Abstract
Selenocysteine incorporation at UGA codons requires cis-acting mRNA secondary structures and several specialized trans-acting factors. The latter include a selenocysteine-specific tRNA, an elongation factor specific for this tRNA and a SECIS-binding protein, SBP2, which recruits the elongation factor to the selenoprotein mRNA. Overexpression of selenoprotein mRNAs in transfected cells results in inefficient selenocysteine incorporation due to limitation of one or more of these factors. Using a transfection-based competition assay employing overexpression of selenoprotein mRNAs to compete for selenoprotein synthesis, we investigated the ability of the trans-acting factors to overcome competition and restore selenocysteine incorporation. We report that co-expression of SBP2 overcomes the limitation produced by selenoprotein mRNA overexpression, whereas selenocysteyl-tRNA and the selenocysteine-specific elongation factor do not. Competition studies indicate that once bound to SECIS elements, SBP2 does not readily exchange between them. Finally, we show that SBP2 preferentially stimulates incorporation directed by the seleno protein P and phospholipid hydroperoxide glutathione peroxidase SECIS elements over those of other selenoproteins. The mechanistic implications of these findings for the hierarchy of selenoprotein synthesis and nonsense-mediated decay are discussed.
Keywords: nonsense-mediated decay/SECIS/selenocysteine/selenoprotein hierarchy
Introduction
Decoding of UGA as selenocysteine occurs in eubacteria, archaea and eukarya. Selenocysteine incorporation requires the presence of specific secondary structures in selenoprotein mRNAs, termed SECIS elements. In the absence of a functional SECIS element, or under conditions of limiting selenocysteine biosynthesis or incorporation, the UGA codons in these messages specify termination. The mechanism of selenocysteine incorporation has largely been worked out through elegant genetic and biochemical studies in prokaryotes. In Escherichia coli, the SECIS elements reside immediately downstream of each of the selenocysteine codons, and their positions are tightly constrained—movement of the stem upstream or downstream by as little as three nucleotides reduces or abolishes incorporation (Heider et al., 1992). In archaea and eukarya, the required signals, also stem–loops but differing in sequence and structure from both their prokaryotic counterparts and from each other, are located in the 3′ untranslated regions (UTR) of the messages, at considerable distances from the selenocysteine codons. These signals serve to recode the entire message, functioning for any upstream in-frame UGA codon (Berry et al., 1993; Hill et al., 1993), providing a minimal spacing requirement of ∼60 nucleotides is met (Martin et al., 1996).
In addition to the cis-acting SECIS elements, selenocysteine incorporation requires several trans-acting factors. These include the selenocysteine-specific tRNA, tRNA[Ser]Sec (Lee et al., 1990), and selenophosphate synthetase, an enzyme required for biosynthesis of selenocysteine on the tRNA (Leinfelder et al., 1988; Low et al., 1995). These two factors have been identified in all three kingdoms. In eubacteria, selenocysteine incorporation is catalyzed by a bifunctional protein, SELB, consisting of an N-terminal selenocysteyl-tRNA-specific elongation factor domain and a C-terminal SECIS RNA-binding domain (Kromayer et al., 1996). Determination of the complete sequence of the genome of the archaeon Methanococcus jannaschii revealed the presence of three EF genes, two bearing homology to EF1 and EF2, and the third unassigned (for review see Dennis, 1997). As selenoprotein-encoding genes and a tRNA[Ser]Sec gene were present in the genome sequence, this suggested that the unassigned EF might function as a selenocysteyl-tRNA-specific elongation factor. This was subsequently shown to be the case (Rother et al., 2000). The sequence contained a short C-terminal extension analogous to that found in E.coli SELB, but did not bind the archaea SECIS elements. Electrophoretic mobility shift assays performed with proteins from M.jannaschii extracts revealed a separate SECIS-binding activity. In parallel, studies in eukaryotes identified a SECIS-binding protein, SBP2, which exhibits specificity for a conserved essential region in eukaryotic SECIS elements and stimulates selenocysteine incorporation in vitro and in vivo (Copeland and Driscoll, 1999; Copeland et al., 2000). Finally, sequence database searches using the archaeal SELB sequence resulted in the identification of EF homologs in Caenorhabditis elegans, Drosophila melanogaster, and the murine and human expressed sequence tag (EST) databases (Fagegaltier et al., 2000; Tujebajeva et al., 2000). The murine selenocysteyl-tRNA-specific elongation factor, designated EFsec, interacts both in vitro and in vivo with the SBP2–SECIS complex and delivers the tRNA to the ribosome (Tujebajeva et al., 2000). Studies of selenocysteine incorporation in transiently transfected mammalian cells have revealed an apparent inefficiency in this process (Berry et al., 1992, 1994). Overexpression of the mRNA for the selenoprotein, type 1 iodothyronine deiodinase (D1), by transfection in human HEK cells resulted in premature termination at the UGA codon, presumably due to limitation of a factor or factors required for selenocysteine incorporation (Berry et al., 1994). Co-transfection of the selenocysteyl-tRNA gene only partially restored incorporation, suggesting that other factors may also be limiting. With the availability of SBP2 and EFsec cDNAs, we could now investigate whether the limitation of either of these factors is responsible for the inefficiency of selenocysteine incorporation in transfected cells. In studies described herein, we report the establishment of an in vivo assay to investigate competition between SECIS elements for the factors required for selenoprotein synthesis. We show that a factor or factors required for selenocysteine incorporation can be made limiting by the expression of an excess of competitor mRNAs containing wild-type SECIS elements. Furthermore, we show that the first selenoprotein P (sel P) SECIS element is a more effective competitor than that of D1, suggesting a differential affinity of the limiting factor for the two elements. Time-course competition studies suggest that once the factor binds, it does not readily exchange between SECIS elements. Co-transfection of the SBP2 cDNA overcomes this competition, whereas transfection of either the selenocysteyl-tRNA gene or the EFsec cDNA does not. These results predict that, under conditions in which SBP2 was limiting in vivo, differential affinity of the protein for different SECIS elements would result in the preferential synthesis of some selenoproteins over others.
