Abstract
Human norepinephrine (NE) deficiency (or dopamine β-hydroxylase (DBH) deficiency) is a rare congenital disorder of primary autonomic failure, in which neurotransmitters NE and epinephrine are undetectable. Although potential pathogenic mutations, such as a common splice donor site mutation (IVS1+2T→C) and various missense mutations, in NE deficiency patients were identified, molecular mechanisms underlying this disease remain unknown. Here, we show that the IVS1+2T→C mutation results in a non-detectable level of DBH protein production and that all three missense mutations tested lead to the DBH protein being trapped in the endoplasmic reticulum (ER). Supporting the view that mutant DBH induces an ER stress response, exogenous expression of mutant DBH dramatically induced expression of BiP, a master ER chaperone. Furthermore, we found that a pharmacological chaperone, glycerol, significantly rescued defective trafficking of mutant DBH proteins. Taken together, we propose that NE deficiency is caused by the combined abnormal processing of DBH mRNA and defective protein trafficking and that this disease could be treated by a pharmacological chaperone(s).
Keywords: Genetic Diseases, Intracellular Trafficking, Mutant, Neurobiology, Neurotransmitters, Dopamine beta-Hydroxylase, NE Deficiency, Missense Mutations, Splice Mutation
Introduction
Norepinephrine (NE)3 is a key neurotransmitter that is synthesized in both the CNS and the peripheral nervous system and regulates many essential functions, including attention, memory, emotion, and autonomic and cardiovascular function. Like dopamine (DA) and epinephrine, NE is a catecholamine neurotransmitter and is synthesized by oxidative hydroxylation of DA catalyzed by dopamine β-hydroxylase (DBH). DBH is a hallmark protein of noradrenergic cells and is specifically expressed in noradrenergic neurons and chromaffin cells of the adrenal medulla. Among catecholamine-synthesizing enzymes, DBH is unique in that it is localized within large dense core vesicles (LDCV), where it exists in both soluble and membrane-bound forms (1). During neurotransmission, the soluble form of DBH is released into the synaptic cleft together with NE by exocytosis (2). As a result of such synaptic release, both NE and DBH are readily detectable in plasma and cerebrospinal fluid (3).
NE deficiency (or DBH deficiency) is a rare congenital disorder, first described in 1986 (4, 5). Given the fundamental role of NE in CNS and peripheral nervous system functions, the report of adult patients with undetectable NE is both surprising and interesting. The report of frequent miscarriages and spontaneous abortions in mothers of known NE deficiency cases (5, 6) suggests the interesting possibility that there could be many more undiagnosed fetal and neonatal deaths resulting from NE deficiency and that those adult patients are lucky survivors. In line with this, a DBH knock-out mouse study showed less than 5% live births (7), where mortality appeared to be due to cardiovascular failure caused by NE deficiency in utero, which is reminiscent of tyrosine hydroxylase null mice (8). Pathophysiologically, NE deficiency is a severe autonomic disorder exhibiting sympathetic noradrenergic failure and adrenomedullary failure but intact vagal and sympathetic cholinergic functions. Clinically, NE-deficient patients exhibit severe deficits in autonomic regulation of cardiovascular function, predisposing them to orthostatic hypotension. Biochemically, these patients display characteristic perturbations in the level of catecholamines: undetectable NE and its metabolites and highly elevated DA and its metabolites. Consistent with this, the DBH protein is undetectable in these patients' plasma (enzyme assay; see Refs. 4, 5, and 9), cerebrospinal fluid (radioimmunoassay; see Ref. 9), and sympathetic fibers (immunocytochemistry; see Ref. 10).
We and others have previously reported a mutation, in the DBH gene of patients, at the splice donor site of the first exon-intron junction and several missense mutations associated with the NE deficiency syndrome (11, 12). Because these mutations were not found in DNA samples of healthy subjects, we suggested that they caused the observed NE deficiency. The mutation in the splice donor site (IVS1+2T→C) of DBH resulted in abnormal mRNA splicing and generated a transcript containing a premature stop codon as well as a normal transcript (12). Interestingly, all mutations in the coding region so far identified in NE-deficient patients are amino acid substitutions (Table 1). Together, the pathogenic mechanisms underlying NE deficiency remain unclear.
TABLE 1.
Potential pathogenic mutations in the DBH gene identified in NE deficiency
| Mutation designation | Location | Nucleotide substitution | Type of mutation | Source/Reference |
|---|---|---|---|---|
| IVS1+2T→C | Intron 1 | 297 + 2T→C | Splice site | Refs. 11 and 12 |
| D100E | Exon 2 | 300C→A | Missense | Ref. 12 |
| E192fs | Exon 3 | 575delA | Frame shift | Ref. 11 |
| C225F | Exon 4 | 764G→T | Missense | Ref. 11 |
| D331N | Exon 6 | 991G→A | Missense | Ref. 12 |
| A348E | Exon 6 | 1043C→A | Missense | This work |
| Y542C | Exon 11 | 1625A→G | Missense | Ref. 11 |
In this study, we report a previously unidentified missense mutation found in a new NE-deficient patient carrying a mutation in exon 6 resulting in the substitution of glutamic acid for alanine at the amino acid position 348 (A348E) and the same splicing mutation in the first intron (IVS1+2T→C). In order to investigate the molecular basis of the NE deficiency, we performed a systematic functional analysis of these potential pathogenic mutations. First, we analyzed protein expression and found that no detectable DBH protein was produced from the allele containing the IVS1+2T→C splice mutation. Second, we characterized the biochemical properties of the A348E as well as other amino acid-substituted DBH proteins in Chinese hamster ovary (CHO) cell lines. Strikingly, we found normal expression of A348E and other substituted (D100E and D331N) mutant proteins but found that they are exclusively retained inside cells in a premature ER form, whereas the wild-type mature protein is secreted in the medium. Furthermore, when tested in the rat pheochromocytoma cell line (PC12) containing secretary granules, the wild-type DBH protein completely colocalized with chromogranin A (a marker of LDCV), whereas the mutant forms colocalized with the ER marker, BiP/GRP78, suggesting abnormal trafficking and mislocation of these mutant DBH proteins. In line with this, we found that defective trafficking and secretion of these mutant proteins can be prominently rescued by treatment with a pharmacological chaperone. Based on our data, we propose that the combined aberrant splicing and protein misfolding represent the molecular basis of NE deficiency disease.
