Abstract
Cerebral blood volume (CBV) has been shown to be an important biomarker in a number of neurological disorders and in the quantitative interpretation of functional MRI. One approach to determine CBV in humans is Vascular-space-occupancy (VASO) MRI and this technique has been applied to the studies of brain glioma, Schizophrenia and Alzheimer’s Disease. However, validation of this technique with a gold standard method has not been reported. In this study, we compared VASO MRI with a radiotracer based PET technique in a group of healthy subjects. It was found that regional CBV measured with VASO MRI was highly correlated with that of the PET data (R=0.79±0.10, N=8). Furthermore, absolute CBV values quantified by VASO were also in excellent agreement with those by PET (slope=1.00±0.15). Due to differences in the labeling principles between the two modalities, systematic CBV differences were observed in large vessel and ventricle regions.
Keywords: cerebral blood volume, VASO MRI, Positron Emission Tomography, Gd-DTPA, brain
INTRODUCTION
Cerebral blood volume (CBV) is an important parameter for understanding the mechanism of blood-oxygenation-level-dependent (BOLD) functional MRI (1) and is also a useful marker in a number of brain disorders (2). We have recently developed a Vascular-Space-Occupancy (VASO) MRI technique to quantify CBV (1,3,4). This technique uses an MR contrast agent, gadolinium diethylenetriamine pentaacetic acid (Gd-DTPA), to differentiate tissue and vessel spaces, and is based on pre/post-contrast signal differences to calculate CBV. The VASO pulse sequence was designed such that the pre-contrast blood signal is nullified while the post-contrast blood signal is at equilibrium magnetization. This feature of the VASO technique provides a number of important advantages compared to other methods such as Dynamic Susceptibility Contrast (DSC) MRI (5) and steady-state contrast enhancement technique (6) in that the signal difference is maximized and that CBV quantification does not require the delineation of pure blood voxels. In addition, owing to the relatively long inversion time used in VASO MRI, the requirement that the post-contrast blood signal is approximately at equilibrium magnetization can be achieved at a relatively large range of contrast agent concentrations, thus rendering the method not affected by small variations in the actual dosage in individual subject.
It has been previously shown that CBV measured by VASO MRI is highly correlated with relative CBV measured by DSC-MRI (3) and is a useful marker for cerebral glioma (7), Schizophrenia (8), and Alzheimer’s disease (9). However, a validation of VASO MRI using a gold standard method has not been reported. The purpose of this work is to validate CBV measured by VASO MRI with a PET technique using 15O-labeled carbon monoxide (C15O). We performed PET and MRI scans on the same group of subjects to directly compare the CBV values measured by the two methods.
MATERIALS AND METHODS
Subjects
We recruited eight human subjects (age 34±13 years old, six men and two women) for this study. The Health Insurance Portability and Accountability Act (HIPAA) compliant protocol was approved by the Institutional Review Board of University of Texas Health Science Center and written informed consents were obtained from all participants. The PET and MRI scan sessions were performed on the same day. We minimized the time gap between the two sessions which was less than one hour for all subjects. Six of the subjects received PET scan first whereas the remaining two received MRI first.
PET experiment
The PET CBV measurement was conducted on a CTI Siemens HR+ scanner (Siemens, Knoxville, TN) in two-dimensional mode using 15O-labeled carbon monoxide (C15O) (half life 2 min). A thermoplastic facial mask was used to restrain head motion during the scan. Before the emission scans, a transmission scan using a 68Ge source was performed for attenuation correction. C15O gas was produced by an on-site cyclotron (Scanditronix AB, Uppsala, Sweden) and delivered to the subject via an air cushion mask (Teleflex Medical, NC). The emission scan started 5 min after a bolus inhalation of C15O gas (dosage 38–60 mCi) when the gas was fully mixed in the systemic blood, and continued for 5 min. PET image was reconstructed from the raw data using a standard attenuation correction and back-projection algorithm. The intrinsic resolution was 4.5 mm full-width-at-half-maximum (FWHM) in-plane and 5.1 mm FWHM axially.
During the PET session, venous blood samples were also collected with an antecubital intravenous line to measure emission activity of whole blood, which was used for absolute CBV quantification. Three samples were collected at the beginning, mid-point and end of the PET scan. The activities of the blood samples were measured by a well counter (Canberra Industries, Meriden, CT) that had been cross-calibrated to the PET scanner. The emission activities were corrected for attenuation based on well-established half life of 15O radiotracer and the data were averaged to improve signal to noise ratio (SNR).
The entire procedure for PET CBV measurement including the determination of full blood activity was conducted twice on each subject for improving SNR and also for assessing intra-modality reproducibility. The second PET scan was started as soon as the radioactivity returned to the background level after the first scan, which took approximately 20 minutes.