Previous studies have shown a hierarchy of selenium retention in different tissues. For example, testes have been shown to retain selenium stores ∼20-fold better than liver or heart upon dietary selenium limitation (Behne et al., 1988). In addition, a hierarchy for synthesis of different selenoproteins within a single tissue, as well as in different tissues and cell lines, has been observed (Behne et al., 1988; Hill et al., 1992; Lei et al., 1995; Mitchell et al., 1997). As examples of this, glutathione peroxidase (GPX) activity was reduced to 1% of normal levels in liver, and to ∼4–9% in kidney, heart and lung of selenium-deficient rats. Phospholipid hydroperoxide glutathione peroxidase (PHGPX) activity was decreased to 25–50% in these tissues, but was unaffected by selenium deprivation in testes. The dramatic decline in GPX activity upon selenium deprivation is due in large part to rapid turnover of the mRNA for this protein (Saedi et al., 1988; Christensen and Burgener, 1992; Lei et al., 1995).
mRNAs containing premature nonsense codons are eliminated from most cells via a pathway known as mRNA surveillance, or nonsense-mediated decay (NMD; Nagy and Maquat, 1998; Hentze and Kulozik, 1999). A critical feature in discrimination between physiological and premature termination codons in mammalian cells appears to be the position of the last intron in the pre-mRNA relative to the termination codon. An intron >∼50 nucleotides downstream from a termination codon will define this stop codon as premature (Nagy and Maquat, 1998; Thermann et al., 1998). Thus, selenoprotein mRNAs whose pre-mRNAs contain introns downstream of the selenocysteine codon should be targeted for NMD when selenocysteine incorporation is inefficient. This has recently been shown to be the case for GPX mRNA (Moriarty et al., 1998; Weiss and Sunde, 1998), whereas PHGPX mRNA is much less sensitive to NMD, despite the presence of appropriately spaced introns in both pre-mRNAs (Lei et al., 1995). To understand further the reasons for this differential sensitivity to NMD, we investigated the responses of a series of reporter constructs containing different SECIS elements to the co-transfection of SBP2. We show that SBP2 stimulates incorporation from different SECIS elements to dramatically different extents, with the first SECIS element from sel P at the top of the hierarchy, followed by that of PHGPX, and the GPX SECIS element at the bottom. As this hierarchy parallels those of both selenoprotein synthesis and sensitivity to NMD, the implications for differential SBP2–SECIS interactions on these pathways are discussed.
Results
SECIS elements compete for a limiting factor in transfected cells
The role of the SECIS element is postulated to be the recruitment of factors required for selenocysteine incorporation, and the delivery of selenocysteyl-tRNA as part of a complex with EFsec, SBP2, and possibly other factors, to the UGA codon at the ribosome. It follows from this prediction that some or all of these factors might be rendered limiting in vivo by the introduction of excess SECIS elements, which would in turn be predicted to decrease the efficiency of selenocysteine incorporation into a co-transfected reporter selenoprotein. To test this, we examined the ability of constructs containing wild-type or mutant SECIS elements to inhibit expression of a reporter selenoprotein construct, in this case the wild-type D1. Initial studies employed wild-type or mutant SECIS elements alone, in the absence of an upstream open reading frame (ORF). Co-transfection of a 6-fold excess of either wild-type D1 SECIS or the corresponding construct from which the nine nucleotide loop had been deleted had no effect on expression of the reporter selenoprotein, while a construct encoding the two sel P SECIS elements produced a slight inhibitory effect of ∼10% (not shown). The lack of efficient competition by the SECIS elements alone might be explained by instability of the RNA elements when expressed without an upstream ORF, failure of these small RNAs to be localized properly for competition, or an inability to fold properly. The latter might be hindered further by the fact that the consensus SECIS sequence contains an AUG, a translation initiation codon, near the 5′ end. Quantitative RT–PCR analysis of the steady-state levels of these small RNAs in cytoplasmic extracts indicated that their stability was comparable to that of the D1 reporter, and that they were predominantly cytoplasmic. However, we could not assess further either subcellular localization, RNA folding, or translation of these small RNAs.
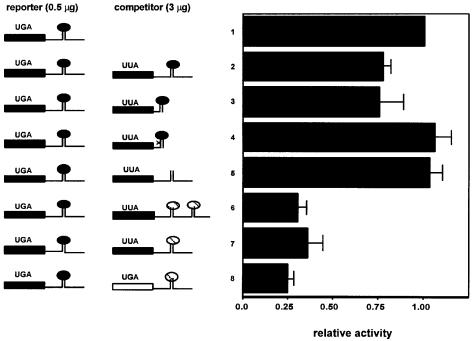
Next, we examined the ability of SECIS elements linked downstream of ORFs to compete for selenocysteine incorporation. The first series of these competitor constructs contained either wild-type or mutant SECIS elements from the D1 or sel P sequences, linked to a mutant D1 ORF. All of the SECIS mutations used have been shown to result in the complete loss of selenocysteine incorporation when linked downstream of the D1 coding region (Berry et al., 1991c; Martin et al., 1998). The D1 ORF linked to the competitors is based on the wild-type D1 sequence, but in place of the native UGA selenocysteine codon we introduced a leucine codon. The protein containing this substitution is enzymatically inactive and thus will not contribute to the measured D1 activity (Berry et al., 1991a). As shown in Figure 1, constructs encoding leucine mutant D1 linked to SECIS elements from either D1 or sel P effectively compete with translation of D1 and thus decrease its activity (lines 2, 3, 6 and 7). Constructs containing the first sel P SECIS element were more effective competitors than those containing the D1 element, consistent with our previous demonstration that the former is ∼3-fold more potent than the latter at directing selenocysteine incorporation (Berry et al., 1993). In contrast to the results with wild-type SECIS competitors, constructs containing mutant SECIS elements had no effect on D1 activity (lines 4 and 5), indicating that they do not compete with the reporter construct. In fact, a single point mutation in the conserved core of the D1 SECIS element (line 4) is sufficient to render it inactive as a competitor. These results indicate that a factor (or factors) that exhibits in vivo specificity for SECIS elements and is required for selenocysteine incorporation becomes limiting under conditions of SECIS excess.
Fig. 1. Competition for selenoprotein synthesis between wild-type selenoprotein constructs and constructs containing wild-type or mutant SECIS elements. Deiodinase activity was assayed in cell homogenates following transfection with 0.5 µg/dish of the wild-type D1 construct and 3 µg/dish of the following competitors: line 1, vector; line 2, leucine mutant D1 coding region/wild-type D1 SECIS; line 3, leucine mutant D1 coding region/minimal wild-type D1 SECIS element; line 4, leucine mutant D1 coding region/AUGA→AUCA point mutant of minimal D1 SECIS element; line 5, leucine mutant D1 coding region/D1 3′ UTR with SECIS loop deleted; line 6, leucine mutant D1 coding region/wild-type sel P SECIS elements 1 and 2; line 7, leucine mutant D1 coding region/wild-type sel P SECIS element 1; line 8, wild-type D2 coding region/wild-type sel P SECIS element 1. Activity shown is the mean ± SE of duplicate assays from at least three independent transfections, normalized to human growth hormone in the medium. In the left panel, black bars and ovals represent D1 sequences, the open bar represents D2 sequences and hatched ovals represent sel P sequences.