EXPERIMENTAL PROCEDURES
Genomic DNA Analysis
The region containing both the 12 exons and intron-exon boundaries of the DBH gene was PCR-amplified. PCRs were performed as follows: initial denaturation at 95 °C for 2 min, followed by 40 cycles of denaturation at 95 °C for 40 s, annealing at 55 °C for 30 s, and extension at 72 °C for 1 min. PCR products were purified by PAGE followed by DNA extraction. Direct sequencing of the double-stranded PCR fragments was performed according to the thermal cycle sequencing protocol (PerkinElmer Life Sciences).
Expression Vector Construction
Human DBH cDNA was amplified by RT-PCR using poly(A+) RNA from the human neuroblastoma SK-N-BE(2)C using forward 5′-GGA TCC GAA TTC ATG CGG GAG GCA GCC TTC AT-3′ and reverse 5′-AAA AAA CTC GAG GCC TTT GCC CCC ACC AAT GCT-3′ oligonucleotides. The amplified PCR fragment was cut with EcoRI and XhoI and then cloned into the plasmid pCDNA3.1/Myc-His (Invitrogen), resulting in plasmid pCMV DBHwt-Myc. The pCMV DBHD100E-Myc, pCMV DBHD331N-Myc, and pCMV DBHA348E-Myc constructs were generated by site-directed mutagenesis from plasmid pCMV DBHwt-Myc, using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The mutagenic primers 5′-CTG CGG CGC TTC AAC GAG GGG ATC ATG GAG CTG-3′ and 5′-CAG CTC CTA GAT CCC CTC GTT GAA GCG CCG CAG-3′ (underlined letters indicate the mismatch) were used to replace alanine at amino acid position 348 with glutamic acid. The EndoFree Plasmid Medi Kit (Qiagen) was used to prepare both the WT and mutant plasmids used in transfection experiments. For construction of retrovirus vectors, WT DBH and each mutant fragment were cloned into the pCL retroviral expression vector (Orbigen, San Diego, CA). All constructs were confirmed by sequence analysis.
Generation of DBH Antibodies
Rabbit anti-DBH antibodies were generated against full-length recombinant human DBH protein. The coding sequence for the mature form of human DBH, freed from signal peptides, was generated by PCR using plasmid pCMV DBHwt-Myc as template and followed by subcloning into pET15bat NdeI/XhoI sites to generate the His6-fused DBH. The recombinant human DBH protein was purified by Ni2+-NTA column chromatography (Qiagen), followed by removal of the His6 by treatment with thrombin (Amersham Biosciences). Immunization and bleeding were carried out by Quality Bioresources, Inc.
Cell Culture and Transfection
CHO cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. PC12 cells were grown in DMEM supplemented with 10% horse serum and 5% fetal bovine serum. All sera were heat-inactivated. All culture media contained 100 units/ml penicillin and 100 μg/ml streptomycin. Transfection consisted of mixing LipofectamineTM (Invitrogen) with 2 μg of plasmid DNA and ∼4 × 105 CHO cells. To establish cells stably expressing DBH, 48 h after transfection, one-one thousandth of cells were seeded and further cultured in the presence of 400 μg/ml G418 (Invitrogen). The expression of DBH from G418-resistant colonies was determined by Western blot analysis.
Retroviral Transduction
To obtain retroviral supernatant, a retroviral plasmid was introduced into the retrovirus packaging cell line 293GPG (13) by transient transfection with LipofectamineTM (Invitrogen). After 48 h, the medium was replaced with fresh medium and harvested 24 h later, filtered through a 0.45-μm pore size filter (Millipore, Bedford, MA), and stored at −80 °C before use. In parallel, empty vector retroviruses were produced as control (mock transduction). PC12 cells were incubated with viral supernatant(s) containing Polybrene (4 μg/ml) for 4 h, followed by a change of medium. The following day, PC12 cells were differentiated by the addition of 200 ng/ml nerve growth factor (NGF) and maintained under differentiation conditions for an additional 2–5 days.
Western Blotting
Culture medium from transformed cells was concentrated using an Amicon Centricon YM-10 centrifugal filtration (Millipore) and mixed with an equal volume of SDS-sample buffer consisting of 125 mm Tris (pH 6.8), 2% SDS, 15% glycerol, 5% β-mercaptoethanol, 0.05% bromphenol blue. Cells were washed twice with phosphate-buffered saline (PBS) and harvested in a solution containing 1% Triton X-100, 20 mm Tris (pH 7.6), 150 mm sodium chloride, 1 mm phenylmethylsulfonyl fluoride (PMSF). The cell suspension was sonicated and boiled in an equal volume of SDS-sample buffer. Samples were subjected to SDS-PAGE and transferred to a nitrocellulose membrane (Hybond-ECL, Amersham Biosciences). After blocking, the membrane was incubated with primary antibodies diluted in PBS containing 0.1% BSA. The following primary antibodies were used: rabbit anti-Myc (Upstate; 1:3000), mouse anti-KDEL (Calbiochem; 1:2000), rabbit anti-chromogranin A (Abcam; 1:5000), and mouse anti-secretogranin II (Abcam; 1:2000). The membrane was incubated with a 1:3000 dilution of horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG antibody (Amersham Biosciences). Detection was achieved using an enhanced chemiluminescent substrate (Amersham Biosciences).
Subcellular Fractionation
PC12 cells infected with retrovirus expressing WT or mutant DBH were collected with a rubber policeman in 1 ml of homogenization medium (130 mm KCl, 25 mm NaCl, 25 mm Tris, 1 mm EGTA, pH 7.4) and homogenized by pushing them three times each through 21-, 24-, and 27-gauge injection needles, respectively. The homogenates were centrifuged for 10 min at 1,000 × g to remove nuclei and cellular debris. The supernatants were then spun for 10 min at 3,000 × g and layered on top of a discontinuous iodixanol gradient consisting of 1 ml each of 30, 25, 20, 15, 12.5, 9, 7.5, and 2.5% (v/v) iodixanol in homogenization buffer. After centrifugation at 126,000 × g for 1 h, 20 fractions were collected from the top of the gradient and analyzed for density and protein concentration (Bradford) (14). A 50-μl aliquot of each fraction was used for immunoblotting. Three independent fractionation experiments were performed for each cell line.