MRI Experiment
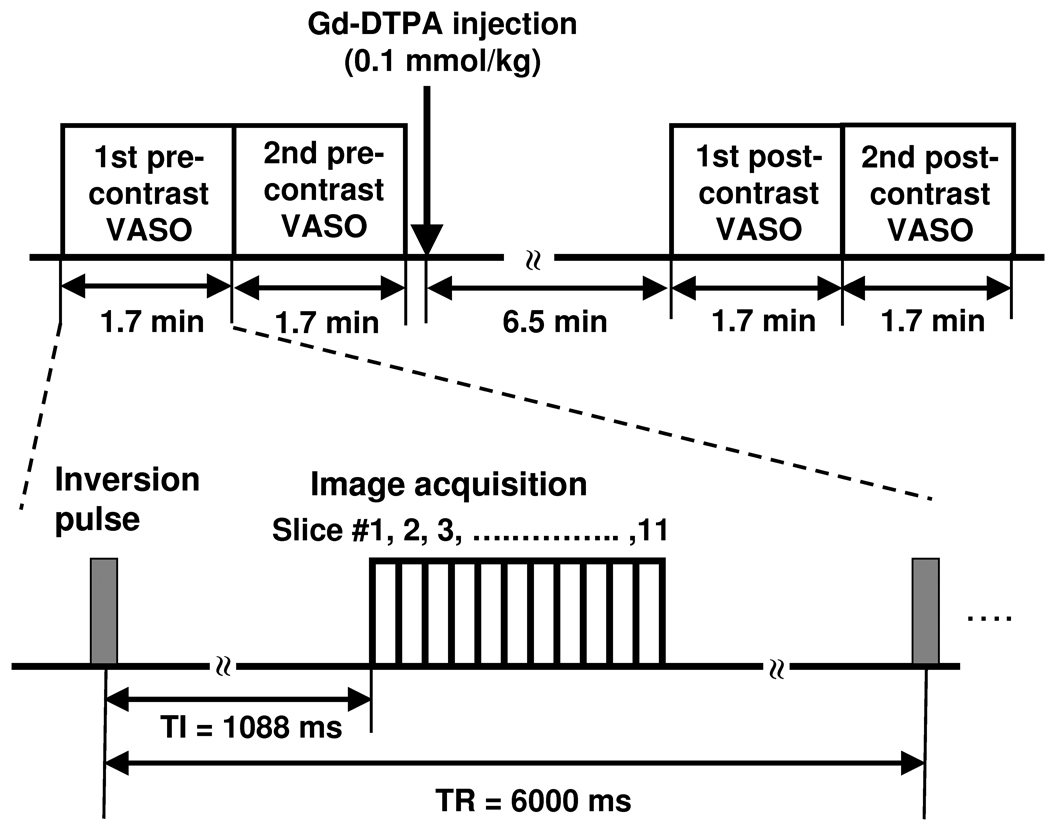
The MRI scans were conducted on a 3T MRI scanner (Siemens TIM Trio, Erlangen, Germany). The imaging parameters of VASO MRI sequence were: TI/TR/TE=1088ms/6000ms/5.9ms, FOV=220×220mm2, in-plane resolution=1.7×1.7mm2, EPI factor=7, 11 transverse slices with slice thickness=5mm (no gap), duration per scan=1min 42sec. The VASO protocol consisted of two pre-contrast VASO scans, injection of contrast agent, and two post-contrast VASO scans (Fig. 1). The post-contrast image acquisition started 6.5 min after the injection to allow the contrast agent to be fully mixed in the systemic blood (4). The Gd-DTPA contrast agent (Omniscan™, GE Healthcare Inc., Princeton, NJ) was administered intravenously by an MRI power injector (MEDRAD, Pittsburgh, PA) with a standard dosage (0.1 mmol/kg) and injection rate (5 ml/s).
Figure 1.
Experiment protocol for the VASO MRI study. A total of four VASO MRI scans were performed at the timing shown. The post-contrast VASO scans were started approximately 6.5 minutes after the Gd-DTPA injection to allow the contrast agent to fully mix with the blood and to reduce water exchange effects (4). The VASO pulse sequence includes a global inversion pulse followed by multi-slice image acquisitions.
We also acquired a proton density weighted image (with a TR of 20s and number of image volume acquisitions = 2) at the same resolution as VASO to estimate MR signal at equilibrium which is necessary for quantification of absolute CBV. A T1-weighted anatomical image with a resolution of 1×1×1 mm3 was acquired for spatial registration.
Data Processing
The PET CBV map was calculated by (10):
| [1] |
where the intensities of PET signal and blood sample are in counts per unit time per unit volume (Bq/ml). R is the ratio of hematocrit levels between small vessels and large vessels and was assumed to be 0.85 (11). This factor is included because C15O labels erythrocytes, the density of which is proportional to hematocrit. The two PET scans yielded two CBV maps, “PET CBV 1” and “PET CBV 2”. An averaged map, “PET CBV”, was also generated. These maps were spatially registered to the T1-weighted MR image using Statistical Parametric Mapping software (SPM, University College London, UK).