All of the competitors described contained a leucine codon instead of the native selenocysteine codon. Thus, the competitive effects, although specific to wild-type SECIS elements, could potentially reflect an inability of the complex assembled at the SECIS element to catalyze selenocysteine insertion, and thus perform its physiological function, due to the lack of a UGA codon. One might envisage, for example, that delivery of the EFsec–tRNA complex to the ribosome might elicit a conformational change in SBP2, allowing dissociation of either the elongation factor–tRNA complex or the empty EFsec, and recruitment of a new EFsec–tRNA complex. Substitution of the leucine codon would prevent discharging, potentially trapping the factor in a non-productive complex with the SECIS element. To test this, competition experiments were performed using a wild-type selenoprotein sequence, that of the type II iodothyronine deiodinase (D2), with the first sel P SECIS element linked downstream (Figure 1, line 8). The D2 enzyme exhibits distinct kinetic properties from D1; thus, under appropriate assay conditions it is possible to distinguish between D1 and D2 activities. We found that the D2–sel P SECIS competitor competed as effectively as the leucine mutant D1–sel P SECIS competitor. Thus, competition between wild-type SECIS elements occurs regardless of the presence or absence of a UGA selenocysteine codon.
The SECIS-binding factor does not readily exchange between elements
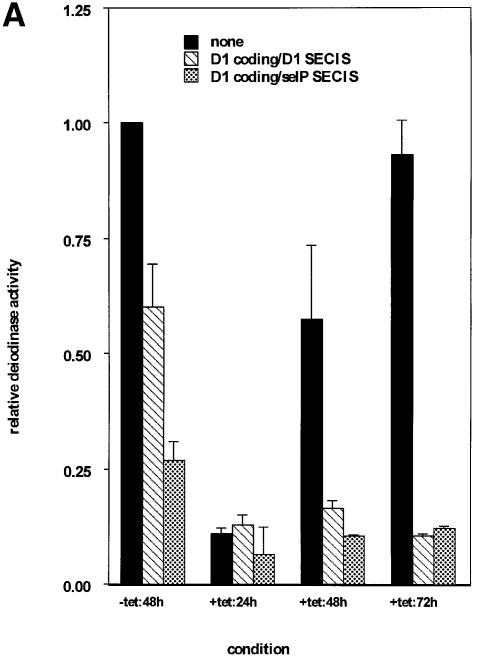
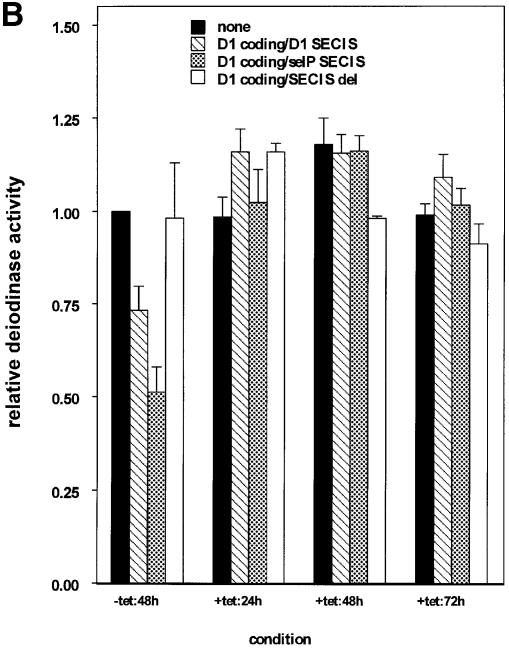
To begin to address the kinetics of binding of the limiting factor to competitors and, in particular, the rate of dissociation once binding has occurred, we employed a tetracycline-repressible expression system to stagger the times of expression of competitor and D1 reporter. Two strategies were used. In the first, expression of D1 reporter was delayed relative to SECIS competitors. Allowing the expression of competitors first would presumably allow the formation of complexes between their SECIS elements and the SECIS-binding factor. Twenty-four hours after transfection, medium was changed and tetracycline removed, allowing induction of D1 expression. Cells were maintained for an additional 24–72 h to allow full expression of D1, then harvested for assay of D1 activity. Results are shown in Figure 2A. In the presence of D1 or sel P competitors without addition of tetracycline, D1 activity was competed to levels comparable to those seen in Figure 1. The addition of tetracycline to the medium repressed D1 expression to background levels in all cases (data not shown). Activity in the absence of competitors recovered only minimally after 24 h, but more dramatically between 48 and 72 h (Figure 2A, black bars). Following repression by tetracycline for 24 h and induction of D1 in the absence of tetracycline for 24 h (+tet:24h), activities in the presence of competitors were low (7–13%) and not significantly different from activity in the absence of competitor. In contrast to the recovery of D1 activity observed in the absence of competitors, incubation for 48 or 72 h in the presence of competitors did not result in increased D1 activity. Activity remained in the 10–16% range with either competitor. This indicates that once the competitors have been expressed for a period of time prior to reporter induction, reporter activity cannot be recovered. In the second strategy, expression of competitors was delayed 24 h relative to D1 reporter. The prediction is that this would allow complex formation between SECIS-binding protein and D1 reporter SECIS elements prior to competitor expression, giving the reporter a ‘head start’. As seen in Figure 2B, this head start allowed D1 to escape competition following subsequent induction of competitors. We interpret the results of these two experiments to indicate that, once binding of the limiting factor to SECIS elements has occurred, the rate of exchange of the factor between elements is very slow.
Fig. 2. Time-staggered expression of competitors and reporter. (A) Expression of SECIS competitors prior to selenoprotein reporter inhibits selenoprotein expression. SECIS competition was performed as described above except that expression of the D1 selenoprotein was delayed 24 h relative to SECIS competitors by use of the tetracycline-repressible vector system. Coding regions and SECIS elements of competitors are indicated in the key. (B) Expression of the selenoprotein reporter prior to SECIS competitors abrogates competition. Expression of SECIS competitors was delayed 24 h relative to D1 expression. Activity shown is the mean ± SE of duplicate assays from at least three independent transfections, normalized to β-galactosidase activity expressed from a co-transfected plasmid.
A SECIS element in an unstable mRNA competes efficiently
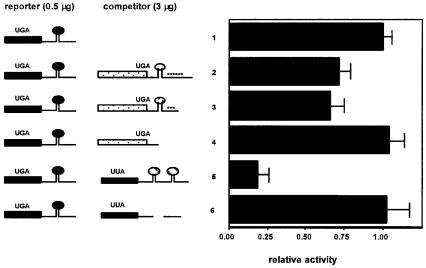
To address further the question of exchange of the limiting factor between elements, we examined the ability of a SECIS element in an unstable mRNA to compete for selenocysteine incorporation, with the thought that if the mRNA were rapidly degraded, the limiting factor might be released to bind another element. The mRNA for human thioredoxin reductase 1 (TR-1) contains seven AUUUA instability elements and is rapidly turned over. In addition, it contains an intron in the 3′ UTR, which would be predicted to target the mRNA for NMD (see Discussion). We have characterized a cDNA containing six of the AU-rich elements, and shown that deletion of either three or all six of the elements confers dramatic increases in mRNA levels (Gasdaska et al., 1999). Co-transfection of either a full-length TR-1 construct, or a construct from which three instability elements were deleted but the SECIS element was retained, resulted in competition for selenocysteine incorporation to the same level as competition by the D1 SECIS element (Figure 3). A TR-1 construct with the SECIS and instability elements deleted did not compete.