Pulse-Chase Experiments and Immunoprecipitation Analysis
Stably transformed cells (2 × 105 cells) in 6-well plates were washed twice with methionine- and cysteine-free DMEM and incubated in methionine- and cysteine-free DMEM supplemented with 5% dialyzed FBS for 30 min. 35S-Labeled methionine and cysteine (200 μCi) were then added to the cells and incubated at 37 °C for 30 min. After labeling, cells were washed with PBS, and the chase was performed by adding 1 ml/well DMEM supplemented with 5% dialyzed FBS, containing a 10-fold excess of methionine and a 5-fold excess of cysteine. Labeled proteins were analyzed at 0, 2, 4, and 6 h. At each time point, cells were disrupted in lysis buffer containing 20 mm Tris (pH 7.6), 100 mm sodium chloride, 0.5% Nonidet P-40, 0.5 mm EDTA, 0.5 mm PMSF, 1× protease inhibitor mixture (Roche Applied Science). One μg of anti-Myc previously absorbed for 4 h with protein A-agarose (Upstate) was added to media and cell lysates and incubated for 16 h at 4 °C. Pellets were collected by centrifugation for 5 min and washed three times with lysis buffer. The immunoprecipitated proteins were released from protein A by boiling for 5 min in SDS-sample buffer. Samples were analyzed by 10% SDS-PAGE. Labeled proteins were visualized by exposing the gels to an x-ray film (Fuji Film).
Immunofluorescent Microscopy
For immunofluorescence staining, cells were fixed in 4% formaldehyde (Electron Microscopy Sciences, Ft. Washington, PA) for 30 min, rinsed with PBS, and then incubated with blocking buffer (PBS, 10% normal donkey serum (NDS)) for 10 min. Cells were then incubated overnight at 4 °C with primary antibodies diluted in PBS containing 2% NDS. The following primary antibodies were used: rabbit anti-Myc (Upstate; 1:200), mouse anti-KDEL (Calbiochem; 1:500), and rabbit anti-chromogranin A (Abcam; 1:100). After additional rinsing in PBS, samples were incubated in fluorescence-labeled secondary antibodies (Alexa 488- or Alexa 594-labeled IgG; Invitrogen) in PBS with 2% NDS for 30 min at room temperature. After rinsing in PBS, Hoechst 33342 (4 μg/ml) was used for counterstaining, and coverslips/tissue sections were mounted onto slides in Mowiol 4-88 (Calbiochem). Confocal analysis was performed using a Zeiss LSM510/Meta Station (Carl Zeiss, Thornwood, NY).
Immunogold Electron Microscopy
PC12 cells infected by retrovirus expressing WT or A348E mutant DBH were fixed for 1 h at 4 °C in PBS containing 0.1% glutaraldehyde, 4% formaldehyde, and 3.5% sucrose. Cells were lifted from the culture dish with a cell scraper and pelleted by centrifugation at 2,000 × g for 2 min at 4 °C. The cell pellets were suspended in warm agar (1% in PBS) and centrifuged. After three washes in PBS, the agar-embedded cell pellets were postfixed with 1% osmium tetroxide on ice for 2 h and washed three times, all in PBS. Cell pellets were then embedded in Epon 812 mixture after dehydration in a series of ethanol and propylene oxide solutions. For immunogold labeling experiments, the ultrathin sections that had been collected on Formvar/carbon-coated nickel grids were floated on drops of freshly prepared 3% sodium metaperiodate for 30 min. The immunogold labeling procedure was modified from the manufacturer's recommended protocol (British Biocell International). After etching and washing, the grids were placed on 50-μl droplets of solution A (PBS solution, pH 8.2, containing 4% normal goat serum, 1% bovine serum albumin, 0.1% Tween 20, 0.1% sodium azide) for 30 min. Grids were then incubated for 2 h at room temperature in a humidified chamber on 50-μl droplets of anti-rabbit α-Myc antibody appropriately diluted in solution B (solution A but with 1% normal goat serum), followed by rinses in solution B. Grids were reacted with 15-nm gold-conjugated goat anti-rabbit IgG diluted in solution A. Controls for the specificity of α-Myc immunogold labeling included 1) omitting the primary antibody and 2) replacing the primary antibody with preimmune serum. After washes in PBS and deionized water, the grids were stained with uranyl acetate (7 min) and lead citrate (2 min) and were viewed with a Phillips 300 electron microscope.
Secretion Assay
For secretion studies, PC12 cells were infected with retrovirus expressing WT or mutant DBH and differentiated with 200 ng/ml NGF for 3 days. Cells were washed three times with PBS without MgCl2 and CaCl2 and preincubated for 30 min in DMEM. After preincubation, cells were incubated with HEPES buffer (pH 7.4) without stimulation at 37 °C for 20 min, and media were collected. Immediately, cells were reincubated with HEPES buffer containing 55 mm KCl for another 20 min. All collected media were precipitated with TCA (10% final concentration) and 100 mg/ml RNase A as a protein carrier overnight at 4 °C. Precipitates were centrifuged for 10 min at 4 °C, and the resulting pellets were washed four times with acetone. Pellets were dried and suspended in SDS-PAGE sample buffer. Equal sample volumes were loaded onto SDS-PAGE, and secretion was determined by Western blotting analysis with anti-Myc antibody.
Chemical Chaperone Treatments
Stable CHO cells expressing WT and mutant DBH were treated with 1.5 m glycerol in DMEM culture media at 37 °C for 24 h. Media were precipitated overnight at 4 °C with 10% TCA and 100 mg/ml RNase A as a protein carrier. Precipitates were subjected to an immunoblot assay.
DBH Enzyme Activity Assay
Stable CHO cells expressing WT and mutant DBH were grown at 27 °C in DMEM medium for 48 h. Media were precipitated overnight at 4 °C with 80% ammonium sulfate, dissolved with 25 mm sodium acetate (pH 5.2), and dialyzed against 50 mm sodium acetate (pH 5.2) and 10 μm CuSO4. Protein concentrations were determined by Bradford assay (14). DBH enzyme activity was determined by the photometric method of Nagatsu and Udenfriend (15) using tyramine as substrate.
RESULTS
Identification of IVS1+2T→C and A348E Mutations in a New Patient
The 20-year-old propositus presented with a history of squatting as a child and severe orthostatic hypotension and passing out beginning in early childhood. Supine and upright blood pressures were 98/69 and 60/47 mm Hg with respective heart rates of 72 and 117 beats/min. Plasma renin was normal. Plasma NE, epinephrine, and dihydroxyphenylglycol were undetectable, whereas plasma dopamines were elevated more than 10-fold at 115 pg/ml supine and 589 pg/ml upright. Treatment with droxidopa raised blood pressure into the normal range.