The MRI CBV map was computed from the VASO data based on algorithms described previously (3):
| [2] |
where Spre and Spost are the MRI signals before and after the injection of contrast agent. M0 is the MRI signal at equilibrium magnetization. Cb is the water density of blood and was assumed to be 0.87 (12). The VASO images were spatially registered using SPM. Two CBV maps were obtained: “VASO CBV 1” was calculated from the 1st pre-contrast and 1st post-contrast images (Fig. 1), and “VASO CBV 2” was calculated from the 2nd pre-contrast and 2nd post-contrast images. An averaged VASO CBV map was also calculated. An ROI covering the lateral ventricles were manually drawn and the mask was applied to the proton density image to obtain M0. We estimated two M0 values from the two acquisitions of the proton density scan and used them respectively for calculating VASO CBV 1 and VASO CBV 2. The mean of the two values was used for the averaged VASO CBV map.
Voxel-wise SNR of the CBV map was calculated as the ratio between the averaged CBV of the whole brain and the background noise. The background noise was estimated by the standard deviation of signal intensities in a region outside the brain. This procedure provides an assessment of the thermal noise but not the physiologic noise.
For a direct comparison of CBV maps across modalities, the PET CBV map was spatially registered to the VASO CBV map using the T1-weighted image as an intermediate. The images were further smoothed (FWHM=10mm) to improve SNR and to account for residual misalignment based on a method reported previously (13). To obtain gray and white matter CBV values, we performed tissue segmentation on the T1-weighted image using SPM with a probability threshold of 95%. The gray and white matter masks were then spatially registered into the CBV space. For statistical analysis, scatter plot between the two modalities was assessed and the Pearson cross correlation oefficient (R) was calculated. The scatter plot used a downsampled (with 10×10×3 voxels) version of the images to improve the SNR of individual points. To quantify the discrepancy between PET CBV and VASO CBV, a mean difference index, D, was calculated as