Fig. 3. A SECIS element in an unstable mRNA competes efficiently. Competition was performed as described in the legend to Figure 1, except that competitor constructs consisted of: line 1, vector alone; line 2, TR-1 coding region (stippled bars), SECIS element (stippled oval) and six AU-rich instability elements (asterisks); line 3, TR-1 coding region, SECIS element and three AU-rich instability elements; line 4, TR-1 coding region; line 5, leucine mutant D1 coding region/wild-type sel P SECIS elements 1 and 2; line 6, leucine mutant D1 coding region/SECIS deleted. Other symbols are as in Figure 1.
What is the identity of the limiting SECIS-binding factor?
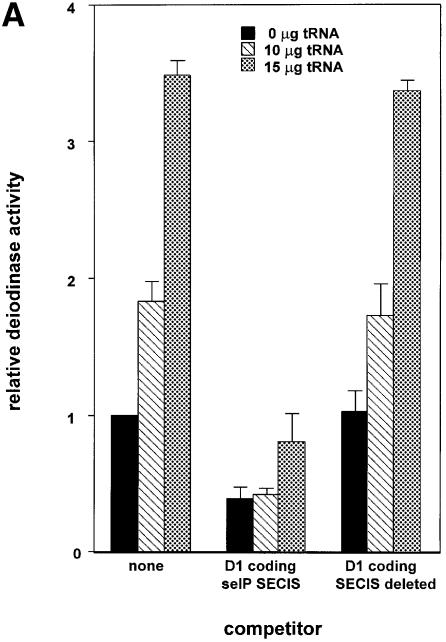
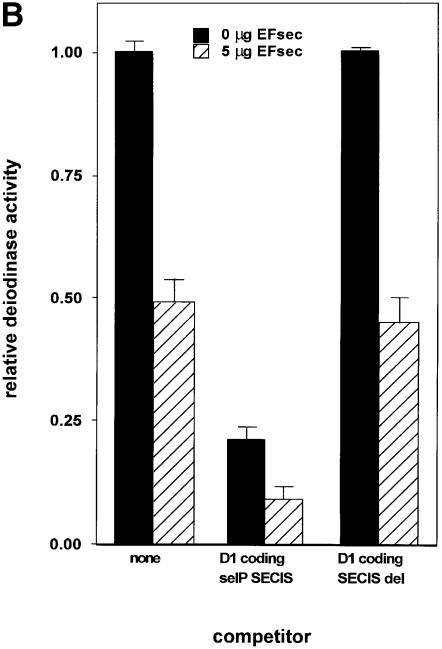
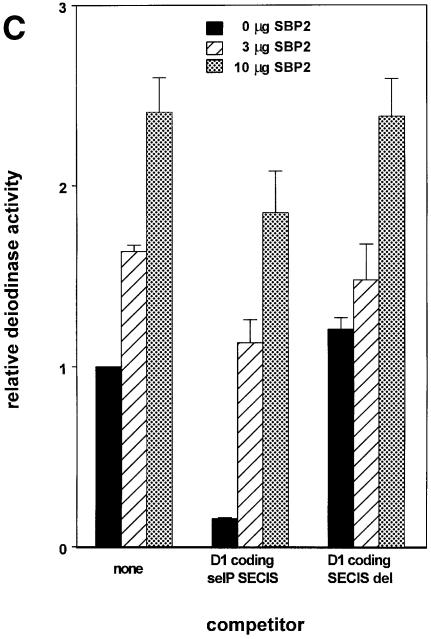
To determine the identity of the limiting factor or factors in SECIS competition studies, we performed competition studies similar to those described above in the presence of plasmids expressing either Xenopus tRNA[Ser]Sec, EFsec or SBP2. As shown in Figure 4A, co-transfection of 10 or 15 µg of Xenopus tRNA[Ser]Sec plasmid increased the activity of D1 by ∼2- and 3.5-fold, respectively, either in the absence of competitor or in the presence of a SECIS deletion competitor. However, 10 µg of the tRNA plasmid had no effect on overcoming the decrease in D1 activity seen in the presence of both sel P SECIS elements. Although 15 µg of tRNA[Ser]Sec plasmid increased activity, levels of enzyme activity were still well below those seen in the absence of competitor. Co-transfection of EFsec resulted in consistent decreases in D1 activity, in either the absence or presence of competitors (Figure 4B). In contrast to the results with the above two factors, co-transfection of SBP2 almost completely reversed the competition effect of the co-transfected D1 coding-sel P SECIS element construct, indicating that this is the factor that is made limiting by excess SECIS elements (Figure 4C).
Fig. 4. Transfection of SBP2 overcomes competition by SECIS elements, but selenocysteyl-tRNA and EFsec do not. Competition was performed as described in the legend to Figure 1. (A) The indicated amounts of selenocysteyl-tRNA-expressing plasmid were co-transfected with the D1 reporter and the indicated competitor constructs. (B) The indicated amounts of EFsec-expressing plasmid were co-transfected with D1 reporter and competitor constructs. (C) The indicated amounts of SBP2-expressing plasmid were co-transfected with D1 and competitor constructs.