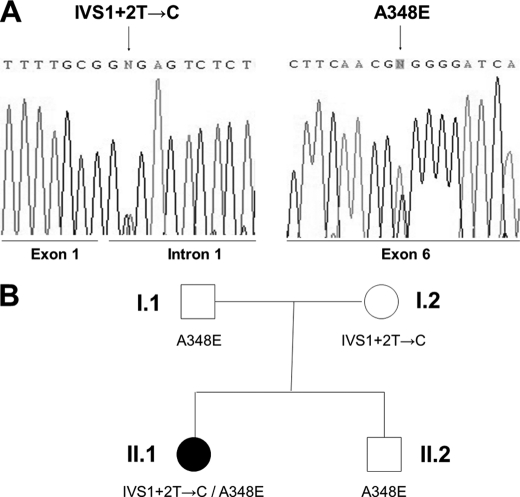
To identify the pathogenic mutations, the whole coding region, including all exon-intron boundaries and ∼1.2 kb of the promoter region of DBH were PCR-amplified from genomic DNAs of the patient and members of his family. Sequence analysis revealed that the patient is a compound heterozygote for a novel, previously unidentified missense mutation and the same splicing mutation in intron 1 that was identified in other patients (Fig. 1A) (12). The novel mutation identified in this patient is a C→A transition occurring in exon 6 at cDNA position 1043, leading to the change of alanine (GCG) to glutamic acid (GAG) at amino acid position 348 (A348E). The same mutation was found in the heterozygous state in the patient's father (Fig. 1B). The other mutation, present in the heterozygous state in the patient's mother, is the previously reported T→C change in the splice donor site of intron 1 of DBH (12). To understand the pathogenic mechanisms underlying NE deficiency, we next performed a systematic functional analysis of these potential pathogenic mutations.
FIGURE 1.
Mutations of the DBH gene in NE deficiency. A, a T→C change in the splice donor site of intron 1 and a C→A transition in exon 6 at cDNA position 1043 bp of the DBH gene were identified in NE deficiency. B, the patient is a compound heterozygote for IVS1+2T→C and A348E inherited from the mother and father, respectively. A filled symbol denotes an affected individual.
The Splice Mutation IVS1+2T→C Results in Undetectable DBH Protein Expression
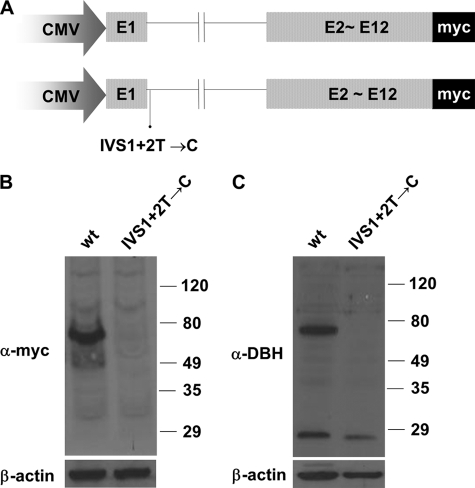
Previous RT-PCR analysis demonstrated that the splice mutation IVS1+2T→C leads to aberrant mRNA processing by disrupting the consensus sequence at the splice donor site of the first intron (12). However, properly spliced transcripts were also detected, suggesting that this splice mutation is leaky to a certain degree. We elected to examine the effect of this mutation on DBH expression at the protein level. Toward this end, we substituted the DBH genomic fragment encompassing exon 1 and intron 1 for the corresponding exon 1 of the intact DBH cDNA sequence tagged with Myc at the C terminus (Fig. 2A). Because it contains only the first intronic sequence, we speculated that this construct would allow us to directly assess the effect of the splice mutation on DBH protein expression. Eukaryotic expression vectors containing the wild-type or mutant DBH allele fragment were transiently transfected into CHO cells. When cell extracts transfected with the normal allele construct were analyzed by Western blot analysis using polyclonal antibodies against Myc, a protein band of 69 kDa was prominently detected (Fig. 2B). This size is consistent with the predicted molecular mass of DBH. Furthermore, we observed similar results using antibodies against the purified full-length DBH protein in Western blot analysis (Fig. 2C). In contrast, no detectable protein was produced from the mutant allele when we used specific antibodies against Myc or DBH (Fig. 2, B and C). These findings strongly suggest that this IVS1+2T→C mutation leads to an undetectable level of DBH protein, thus directly contributing to the NE deficiency.
FIGURE 2.
Effect of the mutation IVS1+2T→C on DBH protein expression. A, schematic representation of the minigene construct. The DBH genomic fragment encompassing exon 1 and intron 1 was substituted with the corresponding exon 1 of the intact DBH cDNA sequence tagged with Myc at the C terminus. B and C, expression vectors containing the wild-type or mutant DBH allele (IVS1+2T→C) were transfected to CHO cells. Western blot analyses using specific antibody against Myc (B) and against DBH (C) revealed that the construct containing the wild-type allele but not the IVS1+2T→C mutation produced the intact DBH protein.
Expression and Characterization of Wild-type and Mutant DBH in CHO Cells
We investigated the potential functional effects of the missense mutations identified in NE-deficient patients. It is noteworthy that the alanine residue at position 348 (site of the novel mutation) of DBH is invariant across different mammalian species as well as in two functionally related proteins (Drosophila melanogaster tyramine β-hydroxylase and human peptidylglycine-amidating mono-oxygenase) (12) (Fig. 3). Interestingly, the aspartic acid residue found at positions 100 and 331 in previously identified mutations is also invariable (12). These observations prompted us to hypothesize that these amino acids are crucial for DBH protein function, such as its stability, activity, and/or trafficking. To address the potential pathogenic mechanism caused by these mutations (i.e. A348E, D100E, and D331N) at the molecular level, we sought to characterize the biochemical properties of these mutant DBH proteins and compare them with wild-type DBH. Toward this goal, we generated mammalian vectors expressing wild-type and three mutant forms of DBH fused to the tagging protein, Myc, at the C terminus (Fig. 4A). Following introduction of these expression vectors into CHO cells, stable cell lines expressing wild-type and mutant DBH proteins were established. Western blot analyses of cell lysates at 48 h following replacement with serum-free media detected comparable levels of DBH proteins, suggesting that these mutations do not affect DBH protein stability/expression (Fig. 4B). Wild-type DBH protein was prominently detected in the medium, demonstrating that this protein represents the secreted form of DBH. In support of this, the DBH secreted into culture media had a molecular mass of 72 kDa, probably due to post-translational modification, mainly N-glycosylation (16). Strikingly, media from all three mutant forms of DBH showed either undetectable (in the case of A348E and D331N) or only marginally detectable (in the case of D100E) amounts of proteins, despite high intracellular expression (Fig. 3B). These results show that the secretion of these mutant DBH proteins is severely impaired.
FIGURE 3.
