Abstract
Repair of DNA damage by homologous recombination has only recently been established as an important mechanism in maintaining genetic stability in mammalian cells. The recently cloned Xrcc2 gene is a member of the mammalian Rad51 gene family, thought to be central to homologous recombination repair. To understand its function in mammals, we have disrupted Xrcc2 in mice. No Xrcc2–/– animals were found alive, with embryonic lethality occurring from mid-gestation. Xrcc2–/– embryos surviving until later stages of embryogenesis commonly showed developmental abnormalities and died at birth. Neonatal lethality, apparently due to respiratory failure, was associated with a high frequency of apoptotic death of post- mitotic neurons in the developing brain, leading to abnormal cortical structure. Embryonic cells showed genetic instability, revealed by a high level of chromosomal aberrations, and were sensitive to γ-rays. Our findings demonstrate that homologous recombination has an important role in endogenous damage repair in the developing embryo. Xrcc2 disruption identifies a range of defects that arise from malfunction of this repair pathway, and establishes a previously unidentified role for homologous recombination repair in correct neuronal development.
Keywords: apoptosis/embryonic lethality/knockout mouse/neuronal development/Rad51-like gene
Introduction
Accurate repair of the genome underpins the correct development and genetic stability of multicellular organisms. Several pathways of DNA repair in mammals have been recognized, but the influence of repair by homologous recombination has only recently been highlighted (Kanaar et al., 1998; Liang et al., 1998). Homologous recombination repair (HRR) entails the invasion of an undamaged DNA molecule by a damaged molecule of identical or very similar sequence, followed by resynthesis of the damaged region using the undamaged molecule as a template. A sister chromatid may be used as the template for repair, or less frequently the paternal and maternal copies of chromosomes provide the required homology. HRR is important for the repair of severe types of DNA damage, such as double-strand breaks, and allows the replacement of damaged regions without loss or alteration of base sequence. However, there are other pathways that can rejoin double-strand breaks in DNA. In mammalian cells, DNA end-joining processes, which do not rely on sequence homology (non-homologous end joining, NHEJ), have an important role in break repair but do not ensure its fidelity (Jeggo, 1998).
HRR has been studied extensively in the yeast Saccharomyces cerevisiae, where the RAD52 group of genes controls this pathway. These genes specify proteins including Rad51p, which is a homologue of the bacterial recA protein and is central to the DNA pairing and strand invasion steps of HRR (Shinohara and Ogawa, 1995; Baumann and West, 1998; Thacker, 1999a). Additionally, the Rad51-like proteins Rad55p and Rad57p form a heterodimer that promotes homologous pairing reactions (Sung, 1997). Other proteins such as Rad52p and Rad54p interact with Rad51p and stimulate recombination. In yeast, none of the HRR genes is essential for survival, but their mutation gives rise to sensitivity to DNA-damaging agents such as X-rays (Game, 1993).
Mammalian counterparts of the RAD52 group of genes, including RAD51, RAD52 and RAD54, have been identified in the last few years (Shinohara et al., 1993; Muris et al., 1994; Kanaar et al., 1996), and recently a number of novel RAD51-like genes have been found (Thacker, 1999b). We recently identified the human RAD51-like gene XRCC2 by positional cloning, making use of its ability to complement a DNA damage-sensitive hamster cell line, irs1 (Tambini et al., 1997). The irs1 line is sensitive to various DNA-damaging agents, including X-rays, UV light and mitomycin-C (Jones et al., 1987), and shows elevated frequencies of spontaneous and radiation-induced gene mutation, chromosomal damage and chromosomal missegregation (Tucker et al., 1991; Thacker et al., 1994; Griffin et al., 2000). Importantly, irs1 has also been shown to be defective in recombination of homologous substrates arranged tandemly in genomic DNA (Johnson et al., 1999; A.M.George and J.Thacker, unpublished). Cloning of the full-length cDNA (Cartwright et al., 1998; Liu et al., 1998) has allowed preliminary functional analysis of the XRCC2 gene product, to show that it interacts (Braybrooke et al., 2000; Schild et al., 2000) and forms a heterodimer (Braybrooke et al., 2000) with the product of another Rad51-like gene, RAD51L3. In multiple alignments, the Xrcc2 protein has greatest homology to Rad55p (Thacker, 1999b), and, therefore, it is possible that Xrcc2 acts with its partner in the same way as the yeast Rad55p–Rad57p heterodimer to facilitate Rad51-dependent recombination processes.