| [3] |
where D is in units of ml blood/100ml brain, CBVPET,i and CBVVASO,i are PET CBV and VASO CBV values, respectively, for a downsampled voxel i, and N is the total number of downsampled voxels in the brain. In addition, similar analysis was performed for PET CBV 1 vs. PET CBV 2 and VASO CBV 1 vs. VASO CBV 2, to assess the intra-modality differences.
We further identified brain regions that showed significant differences between the two modalities by conducting a voxel-wise Student t test for the PET and VASO CBV maps. For this analysis, CBV maps were spatially normalized to a template brain provided in SPM using the T1-weighted image as an intermediate.
RESULTS
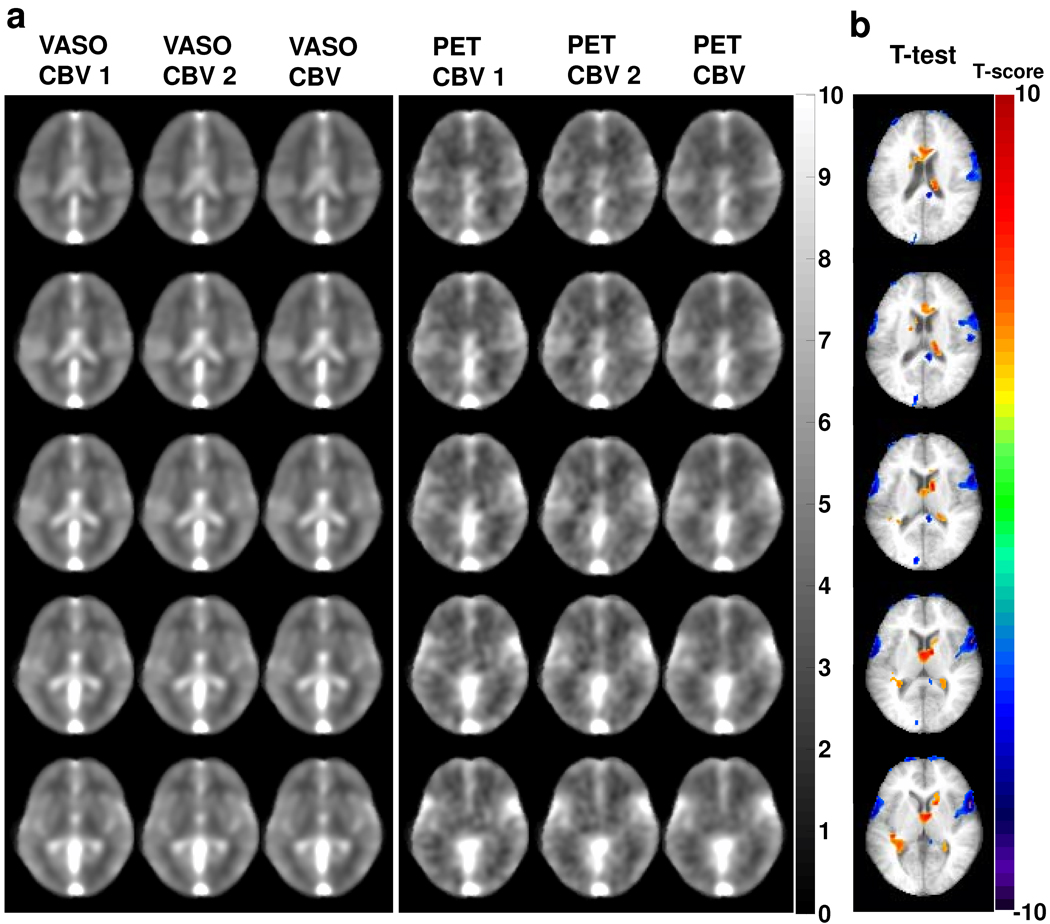
The group averaged CBV maps using VASO and PET methods are shown in Fig. 2a. The image contrasts between the two modalities are similar with gray matter showing higher CBV compared to white matter and large vessel regions having the greatest signals. The whole-brain VASO CBV values were 4.3±0.4 ml/100 ml brain (mean±SD, N=8) and 3.1±0.2 ml/100 ml brain for gray and white matters, respectively. The PET CBV values were 4.4±0.8 ml/100 ml brain and 3.3±0.7 ml/100 ml brain for gray and white matters, respectively. Note that the differences between gray and white matter CBV values were moderate because the spatial smoothing may have introduced some partial volume effects.
Figure 2.
CBV maps from the VASO and PET studies. a) Two repeated measures of VASO and PET CBV maps and their respective averages. The images have been spatially smoothed and averaged across subjects (N=8). b) Thresholded t score maps showing brain regions with significant CBV differences between the two modalities. Group-level paired t test was performed on a voxel-by-voxel basis for the VASO and PET CBV maps. Voxels that meet the threshold of P<0.01 and cluster size>0.8 cm3 are shown in color. Warm color indicates voxels in which VASO CBV is greater than PET CBV. Cold color indicates voxels in which VASO CBV is smaller than PET CBV.