Alignment of the predicted amino acid sequences of DBH orthologs from vertebrate and invertebrate species. Shown are Mus musculus DBH, Rattus norvegicus DBH, Bos Taurus DBH, Homo sapiens DBH, D. melanogaster tyramine β-hydoxylase (TBH), and human peptidylglycine-amidating mono-oxygenase (PAM). The line indicates the amino acid position affected by missense mutation with A348E. The wild-type sequence at Ala348 is highly conserved across all species analyzed.
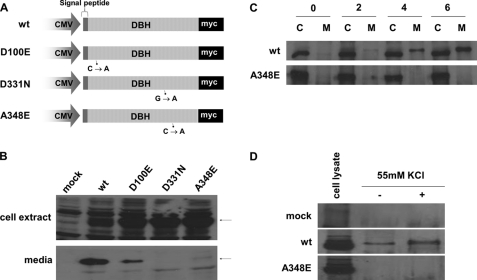
FIGURE 4.
Effect of amino acid substitutions on DBH expression in CHO and PC12 cells. A, a wild-type DBH cDNA was cloned into pCDNA3.1/Myc-His (Invitrogen) in frame with a C-terminal Myc tag. The mutant vectors (D100E, D331N, and A348E) were made by site-directed mutagenesis. B, CHO cells were transfected with either DBH wild-type or DBH mutant vectors. Stably transformed CHO cells were incubated for 48 h in serum-free medium. Proteins from cell lysates (top) and culture media (bottom) were subjected to Western blotting using anti-Myc antibodies. The arrows indicate the DBH proteins at 69 and 72 kDa in cell lysates and culture media, respectively. The wild-type DBH protein was present as a secreted form, whereas DBH mutants were rarely detected in the culture media. C, CHO cells expressing WT (top) and A348E (bottom) were labeled for 30 min with [35S]methionine for pulse-chase experiments. After chasing for the indicated times, immunoprecipitation was performed from either cell extracts (C) or media (M), using anti-Myc antibodies. Immune complexes were analyzed with SDS-PAGE. WT DBH began to appear in the media as a band of 72 kDa after a 2-h chase and reached its peak after a 6-h chase, but mutant DBH did not. D, PC12 cells were infected with retrovirus expressing wild-type (top) or A348E (bottom) DBH and were differentiated with NGF for 4 days. The Western blot shows secretion of wild-type DBH protein in the medium, in the presence (+), or absence (−) of 55 mm KCl.
To establish the time course of processing and secretion of DBH proteins, stable CHO cells transformed with wild-type DBH or the A348E mutant were pulse-labeled with [35S]methionine for 30 min and subsequently chased with excess cold methionine, and aliquots taken at various time points. At each chase time, cell lysates and culture media were immunoprecipitated with anti-Myc antibody and analyzed by SDS-PAGE. Cell extracts from both the pulse-chased wild-type and mutant DBH yielded a single band with a molecular mass of 69 kDa (Fig. 4C). The wild-type DBH began to appear in the media as a band of 72 kDa after a 2-h chase, and its levels significantly increased in the culture medium with time. In contrast, the A348E mutant was undetectable in the medium throughout the chase period despite high expression levels in cell extracts (Fig. 4C). DBH is well known to be exclusively present in secretory granules of the adrenal medulla and LDCV of noradrenergic neurons (17). To further characterize the cellular localization and/or trafficking of the secretion-impaired mutants, we used PC12 cells as a model because they contain secretory granules (SG), equivalent to neuronal LDCV, and respond to secretagogues (18, 19). The PC12 cell line was originally established from a rat neuroendocrine tumor and is able to differentiate into neurons upon NGF stimulation (20). We infected PC12 cells with retroviruses expressing wild-type and mutant DBH (A348E) fused to a Myc tag and differentiated them with NGF. Western blot analysis using Myc antibodies showed that exogenous wild-type and mutant DBH proteins were expressed at comparable levels (Fig. 4D). We next addressed whether wild-type and mutant DBH differentially respond to the regulated secretory pathway. PC12 cells were incubated with control medium or medium containing a secretagogue (55 mm KCl). This treatment significantly increased basal secretion of the wild-type DBH (Fig. 4D). In contrast, the mutant DBH was not detected in the absence or presence of the secretagogue (Fig. 4D). Taken together, our data suggest that the secretion of wild-type DBH is stimulated in response to secretagogue, whereas mutant DBH is not secreted despite receiving a depolarization signal.
Subcellular Localization of Wild-type and Mutant DBH
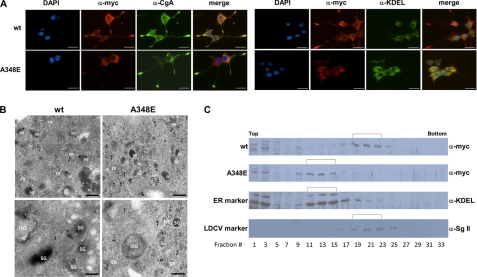
To delineate the intracellular localization of wild-type and mutant DBH, we examined PC12 cells by immunostaining with anti-Myc antibody and compared with the distribution of selected marker proteins, such as chromogranin A (LDCV marker) and BiP/GRP78 (ER marker). As shown in Fig. 5A, double label immunocytochemical analysis demonstrated that the wild-type DBH colocalized with chromogranin A, as evidenced by overlapping punctate staining throughout the cell body, excluding the nuclei. In particular, the DBH-immunopositive granules were typically found within the enlarged endings of the neurite-like extension (Fig. 5A). In sharp contrast, mutant DBH immunoreactivity was located abundantly around the nuclei in differentiated PC12 cells but was very weakly present in the shafts of the neurite-like extension or in the wild-type cone of neurites. Importantly, the mutant DBH did not colocalize with chromogranin A. Instead, it mostly colocalized with BiP/GRP78 (Fig. 5A). We next analyzed immunogold-labeled thin sections of PC12 cells expressing both the wild-type DBH and the A348E mutant by electron microscopy. As shown in Fig. 5B, immunogold-labeled wild-type DBH was predominantly found in the dense core SG (white arrowhead) and occasionally in the rough ER (black arrowhead). In contrast, gold particles for mutant DBH were not observed in the SG but were abundant in association with the rough ER (black arrowhead). These experiments show that the secretion-impaired DBH mutant is retained in the rough ER compartment.
FIGURE 5.