Disruption of the Rad51 gene in mice was shown to give early embryonic lethality (Lim and Hasty, 1996; Tsuzuki et al., 1996), perhaps because of its key role in HRR and the need for high-fidelity repair in replicating cells of the developing embryo. However, other important HRR genes, such as Rad52 and Rad54, do not give embryonic lethality when disrupted (Essers et al., 1997; Rijkers et al., 1998). Given the potential facilitating function of Xrcc2 in HRR, the consequences of loss of Xrcc2 in mice could not be predicted. Therefore, we have assessed the effects of a targeted disruption of the mouse Xrcc2 gene and, surprisingly, found that Xrcc2 has a significant role in ensuring normal embryonic development, especially in the developing nervous system.
Results
Targeted mutagenesis of mouse Xrcc2
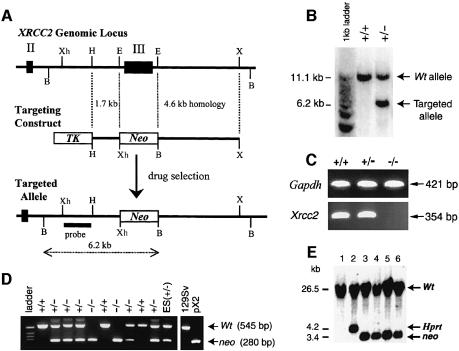
Disruption of Xrcc2 in 129/Sv/Ev embryonic stem cells was achieved as illustrated in Figure 1A. Homologous recombination of the targeting vector with the Xrcc2 gene results in the introduction of a neomycin-resistance cassette deleting exon III, the largest exon of Xrcc2 (86% of the coding region). This deleted region encodes the nucleotide binding motifs, which are conserved in the RecA/Rad51 gene family and essential for their activity (Shinohara et al., 1992), and the whole 3′ end of the gene including poly(A) signal sites (Cartwright et al., 1998). Correctly targeted ES clones were identified by Southern blot analysis (Figure 1B); these were used for blastocyst injections to yield chimeric offspring and transmission through the germ line after crossing with C57BL/6 females.
Fig. 1. Disruption of Xrcc2. (A) Diagram of the wild-type Xrcc2 genomic locus, targeting construct and the targeted locus. Homologous recombination of the targeting vector with the Xrcc2 locus results in the replacement of exon III with a neomycin cassette, and the introduction of a BamHI site. Relevant BamHI (B), EcoRI (E), HindIII (H), XbaI (X) and XhoI (Xh) restriction sites, and the position of the probe used for Southern blot analysis are shown. Exons II and III are indicated by numbered solid boxes. (B) Genomic DNA from ES cell colonies digested with BamHI, and analysed by Southern blotting and hybridization with the probe indicated in (A). Truncation of wild-type 11.1 kb fragment to 6.2 kb confirms correct targeting of an allele. +/+ and +/– represent the genotypes of wild-type (Xrcc2+/+) and targeted (Xrcc2+/–) ES lines, respectively. (C) RT–PCR on total RNA extracts from embryos using Xrcc2 exon II to exon III specific primers, with primers to Gapdh as a control. Xrcc2+/+ and Xrcc2+/–, but not Xrcc2–/–, generated a band corresponding to Xrcc2 mRNA. (D) A representative example of PCR genotyping for a litter of E8.5 embryos from an Xrcc2+/– intercross. PCR primers detect neo or exon III in the same reaction, giving products of 280 and 545 bp, respectively. Control lanes shown are a targeted ES cell line (ES +/–), wild-type genomic DNA (129Sv) and a plasmid constructed to mimic the targeted locus (pX2); –/– denotes homozygous disruption of Xrcc2. (E) Double targeting of Xrcc2 in ES cells shown by Southern blot analysis of XhoI-digested genomic DNA. HM-1 ES cells (lane 1) underwent sequential gene targeting steps, first with an Hprt-based vector (lane 2) and then, following confirmation of correct targeting of an Xrcc2 allele, with the neo-based construct as in (A). Both strategies introduce discernible XhoI restriction sites, reducing a wild-type fragment from 26.5 kb to 4.2 or 3.4 kb for Hprt or neo, respectively. All targeted clones isolated following transfection with the neo vector (shown for four clones; lanes 3, 4, 5 and 6) had re-targeted the already disrupted allele (n = 8).