The voxel-wise SNR of the VASO and PET images (without smoothing) were 7.6±1.1 (mean±SD, N=8) and 2.0±0.4, respectively. Using Monte Carlo simulation, SNR of the smoothed images was estimated to be approximately 67±10 and 16±3 for VASO and PET, respectively.
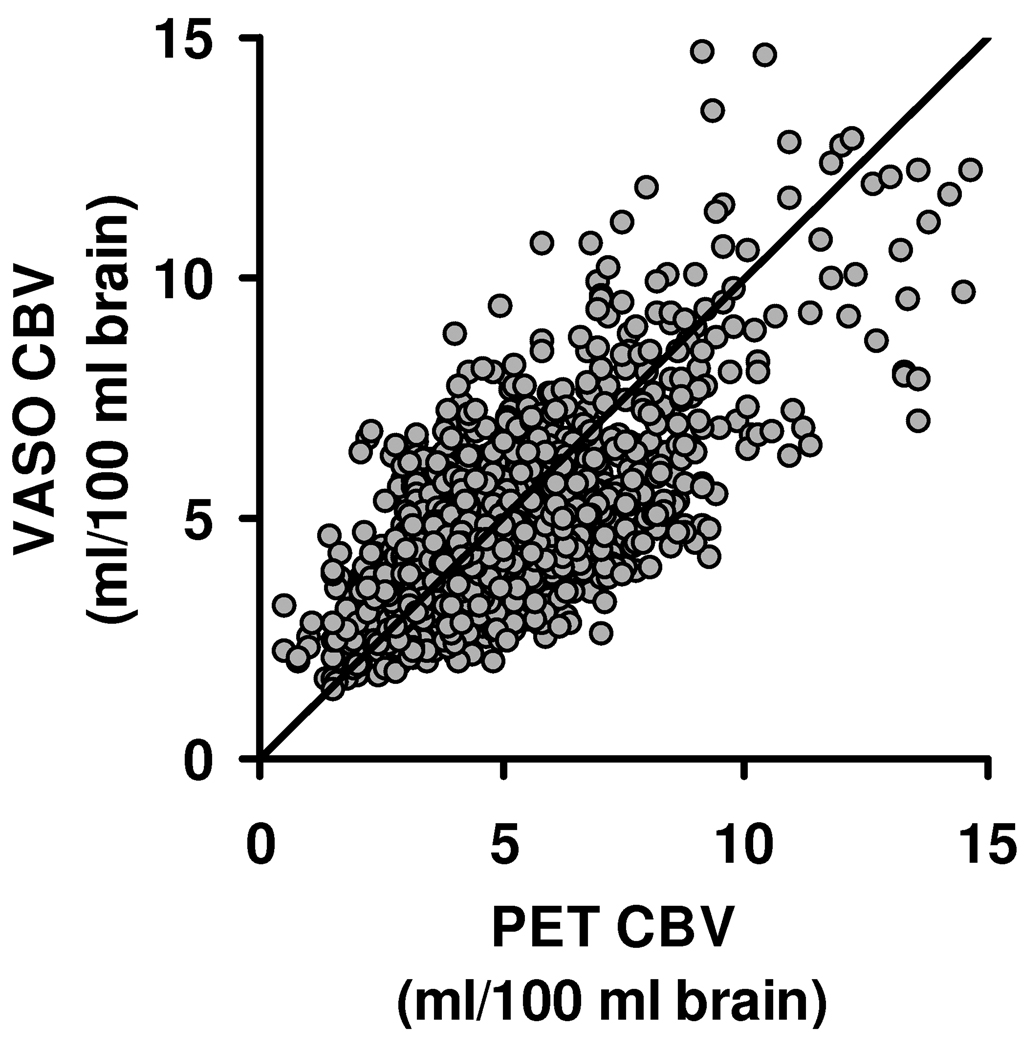
Quantitative analysis revealed that regional CBV values between VASO and PET are highly correlated (R=0.79±0.10, mean±SD, N=8, P<0.0001). Figure 3 shows a scatter plot of regional values (from the downsampled images) for all participants. The slopes of the linear fitting were 1.00±0.15, suggesting that the quantification of absolute CBV using VASO MRI is in excellent agreement with the gold standard method. The mean difference, D, between VASO and PET CBV maps was 1.39±0.36ml/100 ml brain.
Figure 3.
Scatter plot between VASO and PET regional CBV values. The regional values were obtained from the downsampled images. Each circle represents one voxel in the downsampled images. The plot includes data from all subjects studied. The diagonal line is the unity curve.
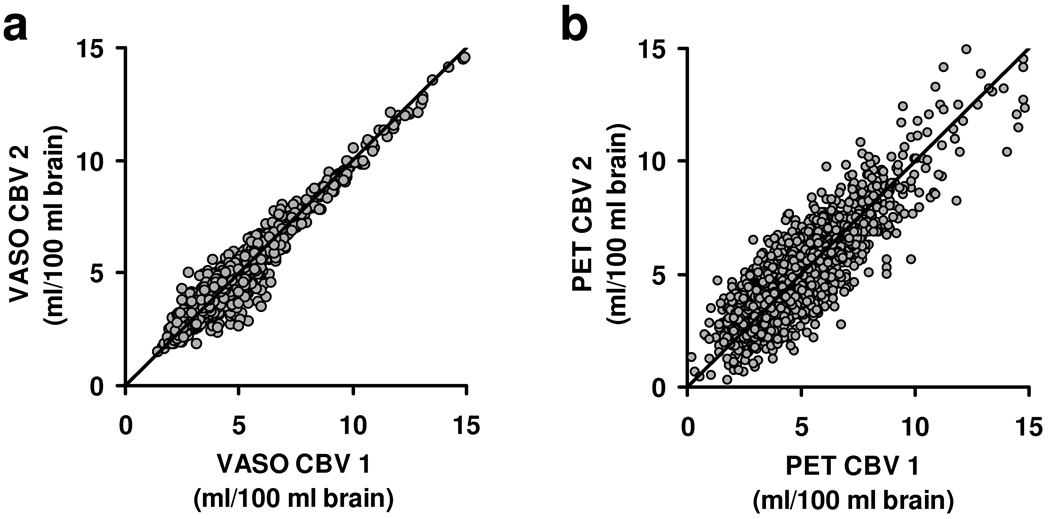
We also assessed intra-modality consistency in CBV maps. Figures 4a and 4b show the scatter plots between the two VASO scans and between the two PET scans, respectively. The slopes of these scatter plots were 0.99±0.02 and 1.00±0.11 for VASO and PET CBV, respectively. The VASO CBV maps had higher reproducibility compared to PET (P<0.0001). The correlation coefficient and the mean difference between the two VASO measurements were 0.97±0.02 and 0.41±0.17ml/100 ml brain, respectively, while they were 0.87±0.05 and 1.12±0.18ml/100 ml brain between the two PET measurements.
Figure 4.
Scatter plots of intra-modality comparisons. a) Relationship between VASO CBV 1 and VASO CBV 2. b) Relationship between PET CBV 1 and PET CBV 2.