Effect of missense mutations on cellular localization of DBH protein. A, PC12 cells expressing wild-type (top) or A348E (bottom) DBH were differentiated with NGF for 4 days and immunostained with anti-Myc antibodies, followed by reaction with an Alexa Fluor 594 chicken anti-rabbit IgG. The same cells were also stained with anti-chromogranin A (CgA) monoclonal antibodies (left) and anti-BiP/GRP78 (KDEL) monoclonal antibodies (right). Wild-type DBH co-localizes with chromogranin A, whereas mutant DBH co-localizes with BiP/GRP78. Scale bars, 10 μm. B, electron microscopic analysis of thin cryosections of PC12 cells expressing wild-type (left) and A348E (right) DBH. A 10-nm immunogold label of wild type is abundantly found on the SG (white arrowhead) and rarely on the ER (black arrowhead). In contrast, immunogold label for mutant DBH is absent from the SG and is mostly found in the ER. Scale bars, 200 nm. C, subcellular fractionation of PC12 cells expressing wild-type and A348E DBH was analyzed by Western blot assay using anti-Myc antibodies (for DBH protein), anti-KDEL (for ER marker), and anti-secretogranin II (for LDCV marker). Wild-type DBH protein was present in fractions 19–23 together with secretogranin, whereas mutant DBH A348E was present in fractions 11–15 together with KDEL.
We also performed subcellular fractionation using discontinuous iodixanol gradient centrifugation. PC12 cells infected with wild-type or A348E-expressing retroviruses were differentiated with NGF and subjected to ultracentrifugation. Protein fractions from the gradient were analyzed by Western blot analyses for the presence of DBH in different subcellular fractions. As shown in Fig. 5C, fractions 17–25 were enriched for the LDCV marker secretogranin II (Sg II) as well as the wild-type DBH migrating as a typical band of ∼69 kDa. In contrast, the A348E mutant DBH was predominantly present between protein fractions 9 and 17, which were enriched for the ER marker. Taken together, our data demonstrate that wild-type DBH colocalizes with secretory granules, showing normal trafficking, whereas the mutant DBH is predominantly present in the ER, indicating abnormal trafficking.
Defective Trafficking of Mutant DBH Proteins Can Be Relieved by a Pharmacological Chaperone
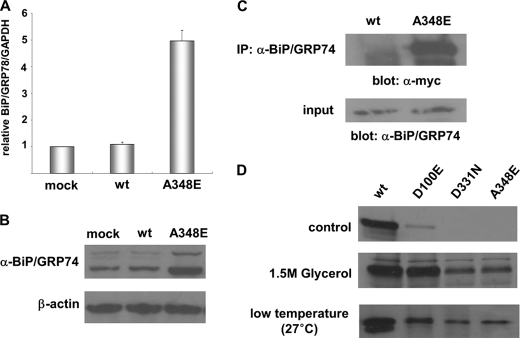
Our data prompted us to hypothesize that mutant DBH proteins may not be properly folded and thus become trapped in the ER. If true, this situation may trigger an ER stress response (unfolded protein stress). To test this, we analyzed the induction of the prototype ER chaperone, BiP/GRP78, in CHO cells expressing either wild-type or mutant DBH. Quantitative RT-PCR indeed showed that BiP mRNA was robustly up-regulated in CHO cells when the mutant DBH was overexpressed, whereas overexpression of wild-type DBH did not affect it (Fig. 6A). Western blot analyses confirmed that BiP protein expression was robustly up-regulated in CHO cells expressing mutant DBH but not in those expressing wild-type DBH (Fig. 6B). We next analyzed the interaction between DBH and BiP. DBH was first immunoprecipitated with anti-BiP antibodies, and then the immunoprecipitates were further analyzed by Western blot analysis with antisera against Myc. In mutant DBH-expressing cells, robust interactions between BiP and mutant DBH were detected, whereas BiP barely formed any complex with wild-type DBH (Fig. 6C). Together, these results suggest that misfolding of the mutant DBH in the ER induces up-regulation of BiP mRNA and protein, resulting in the formation of stable complexes consisting of misfolded proteins and chaperone molecules being retained in the ER. Based on this, we speculated that pharmacological chaperones or lower growth temperature may rescue this pathological situation. To address this, wild-type and mutant-expressing CHO cells were incubated at 37 °C with glycerol, one of the well known chemical chaperones, and analyzed for the presence of secreted DBH proteins. When cells were treated with 1.5 m glycerol for 24 h, significant levels of all three mutant DBH proteins were secreted into the culture medium (Fig. 6D). This effect was particularly dramatic in the case of D100E, in which the efficiency of secretion was almost comparable with that of wild-type DBH. When cells were grown at lower temperature (e.g. 27 °C), a similar effect was observed (Fig. 6D). To investigate whether the rescued DBH is functional, we performed a photometric enzyme activity assay (15). We found that the enzyme activity of all secreted mutants at 27 °C was comparable with that of wild-type DBH (supplemental Fig. 1). These results further support the idea that these mutant proteins are trapped in the ER due to protein misfolding and ER stress responses and that a pharmacological chaperone is able to rescue the processing and secretion of these mutant DBH proteins.
FIGURE 6.
DBH·BiP complex formation and the effect of pharmacological chaperone. Forced expression of A348E but not wild-type DBH robustly induced BiP mRNA (A) and protein (B) expression in CHO cells. C, immunoprecipitation (IP) with anti-BiP/GRP78 antibodies shows that expression of A348E but not wild-type DBH dramatically increased formation of a DBH and BiP/GRP78 complex. D, effect of a chemical chaperone, glycerol, and lower growth temperature (27 °C) on secretion of DBH proteins in culture medium of CHO cells. Error bars, S.D.
DISCUSSION
NE deficiency is an autosomal recessive disorder and is quite a rare form of primary autonomic failure, which is biochemically characterized by undetectable levels of NE and epinephrine in plasma and tissues, increased levels of dopamine, and the absence of serum DBH protein (4, 5). These patients clinically exhibit severe orthostatic hypotension with low blood pressure at standing and normal positions when seated. Long term administration of dihydroxyphenylserine results in the restoration of plasma and urinary levels of NE and increases blood pressure (21). We have previously identified mutations in the DBH gene of NE deficiency patients, including a common splice donor site mutation in intron 1 (IVS1+2T→C) and three missense mutations (11, 12) (Table 1). Here, we describe another NE deficiency patient with compound heterozygous alleles for the same IVS1+2T→C mutation and a novel missense mutation (A348E) in the DBH gene. Thus, in addition to the splice donor site mutation, at least six missense mutations as well as one frameshift mutation have been identified in the DBH gene so far (Table 1). However, the molecular basis of the pathogenic mechanisms of these mutations was unknown.
