind L. (1997) Curr. Biol. 7, R674–R677 [DOI] [PubMed] [Google Scholar]
- 35. Stevens A. M., Dolan K. M., Greenberg E. P. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 12619–12623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ellison D. W., McCleary W. R. (2000) J. Bacteriol. 182, 6592–6597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pérez-Martín J., Rojo F., de Lorenzo V. (1994) Microbiol. Rev. 58, 268–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Browning D. F., Busby S. J. (2004) Nat. Rev. Microbiol. 2, 57–65 [DOI] [PubMed] [Google Scholar]
- 39. Schneider T. D. (1996) Methods Enzymol. 274, 445–455 [DOI] [PubMed] [Google Scholar]
- 40. Pompeani A. J., Irgon J. J., Berger M. F., Bulyk M. L., Wingreen N. S., Bassler B. L. (2008) Mol. Microbiol. 70, 76–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Weingart C. L., White C. E., Liu S., Chai Y., Cho H., Tsai C. S., Wei Y., Delay N. R., Gronquist M. R., Eberhard A., Winans S. C. (2005) Mol. Microbiol. 57, 452–467 [DOI] [PubMed] [Google Scholar]
- 42. Vannini A., Volpari C., Gargioli C., Muraglia E., Cortese R., De Francesco R., Neddermann P., Marco S. D. (2002) EMBO J. 21, 4393–4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zelzer E., Wappner P., Shilo B. Z. (1997) Genes Dev. 11, 2079–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.