SBP2 dictates a hierarchy amongst SECIS elements
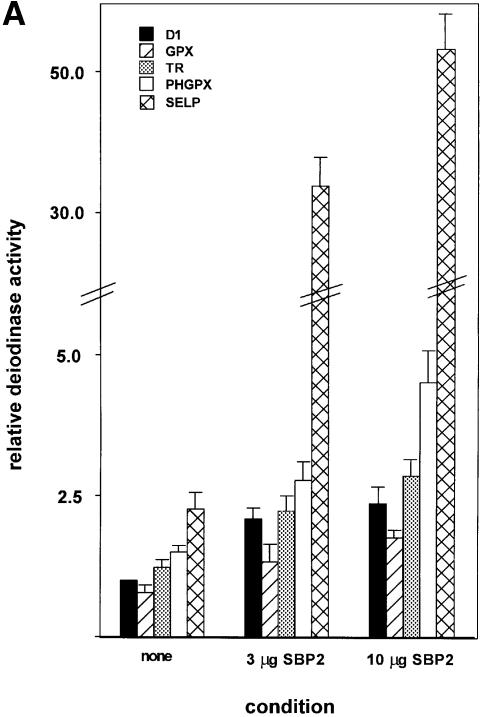
We next assessed the effects of SBP2 on expression from constructs containing SECIS elements from different selenoproteins, linked to a common selenoprotein ORF, that of D1. In all cases, the minimal SECIS elements were used (Martin et al., 1996, 1998), so that other sequences that might possibly affect mRNA stability or translation would not be included. We have previously shown that the GPX and PHGPX elements exert minimal effects on mRNA turnover in the D1 chimeric constructs (Martin et al., 1998; Martin, 1999). This was further verified for the TR-1 and sel P elements (see below). The relative activities of the SECIS elements in the absence of co-transfected SBP2 were slightly lower for cytoplasmic GPX, and slightly higher for TR-1, relative to D1 (Figure 5A), in agreement with previous findings (Berry et al., 1991c; Gasdaska et al., 1999). The PHGPX and first sel P SECIS elements were ∼50% and 2.5-fold more active, respectively, as previously observed (Berry et al., 1993; Kollmus et al., 1996; Lesoon et al., 1997). Strikingly, co-transfection of SBP2 at either 3 or 10 µg per 60 mm dish dramatically increased these differences for the latter two constructs. The enhancement of expression from the mRNA containing the PHGPX SECIS element was ∼3- to 4.5-fold, versus ∼2- to 2.5-fold for D1, GPX and TR-1. The effects on the sel P SECIS element were even more pronounced: ∼14- and 22-fold at the two concentrations of SBP2 tested.
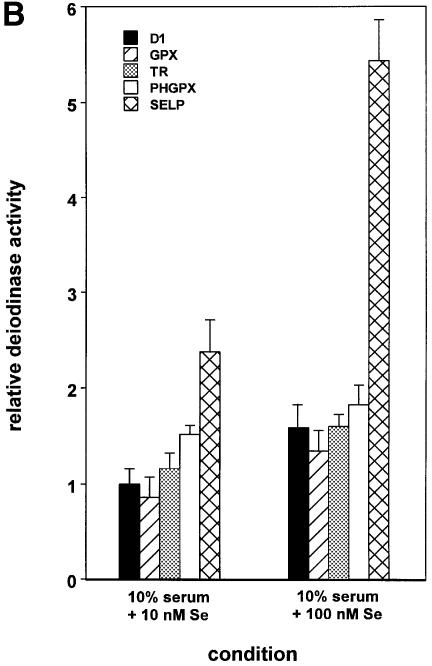
Fig. 5. Differential SECIS activities and responses to SBP2 and selenium dictate selenoprotein synthesis hierarchy. (A) Deiodinase activities expressed from constructs containing the D1 coding region, linked to the indicated SECIS elements, were assayed in the absence and presence of co-transfected SBP2, as described in Materials and methods. (B) Deiodinase activities expressed from the indicated constructs were assayed following culturing in the presence of the indicated concentrations of selenium added to the medium.
To determine whether differential mRNA turnover contributed to the differences in competition and response to SBP2, mRNA half-lives were determined. Cells were transfected with constructs encoding the D1 ORF plus various SECIS elements, treated with actinomycin D 48 h after transfection, and RNA harvested 6, 12 and 24 h thereafter. Half-lives for mRNAs containing the minimal D1 and TR-1 SECIS elements were 14–18 h, whereas the mRNAs containing the minimal GPX or first sel P SECIS both exhibited half-lives of ∼24 h. These differences do not correlate with the widely differing competitiveness and SBP2 responses of the GPX and first sel P SECIS elements.
Response of different SECIS elements to Se supplementation
Finally, we assessed the effects of selenium supplementation on expression from this same series of constructs. Expression from these constructs followed the same general trends as in the SBP2 supplementation studies above (Figure 5B), but the magnitude of the responses was less pronounced, possibly due to the inability of the translation machinery to respond fully to selenium supplementation, due to limiting SBP2.
Discussion
The mechanism of selenocysteine incorporation into proteins remains the most elusive aspect of selenoprotein biology. In prokaryotes, the location of the stem–loop structures directing selenocysteine incorporation in the coding region requires that the SELB protein dissociate and the stem–loop structure be melted so that ribosomes can translate through this region. In eukaryotes, because the SECIS elements are located in the 3′ UTR, there is no inherent mechanistic requirement for the dissociation of SBP2. In fact, we would predict that the efficiency of incorporation would increase if the protein were to stay bound through repeated cycles of selenocysteine incorporation, and this scenario would be particularly advantageous for the translation of sel P. The results reported herein provide the first demonstration that a SECIS element-specific factor required for selenoprotein translation, presumably SBP2, does not readily exchange between elements during the time course of the experiments. The lack of exchange of the factor between mRNAs suggests that once binding has taken place, SBP2 either dissociates very slowly or not at all, and may stay bound until either the mRNA or the factor itself is degraded. In fact, the protein may stay bound to the core of a SECIS element even when the rest of the mRNA is degraded, as in the case of the rapidly turned over TR-1 mRNA.
The fact that SBP2 overcomes SECIS element competition, but that EFsec does not and the tRNA has only a minor effect, indicates that the first of these is the factor rendered most limiting by this competition. Other recent in vivo and in vitro studies support the fact that EFsec is not limiting either in HEK cells (our unpublished studies) or in rabbit reticulocyte lysates (P.R.Copeland, personal communication). In E.coli, overexpression of the selenocysteyl-tRNA-specific elongation factor, SELB, resulted in inhibition of selenocysteine incorporation, possibly due to perturbation of the normal balance between this and other factors (Tormay et al., 1996). A similar effect may explain the inhibition of selenoprotein expression in the presence of co-transfected EFsec.
As discussed above, recent studies have shown that the mRNA for GPX undergoes NMD when selenium is limiting (Moriarty et al., 1998; Weiss and Sunde, 1998). However, other selenoprotein mRNAs, including those for D1, PHGPX and selenoprotein P, appear to be much less sensitive to this pathway under conditions of selenium deprivation, despite the presence of introns downstream of the UGA codons in these mRNAs (Hill et al., 1992; Lei et al., 1995). The thioredoxin reductase mRNAs, on the other hand, would be predicted to be sensitive to NMD independent of selenium status, as the selenocysteine codons and authentic termination codons are not separated by introns, and the final intron in at least one of these mRNAs is located in the 3′ UTR, ∼70 nucleotides downstream of the termination codon.