Figure 2b shows the result of voxel-wise comparison. It can be seen that the regions where VASO CBV is significantly lower than PET CBV (marked by cold color) are primarily large vessel areas, while the regions where VASO CBV is higher than PET CBV are mainly in ventricle areas (marked by warm color).
DISCUSSION
In this study, we validated a recently developed CBV technique, VASO MRI, with a gold standard PET method. Our data suggested that VASO CBV showed good agreement with PET CBV, both in terms of regional correlation and in absolute CBV values. In addition, reproducibility assessment and SNR analysis suggested that VASO CBV maps were more reproducible compared to PET CBV.
CBV is a useful biomarker in a number of clinical applications. We and others have shown that CBV can be used as a sensitive index for differentiating between low grade and high grade brain tumors (2,7,14). Our previous study has also shown that patients with Alzheimer’s Disease have significantly lower CBV in frontal and parietal regions (9). CBV changes associated with neural activation have also been exploited for functional brain imaging (1). There are also evidences that CBV may be a useful marker for delineation of tissues at risk in stroke (15). Therefore, validation of the VASO CBV method will provide an important foundation for future applications of this technique.
It should also be noted that several previous studies have provided similar PET validations for other MRI techniques. Grandin and colleagues compared CBV determined by DSC MRI to that using 11CO PET and observed that DSC MRI over-estimated CBV values but the effect could be corrected with a scaling factor (16). Carroll et al. conducted comparison between CBF measured by DSC MRI and that by H2 15O PET, and concluded the vascular artifacts are major confounding factors in DSC results (17). Similar observations on vascular contributions were noted by Ibaraki and coworkers in a DSC/PET comparison (18). In addition, several laboratories have conducted validation studies for a non-contrast-based CBF technique, arterial spin labeling (ASL) MRI (19). The present study is the first to validate the VASO MRI method.
Despite a significant correlation between VASO and PET CBV values (Fig. 3), we noted that not all points in the scatter plot are located on the unity line and there is considerable spread in the data. This discrepancy may be attributed to either systematic differences between the two modalities or measurement uncertainty in each modality (i.e. random noise). We therefore assessed the intra-modality correlation for each technique. It is apparent that the inter-modality difference is greater than the intra-modality difference. Note that the inter-modality D values reported above contained both the measurement errors of each modality and systematic differences between the two modalities, whereas the intra-modality D values primarily reflect the measurement errors. Thus we can further calculate the systematic differences between VASO and PET using the three D values (see Appendix for derivations). The differences due to modalities were found to be 1.23±0.44ml/100 ml brain, which corresponds to the majority (1.23 out of 1.39 ml/100ml brain) of the spread in Fig. 3. One important mechanistic difference between the two techniques is that C15O PET labels erythrocytes while VASO MRI uses a plasma agent Gd-DTPA. This difference may explain the observed CBV discrepancies in both large vessels and ventricles (Fig. 2b). Blood in large vessels is known to have higher erythrocyte fraction (i.e. hematocrit) compared to microvessels. Thus the tracer signal in large vessels will appear higher than the actual blood volume, which causes the estimated PET CBV value to be greater. In the ventricle areas, the vascular structure is called choroid plexus. Choroid plexus does not have blood-brain-barrier (BBB) unlike the brain parenchyma, thus the Gd-DTPA molecules can diffuse to the surrounding CSF space, causing signal enhancement that is not associated with blood volume (20). Consequently, CBV measured by MRI may contain some over-estimation in ventricle areas while the PET method does not have this confound as erythrocytes do not leak into CSF space under normal conditions (18,20).
In the present study, we used smoothed images for quantitative comparison between PET and VASO CBV maps. However, a disadvantage of this processing is that the spatial information at local regions was lost. The smoothed images were used as a result of a tradeoff between resolution and SNR. We have tested the feasibility of using the unsmoothed images. For this test, the original VASO CBV map and the spatially registered PET CBV map (voxel size of both maps =1.7×1.7×5 mm3) were not smoothed but only downsampled with 3×3×1 voxels so that the downsampled voxel size is comparable to the intrinsic resolution of PET (4.5×4.5×5.1 mm3). The comparison using the unsmoothed images confirmed that quantitative PET and VASO CBV values agree with each other on the global level. The scatter plot of CBV values showed slopes of 1.05±0.16 which were not different from unity (p=0.40). On the other hand, the regional CBV values were only moderately correlated (R=0.47±0.07). Intra-modality comparison revealed that this moderate correlation is primarily attributed to the poor reproducibility of the unsmoothed PET images. The correlation coefficient between the two unsmoothed PET measurements (0.38±0.08) was significantly lower than that of the smoothed images (0.87±0.05), while the two VASO measurements showed high correlation coefficient even in the unsmoothed version (0.97±0.02). This result indicates that the moderate correlation is not necessarily due to mismatch between PET and VASO CBV at local regions. Instead, it appears that the poor SNR of PET measurement precludes a reliable comparison at a higher resolution.