Disruption of Xrcc2 results in embryonic lethality
Mice heterozygous for the Xrcc2 disruption (Xrcc2+/–) appear indistinguishable from wild-type littermates at 9 months. However, heterozygote intercrosses failed to produce mice homozygous for the mutation (Xrcc2–/–) in >300 offspring (Table I). Xrcc2+/– and Xrcc2+/+ were found in normal Mendelian ratios of ∼2:1, indicating that Xrcc2–/– is lethal.
Table I. Genotypes of progeny from Xrcc2+/– intercrosses.
| Age (days) | Litters (decidua) |
Xrcc2 genotype: live (dead) |
Resorbed | % –/– | ||
|---|---|---|---|---|---|---|
| +/+ | +/– | –/– | ||||
| E9.5 | 5 (46) | 13 | 18 (1) | 14 | 0 | 100 |
| E10.5 | 8 (66) | 18 | 35 | 8 (3) | 2 | 45 |
| E11.5 | 5 (49) | 12 (1) | 22 (2) | 4 (4) | 4 | 35 |
| E12.5 | 3 (34) | 6 (2) | 15 (1) | 2 (7) | 1 | 29 |
| E14.5–15.5 | 12 (107) | 21 | 54 (1) | 6 (8) | 17 | 24 |
| E18.5 | 9 (77) | 16 | 37 | 5 (1) | 18 | 28 |
| 3 weeks | 50 | 95 | 228 | 0 | NA | 0 |
The numbers of live embryos for each genotype at the indicated stages of development are shown. Genotypes in parenthesis are for necrotic or partially resorbed embryos where sufficient yolk sac tissue was available to allow genotyping; the numbers of decidua where maternal resorption was complete are indicated. % –/– denotes the number of viable Xrcc2–/– embryos observed as a percentage of that predicted by Mendelian law. NA, not available.
To confirm the effectiveness of the gene disruption, RNA was isolated from embryonic tissue of wild- type, Xrcc2+/– or Xrcc2–/– embryos. RT–PCR analysis (Figure 1C) showed the absence of full-length Xrcc2 transcript in Xrcc2–/– embryos.
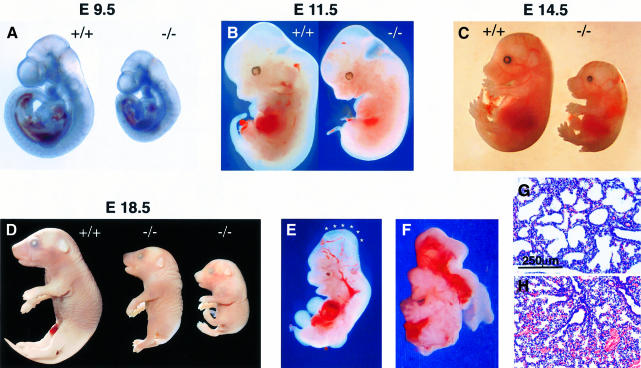
The stage of development attained by Xrcc2–/– embryos was determined through the analysis of litters from timed matings between heterozygotes. Embryos were dissected at various stages of gestation and genotype determined by PCR of yolk-sac tissue (Figure 1D). The results of genotyping are summarized in Table I, with embryo stages shown in Figure 2. Until E8.5 (E = days post-coitum), the development of Xrcc2–/– embryos was overtly normal. At E9.5, while embryos were recovered at the frequencies predicted by Mendelian law, Xrcc2–/– embryos were reduced in size by comparison with littermates. The retardation in growth and development was increasingly apparent at later stages, with Xrcc2–/– embryos also displaying consistent morphological abnormalities including dorsal truncation and a stunted tail, and cranial defects. Additional, more severe cranial malformation was evident in three of 10 Xrcc2–/– embryos at E11.5–14.5. These included an E11.5 embryo with incomplete closure of the neural tube and an E14.5 embryo with exencephaly, a phenotype likely to arise from the former defect (Figure 2E and F). No such abnormalities were evident in wild-type or Xrcc2+/– littermates.