The requirement for the presence of a SECIS element for UGA decoding indicates that factor binding to the SECIS element should precede arrival of the first ribosome at the first UGA codon to circumvent termination and NMD. The NMD pathway may thus serve as a regulatory step to direct available selenium to some selenoproteins, i.e. those higher up in the hierarchy, at the expense of others. If, for example, the selenoprotein mRNA pool exceeded the pool of available SBP2, such that some selenoprotein mRNAs were to undergo translation without recruiting the SBP2–EFsec–tRNA complex, those mRNAs would be degraded via the NMD pathway. The hierarchy of SBP2 effects reported herein indicates that differential SBP2 binding may contribute to the increased sensitivity of GPX versus, for example, D1 to NMD. However, the differences in response were <2-fold, indicating that other factors inherent in the GPX mRNA are likely to play a greater role in the high sensitivity of this mRNA to NMD under conditions of selenium limitation. The high affinity of SBP2 for the first sel P SECIS element and the PHGPX element may also contribute to the relative resistance of these two mRNAs to NMD (Hill et al., 1992; Lei et al., 1995). Preliminary in vitro binding and competition studies indicate that the first sel P SECIS does in fact compete more effectively for SBP2 binding than that of D1. The relative lack of exchange of SBP2 between SECIS elements may further contribute to the apparent SECIS-specific hierarchy of selenoprotein synthesis and NMD sensitivity.
The tissue-specific differences in selenoprotein synthesis and sensitivity to selenium deprivation are likely to be due to several factors, including the efficiency of selenium retention, levels of SBP2 (Copeland et al., 2000) and EFsec (Tujebajeva et al., 2000), all of which are highest in testes, and effects on the selenocysteyl-tRNA population. Selenium supplementation has been shown to effect a specific modification of one of the nucleosides in selenocysteyl-tRNA (Diamond et al., 1993), and this modification occurs to different extents in different tissues (Chittum et al., 1997). The differential stimulation seen herein upon selenium addition may reflect differential utilization of newly synthesized selenocysteyl-tRNA, dictated by SBP2 affinity for different SECIS elements. However, we cannot rule out the possibility that the selenium-induced modification of the tRNA also results in its differential utilization by different SECIS elements. The combination of all of these factors is likely to play important regulatory roles in contributing to the selenoprotein synthesis hierarchy and differential sensitivities to NMD.
Materials and methods
Constructs
Reporter and competitor constructs. Reporter constructs consisted of the full-length rat D1 cDNA sequence (Berry et al., 1991a) in either the vector pUHD10-3 (Gossen and Bujard, 1992) or CDM-8 (Aruffo and Seed, 1987). Competitor constructs are derived from portions of the rat D1 sequence, the human D2 sequence, the rat sel P 3′ UTR, or the human TR-1 sequence. Competitors consisted of either wild-type or mutant SECIS elements alone, or SECIS elements linked downstream of ORFs. For studies of SBP2 or selenium effects, constructs consisted of the wild-type D1 coding region followed by the SECIS elements from rat D1, rat GPX (gift of Dr Ye Shih Ho), human TR-1 (Gasdaska et al., 1999), rat PHGPX (gift of Roger Sunde) or rat sel P (gift of Kris Hill and Ray Burk).
ORF-less competitors. A region of the wild-type D1 3′ UTR consisting of the SECIS element and flanking sequences (∼250 nucleotides) or the two wild-type sel P elements and flanking sequences was subcloned into CDM-8 immediately downstream of the CMV promoter. As a control competitor, a construct containing the D1 SECIS stem but lacking the nine nucleotide loop was subcloned into the same vector.
Competitors linked to ORFs. For competitors linked to an ORF, either the D1, D2 or TR-1 ORF was used. PCR mutagenesis was used to create a leucine (UUA) codon in place of the native selenocysteine (UGA) codon at amino acid 126 in D1. The leucine mutant has no D1 activity and, therefore, will not contribute to activity measured in D1 assays (Berry et al., 1991a). Wild-type or mutant D1 or sel P SECIS elements were cloned downstream of the leucine mutant D1 coding region, as indicated in Figure 1. These include: the wild-type D1 SECIS and flanking sequences (line 2); the minimal D1 SECIS element (line 3) (Martin et al., 1996); a point mutant of the minimal D1 SECIS element (line 4, AUGA→AUCA); the D1 SECIS nine nucleotide loop deletion mutant (line 5; Berry et al., 1991c); the two wild-type sel P elements and flanking sequences (line 6; Berry et al., 1993); or the first sel P element (line 7). The mutant SECIS elements (lines 4 and 5) have previously been shown to result in the complete loss of selenocysteine incorporation into D1 (Berry et al., 1991c). A final competitor in this series consisted of the coding region of D2 linked to the first sel P element (Figure 1, line 8) (Salvatore et al., 1996). Using appropriate reaction conditions [i.e. 1 µM reverse T3 (rT3)], D1 activity can be assayed without any contribution from the D2 enzyme.
TR-1 competitors. The TR-1 selenoprotein sequence encodes seven AU-rich elements (AREs) in the 3′ UTR, conferring rapid turnover of the mRNA product (Gasdaska et al., 1999). TR-1 competitors consisted of the nearly full-length cDNA (containing six of the seven AREs) (Figure 3, line 2), the cDNA truncated at nucleotide 3326 (numbering according to Gasdaska et al., 1999), which removes three of the AREs but retains the SECIS element and three AREs (line 3), and the cDNA truncated at nucleotide 1975, which removes the SECIS element and all six AREs (line 4).
Selenocysteine incorporation factors. The Xenopus laevis selenocysteyl-tRNA gene was generously provided by Dolph Hatfield. SBP2 in PCR3.1 was generously provided by Paul Copeland and Donna Driscoll. Murine EFsec was recently identified and characterized in this laboratory (Tujebajeva et al., 2000).
Transient transfections
Transient transfections were carried out in human embryonic kidney (HEK 293) cells using the calcium phosphate method of transfection as described previously (Berry et al., 1991b). Three days prior to transfection, HEK 293 cells were plated onto 60 mm culture dishes in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum. Cells were transfected with amounts of plasmid indicated in the figure legends. To monitor transfection efficiencies, cells were co-transfected with 1–3 µg of a plasmid encoding the β-galactosidase gene or with 3 µg of an expression vector containing the human growth hormone cDNA under the control of the HSV thymidine kinase promoter. For experiments utilizing pUHD10-3 constructs, cells were also transfected with 4 µg of the pUHD-15 plasmid (Gossen and Bujard, 1992), which encodes a protein necessary for transcriptional activation of the pUHD10-3 promoter. Experiments to identify a limiting factor in the competition studies were performed with the indicated amounts of co-transfected X.laevis tRNA[Ser]Sec plasmid (Lee et al., 1990), EFsec expression plasmid (Tujebajeva et al., 2000), or SBP-2 expression plasmid (Copeland and Driscoll, 1999). Medium was changed 1 day following transfection. Two days after transfection, cells were harvested, washed, and resuspended in 0.1 M potassium phosphate pH 6.9, 1 mM EDTA containing 0.25 M sucrose. All experiments were performed in duplicate and carried out at least three times with similar results. For selenium supplementation studies, the indicated concentrations of sodium selenite were added to the medium the day before transfection, with fresh medium supplemented with sodium selenite added the day after transfection.