The findings from the present study need to be interpreted in the context of a few limitations. This validation study only used healthy control subjects, but no patient populations were included. Patients with neurological disorders may have considerable BBB leakage which may cause VASO MRI to over-estimate CBV, similar to the situation of choroid plexus described earlier. Therefore, a validation in the presence of BBB breakdown shall be considered in future studies. In addition, the VASO experiments conducted in the present study are based the T1 shortening effect of Gd-DTPA, but the T2 effect was not considered. This is because a relatively short TE of 5.9ms was used. However, in large vessels where the apparent T2 is short due to spin outflow, the T2 effect may cause an under-estimation of CBV, which may explain part of the discrepancy between VASO CBV and PET CBV in these regions. Multi-echo acquisition schemes may be useful in minimizing such effects.
CONCLUSIONS
Comparison of VASO MRI with a gold standard PET method suggests that this technique can provide a quantitative estimation of regional CBV in humans. Certain discrepancies were observed in large vessels and ventricle regions which were attributed to the different labeling methods between the two modalities.
Acknowledgments
Grant sponsors: NIH R21 NS054916, NIH R01 MH084021, NIH R21 EB007821, American Heart Association 0865003F
APPENDIX
Relationship between intra- and inter-modality differences in CBV measures
The following derivation will show that the systematic differences between VASO and PET can be calculated from the intra- and inter-modality differences. We first define VASO CBV 1 and VASO CBV 2 of a voxel i (denoted by CBVV1,i and CBVV2,i) as:
| [A1] |
where TV,i is the expected VASO CBV value and εV1,i and εV2,i are measurement errors. The mean difference index, D, between VASO CBV 1 and VASO CBV 2 is:
| [A2] |
where the measurement errors were assumed to be random variables and are independent across voxels.
Similar derivation can be made for PET CBV 1 and PET CBV 2, and the mean difference between the repeated PET measures can be written as:
| [A3] |
where εP1,i and εP2,i are measurement errors in PET.
Finally, the mean difference between VASO CBV (an average of VASO CBV 1 and VASO CBV 2) and PET CBV (an average of PET CBV 1 and PET CBV 2) as defined in Eq. [3] can be written as
| [A4] |
If the measurement errors were again assumed to be random variables and are independent across voxels, the terms with linear errors will vanish but those with square of error will remain. Therefore,
| [A5] |
By comparing Eqs. [A2–5], the index for systematic difference between VASO and PET can be calculated by:
| [A6] |
REFERENCES
- 1.Lu H, Golay X, Pekar JJ, Van Zijl PC. Functional magnetic resonance imaging based on changes in vascular space occupancy. Magn Reson Med. 2003;50(2):263–274. doi: 10.1002/mrm.10519. [DOI] [PubMed] [Google Scholar]
- 2.Law M, Yang S, Babb JS, Knopp EA, Golfinos JG, Zagzag D, Johnson G. Comparison of cerebral blood volume and vascular permeability from dynamic susceptibility contrast-enhanced perfusion MR imaging with glioma grade. AJNR Am J Neuroradiol. 2004;25(5):746–755. [PMC free article] [PubMed] [Google Scholar]
- 3.Lu H, Law M, Johnson G, Ge Y, van Zijl PC, Helpern JA. Novel approach to the measurement of absolute cerebral blood volume using vascular-space-occupancy magnetic resonance imaging. Magn Reson Med. 2005;54(6):1403–1411. doi: 10.1002/mrm.20705. [DOI] [PubMed] [Google Scholar]
- 4.Uh J, Lewis-Amezcua K, Varghese R, Lu H. On the measurement of absolute cerebral blood volume (CBV) using vascular-space-occupancy (VASO) MRI. Magn Reson Med. 2009;61(3):659–667. doi: 10.1002/mrm.21872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Osch MJ, Vonken EJ, Viergever MA, van der Grond J, Bakker CJ. Measuring the arterial input function with gradient echo sequences. Magn Reson Med. 2003;49(6):1067–1076. doi: 10.1002/mrm.10461. [DOI] [PubMed] [Google Scholar]
- 6.Shin W, Cashen TA, Horowitz SW, Sawlani R, Carroll TJ. Quantitative CBV measurement from static T1 changes in tissue and correction for intravascular water exchange. Magn Reson Med. 2006;56(1):138–145. doi: 10.1002/mrm.20937. [DOI] [PubMed] [Google Scholar]