In this study, we performed functional characterization of DBH mutations associated with the human NE deficiency and elucidated how they affect normal DBH expression and processing, which we propose as the molecular and cellular basis of NE deficiency disease. First, we found that the mutation in the splice donor site is unable to produce detectable levels of DBH protein, as examined by Western blot analysis. The point mutation in the splice donor site of the intron occurs in one or two invariable dinucleotides that are required for intron splicing (22). In humans, mutations affecting pre-mRNA splicing have been shown to account for up to a half of disease-causing gene alterations, potentially representing the most frequent cause of hereditary disorders (23). The most common consequence of splicing mutations is skipping of one or more exons, whereas other possible outcomes are the activation of aberrant 5′ or 3′ splice sites and the retention of full introns into mature mRNA (24). The mutation IVS1+2T→C identified in all known NE deficiency patients is located within the invariant GT of the splice donor site of intron 1 (Table 1). Our previous functional analysis indicated that this mutation alters normal splicing of mRNA, generating a transcript with a premature stop codon as well as a normal transcript (12). This leaky expression of normal transcript raised the question of whether this mutation underlies the pathological mechanism. When we analyzed the functional outcome of the minigene containing this mutation, IVS1+2T→C, at the protein level in CHO cells, no DBH proteins could be detected in Western blot analysis using two specific antibodies against DBH and tagging protein Myc. Thus, we conclude that the mutation IVS1+2T→C is a direct cause of the NE deficiency.
More strikingly, we found that all three DBH missense mutations tested in this study (D100E, D331N, and A348E) lead to defective protein trafficking and mislocation, probably due to protein misfolding. Several lines of evidence support this novel idea of NE deficiency disease. First, when exogenously expressed in CHO cells, the wild-type DBH protein was prominently secreted into the medium in its fully glycosylated form, consistent with a previous report (16). In contrast, DBH mutant proteins were not secreted but were found predominantly within the cells. Second, when wild-type DBH and its mutants were expressed in neuronal PC12 cells, wild-type DBH was released upon secretagogue stimulation, strongly indicating that wild-type DBH reside in regulated secretory granules, which represent physiological catecholamine storage vesicles. In contrast, the DBH mutant was not subject to the regulated secretory pathway. Third, confocal and electron microscopy analyses demonstrated that wild-type DBH faithfully colocalized with chromogranin A, an LDCV marker, whereas mutant DBH was predominantly present in the ER. Subcellular fractionation experiments further confirmed that most wild-type DBH was targeted to LDCV, whereas mutant DBH remained in the ER. Taken together, our results strongly suggest that DBH proteins encoded by missense mutations are retained in the ER and fail to be targeted to the LDCV, indicating abnormal trafficking.
To perform their specific cellular functions, newly synthesized proteins must fold into their unique three-dimensional structures. Numerous human diseases, including amyloidosis and cystic fibrosis, are associated with protein misfolding (25, 26). The ER is the principal site of protein folding (27). In the ER, misfolded or unfolded proteins initially display a prolonged association with classical ER chaperones like BiP/GRP78. Accumulation of these complexes may trigger the unfolded protein response or ER stress, which has been observed in several human diseases, such as neurodegenerative diseases and osteogenesis imperfecta (28, 29). To address if mutant DBH proteins trigger the ER stress response, we tested the expression of the Hsp70 family member BiP, a master ER chaperone, whose expression is specifically induced upon ER stress (30). Indeed, expression of mutant DBH dramatically induced expression of both BiP mRNA and proteins, suggesting that accumulation of misfolded DBH proteins activates the ER stress response. Despite enhanced ER chaperone synthesis, mutant DBH proteins could not be properly folded and remained stuck in the ER. Thus, we tested the possibility that chemical or pharmacological chaperones (25, 31) could rescue defective trafficking of mutant DBH proteins. Chemical/pharmacological chaperones are small molecules that can alleviate misfolding, aggregation, and mislocalization of human disease-causing proteins affected by amino acid substitution (25, 31). Our results showed that one of the well known chemical chaperones, glycerol (32), could partially rescue defective secretion of all three DBH mutant proteins tested in this study. In particular, secretion of D100E was almost completely rescued by glycerol treatment, which is in agreement with earlier findings showing that D100E could be partially secreted without any treatment. Thus, our findings open up the possibility that NE deficiency disease could be more fundamentally treated with pharmacological chaperones.
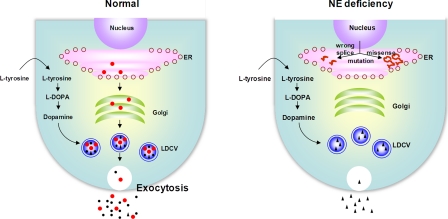
NE is synthesized by DBH, which converts dopamine to NE inside synaptic vesicles, such as LDCV. Under normal conditions, both DBH and NE are secreted into plasma by exocytosis. Based on our results, we propose that the combined abnormal processing of DBH mRNA and its defective protein trafficking causes NE deficiency (Fig. 7). Whereas the IVS1+2T→C mutation in the splice donor site interferes with normal mRNA splicing and protein production, single amino acid substitutions encoded by missense mutations lead to misfolding of newly synthesized DBH protein, trapping it in the ER and blocking proper transport to the LDCV. Thus, we propose that NE deficiency represents a new example of protein misfolding disease where a vesicular protein is misfolded and mislocated, resulting in plasma and cerebrospinal fluid deficiency of both DBH and NE. At present, the prevalence of NE deficiency is unknown. In addition, given that NE critically controls autonomic and cardiovascular function, it is of great interest to investigate how widely abnormal trafficking of DBH affects these pathological conditions.
FIGURE 7.
Schematic representation of functional effects of pathogenic mutations on DBH synthesis and trafficking. Schematic representation of functional effects of pathogenic mutations on DBH synthesis and trafficking. WT DBH enzymes (red) (●) are synthesized in the ER and transported to the Golgi/trans-Golgi network, where they become post-translationally modified and are packaged into the LDCV. In LDCV, DBH converts DA (black) (▴) to NE (black) (●). Both DBH and NE are released by exocytosis upon stimulation of the nerve terminal and are readily detectable in plasma. In NE deficiency, the splicing mutation IVS1+2T→C results in disruption of appropriate splicing of DBH mRNA (short red s shape) and leads to undetectable levels of DBH protein. DBH proteins (long red s shape) encoded by missense mutations are retained in the ER and fail to be targeted to the LDCV due to their misfolding. As a result, NE is not synthesized in LDCV, leading to the absence of both NE and DBH in plasma. Instead, DA is dramatically up-regulated. The open circles in the ER indicate the ribosome.