Experiments in which expression was staggered using the tetracycline-repressible system were carried out essentially as above, except that tetracycline was added to the medium at the time of transfection. For expression of competitors before reporter, the D1 reporter in the tetracycline-repressible vector pUHD-10 (Gossen and Bujard, 1992) and the various competitors in the constitutive vector CDM-8 (Aruffo and Seed, 1987) were co-transfected at the same time, and tetracycline was included to repress D1 expression. Twenty-four hours after transfection, medium was changed and cells rinsed once with fresh medium to remove tetracycline, allowing the induction of D1 expression. Cells were maintained for an additional 24–72 h to allow full expression of D1, then harvested for assay of D1 activity. For expression of reporters before competitor, competitors in the tetracycline-repressible vector were co-transfected with D1 reporter in CDM-8. Again, medium was changed after 24 h to remove tetracycline, and expression allowed to proceed for 24–72 h.
Deiodinase assays
Cell sonicates were assayed for 5′ deiodinase activity as previously described (Berry et al., 1991b). Briefly, cells were harvested, washed, and resuspended in 0.1 M potassium phosphate pH 6.9, 1 mM EDTA containing 0.25 M sucrose and 10 mM dithiothreitol. Cells were then sonicated and assayed for the ability to 5′ deiodinate [125I]rT3. Reactions contained 10–250 µg of protein, 1 µM [125I]rT3 and 10 mM dithiothreitol in a reaction volume of 300 µl. Reactions were incubated at 37°C for 30 min. 125I release was quantitated as described previously (Berry et al., 1991b). Deiodinase activities were calculated per microgram of protein and normalized to either the amount of growth hormone secreted into the medium (Larsen et al., 1986), or the amount of β-galactosidase activity in the cell sonicate. All constructs were tested in at least three separate transfections and deiodinase assays were performed in duplicate from each transfection.
β-galactosidase assays
Following transfection, cells were harvested and resuspended in 0.1 M potassium phosphate pH 6.9 containing 0.25 M sucrose. Aliquots of cell extract were incubated in 0.1 M sodium phosphate pH 7.5 containing 1 mg/ml ONPG (O-nitrophenyl-β-d-galactopyranoside), 1 mM MgCl2 and 45 mM β-mercaptoethanol at 37°C, until a yellow color developed. The reaction was stopped by adding 2 vols of 1 M Na2CO3 to each reaction. The optical density of the reactions was measured at a wavelength of 420 nm. From these absorbance readings, each transfection result could be corrected depending on the transfection efficiency.
RNA turnover
The half-lives of SECIS-containing mRNAs were determined by transfecting HEK cells with constructs encoding the D1 ORF plus various SECIS elements, followed by treatment with actinomycin D 48 h after transfection, and RNA harvest 6, 12 or 24 h thereafter. RNAs were detected by northern blotting using a D1 coding region-specific probe. Hybridization and washes were carried out at high stringency, followed by autoradiography and densitometric quantitation.
Acknowledgments
Acknowledgements
This work was supported by Public Health Service Grants DK-47320 and DK-52963 from the NIH (M.J.B.), and by an EMBO postdoctoral fellowship (S.C.L.).
References
- Aruffo A. and Seed,B. (1987) Molecular cloning of a CD28 cDNA by a high-efficiency COS cell expression system. Proc. Natl Acad. Sci. USA, 84, 8573–8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behne D., Hilmert,H., Scheid,S., Gessner,H. and Elger,W. (1988) Evidence for specific selenium target tissues and new biologically important selenoproteins. Biochim. Biophys. Acta, 966, 12–21. [DOI] [PubMed] [Google Scholar]
- Berry M.J., Banu,L. and Larsen,P.R. (1991a) Type I iodothyronine deiodinase is a selenocysteine-containing enzyme. Nature, 349, 438–440. [DOI] [PubMed] [Google Scholar]
- Berry M.J., Kieffer,J.D., Harney,J.W. and Larsen,P.R. (1991b) Selenocysteine confers the biochemical properties of the type I iodothyronine deiodinase. J. Biol. Chem., 266, 14155–14158. [PubMed] [Google Scholar]
- Berry M.J., Banu,L., Chen,Y., Mandel,S.J., Kieffer,J.D., Harney,J.W. and Larsen,P.R. (1991c) Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature, 353, 273–276. [DOI] [PubMed] [Google Scholar]
- Berry M.J., Maia,A.L., Kieffer,J.D. and Larsen,P.R. (1992) Substitution of cysteine for selenocysteine in type I iodothyronine deiodinase reduces the catalytic efficiency of the protein but enhances its translation. Endocrinology, 31, 1848–1852. [DOI] [PubMed] [Google Scholar]
- Berry M.J., Banu,L., Harney,J.W. and Larsen,P.R. (1993) Functional characterization of the eukaryotic SECIS elements which direct selenocysteine insertion at UGA codons. EMBO J., 12, 3315–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry M.J., Harney,J.W., Ohama,T. and Hatfield,D.L. (1994) Selenocysteine insertion or termination: factors affecting UGA codon fate and complementary anticodon:codon mutations. Nucleic Acids Res., 22, 3753–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittum H.S., Hill,K.E., Carlson,B.A., Lee,B.J., Burk,R.F. and Hatfield,D.L. (1997) Replenishment of selenium deficient rats with selenium results in redistribution of the selenocysteine tRNA population in a tissue specific manner. Biochim. Biophys. Acta, 1359, 25–34. [DOI] [PubMed] [Google Scholar]
- Christensen M.J. and Burgener,K.W. (1992) Dietary selenium stabilizes glutathione peroxidase mRNA in rat liver. J. Nutr., 122, 1620–1626. [DOI] [PubMed] [Google Scholar]
- Copeland P.R. and Driscoll,D.M. (1999) Purification, redox sensitivity and RNA binding properties of SECIS-binding protein 2, a protein involved in selenoprotein biosynthesis. J. Biol. Chem., 274, 25447–25454. [DOI] [PubMed] [Google Scholar]
- Copeland P.R., Fletcher,J.E., Carlson,B.A., Hatfield,D.L. and Driscoll,D.M. (2000) A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J., 19, 306–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis P.P. (1997) Ancient ciphers: translation in Archaea. Cell, 89, 1007–1010. [DOI] [PubMed] [Google Scholar]
- Diamond A.M. et al. (1993) Dietary selenium affects methylation of the wobble nucleoside in the anticodon of selenocysteine tRNA. J. Biol. Chem., 268, 14215–14223. [PubMed] [Google Scholar]