Fig. 2. Xrcc2–/– embryos display growth retardation and morphological defects. Xrcc2 null (–/–) and wild-type littermates (+/+) are shown at (A) E9.5, (B) E11.5, (C) E14.5 and (D) E18.5 stages of development. An additional smaller Xrcc2–/– fetus, from a separate litter, illustrates the extremities of size observed at E18.5. (E) Xrcc2–/– E11.5 embryo with failed neural tube closure, denoted by asterisks. (F) Xrcc2-null E14.5 embryo with exencephaly. (G and H) The lungs of Xrcc2-null neonates fail to inflate. In comparison with wild-type littermates (G), alveolar size is reduced in haematoxylin–eosin-stained lungs of Xrcc2–/– (H) neonates.
Loss of viability of Xrcc2–/– embryos, indicated by necrosis and resorption, occurred from E10.5 during the stages of organogenesis, with survival reduced at E12.5 to ∼30% of that expected (Table I). A corresponding increase in the number of non-surviving, partially resorbed Xrcc2–/– embryos was observed. There was no further reduction in viability by E18.5, indicating that the majority of Xrcc2–/– embryos reaching E12.5 were likely to survive to birth. Observations at birth showed that, while initially alive and attempting to breathe, Xrcc2–/– neonates (n = 3) failed to survive, with cyanosis and death within 20 min, while wild-type and heterozygous littermates flushed pink and were breathing strongly. No live or dead Xrcc2–/– offspring were found beyond post-natal day 1, suggesting rapid cannibalization of deceased or runted mutants.
Anatomical and histological examination of the Xrcc2–/– neonates indicated all organs to be present, although reduced in size when compared with wild-type or Xrcc2+/– littermates. The lungs of post-natal Xrcc2–/– pups showed a reduction in alveolar volume when compared with wild-type littermates, indicating a failure to inflate with air (Figure 2G and H). There were no apparent cardiovascular abnormalities or defects in liver, thymus, spleen or kidneys, although in one Xrcc2–/– neonate, haemorrhaging was evident in the brain. Therefore, it appears likely that lethality in Xrcc2–/– mice results from respiratory failure in the transition from fetus in utero to neonate at birth.
Xrcc2–/– cells fail to proliferate in culture
ES cells were subjected to sequential gene targeting steps: the first to disrupt one allele with Hprt and then the other with neo. As shown by Southern blot analysis, all correct targeting events constituted a re-targeting of the already disrupted allele (n = 8) (Figure 1E). Given an equal chance of targeting either allele, the absence of double-targeted (Xrcc2–/–) cells is highly significant (p <0.005). Furthermore, in contrast to cells derived from Xrcc2+/+ or Xrcc2+/– littermates, primary embryonic fibroblast cells isolated from E10.5–E15.5 Xrcc2–/– embryos (n = 6) underwent premature senescence with complete growth arrest at or before passage 4 in culture (data not shown). Together these results indicate that Xrcc2 is essential for normal proliferation in culture.