- 7.Lu H, Pollack E, Young R, Babb JS, Johnson G, Zagzag D, Carson R, Jensen JH, Helpern JA, Law M. Predicting grade of cerebral glioma using vascular-space occupancy MR imaging. AJNR Am J Neuroradiol. 2008;29(2):373–378. doi: 10.3174/ajnr.A0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uh J, Mihalakos P, Tamminga CA, Lu H. Perfusion deficit in Schizophrenia and correlation with psychopathological symptoms. Proceedings of the 17th Annual Meeting of ISMRM; Honolulu, Hawaii. 2009. p. 3452. [Google Scholar]
- 9.Uh J, Lewis-Amezcua K, Martin-Cook K, Cheng Y, Weiner M, Diaz-Arrastia R, Devous M, Sr, Shen D, Lu H. Cerebral blood volume in Alzheimer's disease and correlation with tissue structural integrity. Neurobiol Aging. 2009 doi: 10.1016/j.neurobiolaging.2008.12.010. in press, doi:10.1016/j.neurobiolaging.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin WR, Powers WJ, Raichle ME. Cerebral blood volume measured with inhaled C15O and positron emission tomography. J Cereb Blood Flow Metab. 1987;7(4):421–426. doi: 10.1038/jcbfm.1987.85. [DOI] [PubMed] [Google Scholar]
- 11.Grubb RL, Jr, Phelps ME, Ter-Pogossian MM. Regional cerebral blood volume in humans. X-ray fluorescence studies. Archives of neurology. 1973;28(1):38–44. doi: 10.1001/archneur.1973.00490190056006. [DOI] [PubMed] [Google Scholar]
- 12.Herscovitch P, Raichle ME. What is the correct value for the brain--blood partition coefficient for water? J Cereb Blood Flow Metab. 1985;5(1):65–69. doi: 10.1038/jcbfm.1985.9. [DOI] [PubMed] [Google Scholar]
- 13.Ashburner J, Friston KJ. Voxel-based morphometry--the methods. NeuroImage. 2000;11(6 Pt 1):805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- 14.Donahue MJ, Blakeley JO, Zhou J, Pomper MG, Laterra J, van Zijl PC. Evaluation of human brain tumor heterogeneity using multiple T1-based MRI signal weighting approaches. Magn Reson Med. 2008;59(2):336–344. doi: 10.1002/mrm.21467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Derdeyn CP, Videen TO, Yundt KD, Fritsch SM, Carpenter DA, Grubb RL, Powers WJ. Variability of cerebral blood volume and oxygen extraction: stages of cerebral haemodynamic impairment revisited. Brain. 2002;125(Pt 3):595–607. doi: 10.1093/brain/awf047. [DOI] [PubMed] [Google Scholar]
- 16.Grandin CB, Bol A, Smith AM, Michel C, Cosnard G. Absolute CBF and CBV measurements by MRI bolus tracking before and after acetazolamide challenge: repeatabilily and comparison with PET in humans. NeuroImage. 2005;26(2):525–535. doi: 10.1016/j.neuroimage.2005.02.028. [DOI] [PubMed] [Google Scholar]
- 17.Carroll TJ, Teneggi V, Jobin M, Squassante L, Treyer V, Hany TF, Burger C, Wang L, Bye A, Von Schulthess GK, Buck A. Absolute quantification of cerebral blood flow with magnetic resonance, reproducibility of the method, and comparison with H2(15)O positron emission tomography. J Cereb Blood Flow Metab. 2002;22(9):1149–1156. doi: 10.1097/00004647-200209000-00013. [DOI] [PubMed] [Google Scholar]
- 18.Ibaraki M, Ito H, Shimosegawa E, Toyoshima H, Ishigame K, Takahashi K, Kanno I, Miura S. Cerebral vascular mean transit time in healthy humans: a comparative study with PET and dynamic susceptibility contrast-enhanced MRI. J Cereb Blood Flow Metab. 2007;27(2):404–413. doi: 10.1038/sj.jcbfm.9600337. [DOI] [PubMed] [Google Scholar]
- 19.Ye FQ, Berman KF, Ellmore T, Esposito G, van Horn JD, Yang Y, Duyn J, Smith AM, Frank JA, Weinberger DR, McLaughlin AC. H(2)(15)O PET validation of steady-state arterial spin tagging cerebral blood flow measurements in humans. Magn Reson Med. 2000;44(3):450–456. doi: 10.1002/1522-2594(200009)44:3<450::aid-mrm16>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 20.Kao YH, Guo WY, Wu YT, Liu KC, Chai WY, Lin CY, Hwang YS, Jy-Kang Liou A, Wu HM, Cheng HC, Yeh TC, Hsieh JC, Mu Huo Teng M. Hemodynamic segmentation of MR brain perfusion images using independent component analysis, thresholding, and Bayesian estimation. Magn Reson Med. 2003;49(5):885–894. doi: 10.1002/mrm.10440. [DOI] [PubMed] [Google Scholar]