- Fagegaltier D., Hubert,N., Yamada,K., Mizutani,T., Carbon,P. and Krol,A. (2000) Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J., 19, 4796–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasdaska J.R., Harney,J.W., Gasdaska,P.Y., Powis,G. and Berry,M.J. (1999) Regulation of human thioredoxin reductase expression and activity by 3′ untranslated region selenocysteine insertion sequence and mRNA instability elements. J. Biol. Chem., 274, 25379–25385. [DOI] [PubMed] [Google Scholar]
- Gossen M. and Bujard,H. (1992) Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl Acad. Sci. USA, 89, 5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heider J., Baron,C. and Böck,A. (1992) Coding from a distance: dissection of the mRNA determinants required for the incorporation of selenocysteine into protein. EMBO J., 11, 3759–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze M.W. and Kulozik,A.E. (1999) A perfect message: RNA surveillance and nonsense-mediated decay. Cell, 96, 307–310. [DOI] [PubMed] [Google Scholar]
- Hill K.E., Lyons,P.R. and Burk,R.F. (1992) Differential regulation of rat liver selenoprotein mRNAs in selenium deficiency. Biochem. Biophys. Res. Commun., 185, 260–263. [DOI] [PubMed] [Google Scholar]
- Hill K.E., Lloyd,R.S. and Burk,R.F. (1993) Conserved nucleotide sequences in the open reading frame and 3′ untranslated region of selenoprotein P mRNA. Proc. Natl Acad. Sci., 90, 537–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmus H., Flohé,L. and McCarthy,J.E.G. (1996) Analysis of eukaryotic mRNA structures directing cotranslational incorporation of selenocysteine. Nucleic Acids Res., 24, 1195–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kromayer M., Wilting,R., Tormay,P. and Böck,A. (1996) Domain structure of the prokaryotic selenocysteine-specific elongation factor SelB. J. Mol. Biol., 262, 413–420. [DOI] [PubMed] [Google Scholar]
- Larsen P.R., Harney,J.W. and Moore,D.D. (1986) Repression mediates cell-type-specific expression of the rat growth hormone gene. Proc. Natl Acad. Sci. USA, 83, 8283–8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.J., Rajagopalan,M., Kim,Y.S., You,K.H., Jacobson,K.B. and Hatfield,D. (1990) Selenocysteine tRNA[Ser]Sec gene is ubiquitous within the animal kingdom. Mol. Cell. Biol., 10, 1940–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X.G., Evenson,J.K., Thompson,K.M. and Sunde,R.A. (1995) Glutathione peroxidase and phospholipid hydroperoxide glutathione peroxidase are differentially regulated in rats by dietary selenium. J. Nutr., 125, 1438–1446. [DOI] [PubMed] [Google Scholar]
- Leinfelder W., Forchhammer,K., Zinoni,F., Sawers,G., Mandrand-Berthelot,M.A. and Böck,A. (1988) Escherichia coli genes whose products are involved in selenium metabolism. J. Bacteriol., 170, 540–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesoon A., Mehta,A., Singh,R., Chisolm,G. and Driscoll,D.M. (1997) An RNA-binding protein recognizes a mammalian selenocysteine insertion sequence element required for cotranslational incorporation of selenocysteine. Mol. Cell. Biol., 17, 1977–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low S.C., Harney,J.W. and Berry,M.J. (1995) Cloning and functional characterization of human selenophosphate synthetase, an essential component of selenoprotein synthesis. J. Biol. Chem., 270, 21659–21664. [DOI] [PubMed] [Google Scholar]
- Martin G.W. III (1999) Regulation of selenocysteine insertion in eukaryotes: functional parameters of the SECIS element and in vivo efficiency of selenoprotein synthesis. PhD thesis, Harvard University, Cambridge, MA.
- Martin G.W., Harney,J.W. and Berry,M.J. (1996) Selenocysteine incorporation in eukaryotes: insights into mechanism and efficiency from sequence, structure and spacing proximity studies of the type 1 deiodinase SECIS element. RNA, 2, 171–182. [PMC free article] [PubMed] [Google Scholar]
- Martin G.W., Harney,J.W. and Berry,M.J. (1998) Functionality of mutations at conserved nucleotides in eukaryotic SECIS elements is determined by the identify of a single non-conserved nucleotide. RNA, 4, 65–73. [PMC free article] [PubMed] [Google Scholar]
- Mitchell J.H., Nicol,F., Beckett,G.J. and Arthur,J.R. (1997) Selenium and iodine deficiencies: effects on brain and brown adipose tissue selenoenzyme activity and expression. J. Endocrinol., 155, 255–263. [DOI] [PubMed] [Google Scholar]
- Moriarty P.M., Reddy,C.C and Maquat,L.E. (1998) Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA. Mol. Cell. Biol., 18, 2932–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy E. and Maquat,L.E. (1998) A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci., 23, 198–199. [DOI] [PubMed] [Google Scholar]
- Rother M., Wilting,R., Commans,S. and Bock,A. (2000) Identification and characterisation of the selenocysteine-specific translation factor SelB from the archaeon Methanococcus jannaschii. J. Mol. Biol., 299, 351–358. [DOI] [PubMed] [Google Scholar]
- Saedi M.S., Smith,C.G., Frampton,J., Chambers,I., Harrison,P.R. and Sunde,R.A. (1988) Effects of selenium status on mRNA levels for glutathione peroxidase in rat liver. Biochem. Biophys. Res. Commun., 153, 855–861. [DOI] [PubMed] [Google Scholar]
- Salvatore D., Tu,H., Harney,J.W. and Larsen,P.R. (1996) Type 2 iodothyronine deiodinase is highly expressed in human thyroid. J. Clin. Invest., 98, 962–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermann R., Neu-Yilik,G., Deters,A., Frede,U., Wehr,K., Hagemeier,C., Hentze,M.W. and Kulozik,A.E. (1998) Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J., 17, 3484–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tormay P., Sawers,A. and Bock,A. (1996) Role of stoichiometry between mRNA, translation factor SelB and selenocysteyl-tRNA in selenoprotein synthesis. Mol. Microbiol., 21, 1253–1259. [DOI] [PubMed] [Google Scholar]
- Tujebajeva R.M., Copeland,P.R., Xu,X.M., Carlson,B.A., Harney,J.W., Driscoll,D.M., Hatfield,D.L. and Berry,M.J. (2000) Decoding apparatus for eukaryotic selenocysteine insertion. EMBO rep., 1, 158–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S.L. and Sunde,R.A. (1998) Cis-acting elements are required for selenium regulation of glutathione peroxidase-1 mRNA levels. RNA, 4, 816–827. [DOI] [PMC free article] [PubMed] [Google Scholar]