Supplementary Material
Acknowledgments
We thank Bonnie Black, R.N., and the Vanderbilt Clinical Research Center nursing staff for taking care of this patient.
This work was supported, in whole or in part, by National Institutes of Health Grant MH48866 (to K.-S. K.), MH087903 (to K.-S. K.), P01 HL056693 (to D. R.), HL071784 (to D. R.), and UL1 RR024975D. This work was also supported by international grants from the Brain Research Center and the Dongyang Corporation Co. in the Republic of Korea.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- NE
- norepinephrine
- DBH
- dopamine β-hydroxylase
- DA
- dopamine
- LDCV
- large dense core vesicle(s)
- ER
- endoplasmic reticulum
- NDS
- normal donkey serum
- SG
- secretory granules.
REFERENCES
- 1. Stewart L. C., Klinman J. P. (1988) Annu. Rev. Biochem. 57, 551–592 [DOI] [PubMed] [Google Scholar]
- 2. Weinshilboum R. M., Thoa N. B., Johnson D. G., Kopin I. J., Axelrod J. (1971) Science 174, 1349–1351 [DOI] [PubMed] [Google Scholar]
- 3. Goldstein D. S., Eisenhofer G., Kopin I. J. (2003) J. Pharmacol. Exp. Ther. 305, 800–811 [DOI] [PubMed] [Google Scholar]
- 4. Robertson D., Goldberg M. R., Onrot J., Hollister A. S., Wiley R., Thompson J. G., Jr., Robertson R. M. (1986) N. Engl. J. Med. 314, 1494–1497 [DOI] [PubMed] [Google Scholar]
- 5. Man in 't Veld A. J., Boomsma F., Moleman P., Schalekamp M. A. (1987) Lancet 1, 183–188 [DOI] [PubMed] [Google Scholar]
- 6. Garland E. M., Hahn M. K., Ketch T. P., Keller N. R., Kim C. H., Kim K. S., Biaggioni I., Shannon J. R., Blakely R. D., Robertson D. (2002) Ann. N.Y. Acad. Sci. 971, 506–514 [DOI] [PubMed] [Google Scholar]
- 7. Thomas S. A., Matsumoto A. M., Palmiter R. D. (1995) Nature 374, 643–646 [DOI] [PubMed] [Google Scholar]
- 8. Zhou Q. Y., Quaife C. J., Palmiter R. D. (1995) Nature 374, 640–643 [DOI] [PubMed] [Google Scholar]
- 9. O'Connor D. T., Cervenka J. H., Stone R. A., Levine G. L., Parmer R. J., Franco-Bourland R. E., Madrazo I., Langlais P. J., Robertson D., Biaggioni I. (1994) Clin Sci. 86, 149–158 [DOI] [PubMed] [Google Scholar]
- 10. Mathias C. J., Bannister R. B., Cortelli P., Heslop K., Polak J. M., Raimbach S., Springall D. R., Watson L. (1990) Q. J. Med. 75, 617–633 [PubMed] [Google Scholar]
- 11. Deinum J., Steenbergen-Spanjers G. C., Jansen M., Boomsma F., Lenders J. W., van Ittersum F. J., Hück N., van den Heuvel L. P., Wevers R. A. (2004) J. Med. Genet. 41, e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim C. H., Zabetian C. P., Cubells J. F., Cho S., Biaggioni I., Cohen B. M., Robertson D., Kim K. S. (2002) Am. J. Med. Genet. 108, 140–147 [PubMed] [Google Scholar]
- 13. Ory D. S., Neugeboren B. A., Mulligan R. C. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 11400–11406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 15. Nagatsu T., Udenfriend S. (1972) Clin. Chem. 18, 980–983 [PubMed] [Google Scholar]
- 16. Oyarce A. M., Eipper B. A. (2000) J. Biol. Chem. 275, 3270–3278 [DOI] [PubMed] [Google Scholar]
- 17. Axelrod J., Weinshilboum R. (1972) N. Engl. J. Med. 287, 237–242 [DOI] [PubMed] [Google Scholar]
- 18. Arvan P., Castle D. (1998) Biochem. J. 332, 593–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tooze S. A., Martens G. J., Huttner W. B. (2001) Trends Cell Biol. 11, 116–122 [DOI] [PubMed] [Google Scholar]
- 20. Tischler A. S., Greene L. A. (1975) Nature 258, 341–342 [DOI] [PubMed] [Google Scholar]
- 21. Goldstein D. S. (2006) Cardiovasc. Drug. Rev. 24, 189–203 [DOI] [PubMed] [Google Scholar]
- 22. Breathnach R., Benoist C., O'Hare K., Gannon F., Chambon P. (1978) Proc. Natl. Acad. Sci. U.S.A. 75, 4853–4857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Teraoka S. N., Telatar M., Becker-Catania S., Liang T., Onengüt S., Tolun A., Chessa L., Sanal O., Bernatowska E., Gatti R. A., Concannon P. (1999) Am. J. Hum. Genet. 64, 1617–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vorechovský I. (2006) Nucleic Acids Res. 34, 4630–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cohen F. E., Kelly J. W. (2003) Nature 426, 905–909 [DOI] [PubMed] [Google Scholar]
- 26. Macario A. J., Conway de Macario E. (2005) N. Engl. J. Med. 353, 1489–1501 [DOI] [PubMed] [Google Scholar]
- 27. Rapoport T. A. (1992) Science 258, 931–936 [DOI] [PubMed] [Google Scholar]
- 28. Lamandé S. R., Bateman J. F. (1999) Semin. Cell Dev. Biol. 10, 455–464 [DOI] [PubMed] [Google Scholar]
- 29. Katayama T., Imaizumi K., Sato N., Miyoshi K., Kudo T., Hitomi J., Morihara T., Yoneda T., Gomi F., Mori Y., Nakano Y., Takeda J., Tsuda T., Itoyama Y., Murayama O., Takashima A., St George-Hyslop P., Takeda M., Tohyama M. (1999) Nat. Cell Biol. 1, 479–485 [DOI] [PubMed] [Google Scholar]
- 30. Hendershot L. M. (2004) Mt. Sinai. J. Med. 71, 289–297 [PubMed] [Google Scholar]
- 31. Ulloa-Aguirre A., Janovick J. A., Brothers S. P., Conn P. M. (2004) Traffic 5, 821–837 [DOI] [PubMed] [Google Scholar]
- 32. Bernier V., Lagacé M., Bichet D. G., Bouvier M. (2004) Trends Endocrinol. Metab. 15, 222–228 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.