Xrcc2–/– blastocysts are hypersensitive to γ radiation
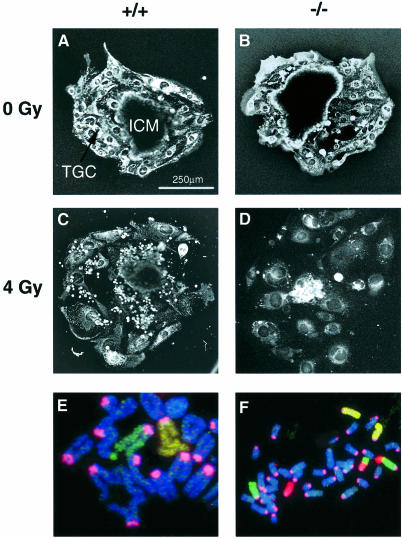
We have shown previously that an established line of Xrcc2-deficient hamster cells (irs1) is ∼3-fold more sensitive to ionizing radiation than wild-type V79 hamster cells (Jones et al., 1987). Therefore, we assessed the impact of disrupting the mouse Xrcc2 gene on sensitivity to radiation using blastocysts isolated at E3.5, following Xrcc2+/– intercrosses. Blastocysts are composed of two cell types: highly proliferative cells in the inner cell mass (ICM) and relatively quiescent trophoblast cells. The blastocysts were given 4 Gy of γ-rays before culture in vitro for 6 days, when the area of the developing ICM was measured. The genotypes of littermates were determined at the end of this period by PCR. All blastocysts attached successfully, and with the exception of one irradiated Xrcc2–/– blastocyst there was outgrowth of the ICM. As shown in Table II, mean ICM areas for untreated control blastocysts were equivalent for each genotype. Following irradiation, the mean ICM areas for both Xrcc2+/+ and Xrcc2+/– blastocysts were reduced to approximately half, while that of Xrcc2–/– mice was reduced to one-fifth of the unirradiated ICM (significantly different from the reduction in Xrcc2+/+ and Xrcc2+/–). Representative photographs of control and irradiated blastocysts are shown in Figure 3A–D. These data suggest that, as in cell culture, the loss of Xrcc2 confers a moderate level of radiation sensitivity in the mouse.
Table II. In vitro culture of blastocysts after γ-radiation.
| Dose (Gy) | Inner cell mass area (mm2) |
||
|---|---|---|---|
|
Xrcc2 genotype |
|||
| +/+ | +/– | –/– | |
| 0 | 0.067 ± 0.012 | 0.054 ± 0.004 | 0.064 ± 0.008 |
| n = 6 | n = 9 | n = 7 | |
| 4 | 0.035 ± 0.006 | 0.033 ± 0.003 | 0.013 ± 0.004a |
| n = 7 | n = 12 | n = 7 | |
aSignificantly different (p <0.01) from irradiated +/+ and +/– groups on a Kruskal–Wallis test.
Fig. 3. Xrcc2–/– embryonic cells are hypersensitive to radiation and have a high level of chromosomal aberrations. Representative photographs of blastocysts cultured for 6 days following either no irradiation [(A) Xrcc2+/+ and (B) Xrcc2–/–] or γ-irradiation (4 Gy) [(C) Xrcc2+/+ and (D) Xrcc2–/–]. ICM, inner cell mass; TGC, trophoblast giant cells. (E) Complex chromatid exchange involving at least six chromosomes, including chromosomes 2 (yellow) and 4 (green); centromeres are stained pink. (F) Reciprocal chromosome exchange involving chromosomes 4 (green) and 11 (red); centromeres are stained pink.
Xrcc2–/– cells have a high level of chromosomal instability
The proliferative capacity of embryonic cells may suffer if the loss of Xrcc2 leads to genetic instability. To check the integrity of chromosomes, disassociated embryonic cells were examined immediately after placing the cells into culture medium. Chromosomal aberration frequencies were scored in ∼100 cells from E15.5 littermates of Xrcc2–/–, Xrcc2+/– and wild-type embryos. Very high frequencies of breaks and exchanges were seen in the Xrcc2–/– cells (approximately two per cell), while very few (∼0.02 per cell) were observed in cells from heterozygotes and wild-type embryos (Table III). The majority of the aberrations involved chromatids, indicating that these were occurring in the current division and that the embryonic cells were experiencing persistent genetic instability (chromatid-type aberrations will become chromosome-type following cell division). Some of the chromatid exchanges in Xrcc2–/– cells were very complex, as illustrated in Figure 3E. Chromosome-type aberrations were also identified using three-colour fluorescence in situ hybridization (FISH) to show rearrangements such as reciprocal translocations, which may be transmissible through somatic cell divisions (Figure 3F).
Table III. Chromosomal aberrations in embryonic cells at E15.5 from littermates of crosses between heterozygous (Xrcc2+/–) mice.
| Embryo genotype | Cell number | Number of chromosomal aberrations |
||||||
|---|---|---|---|---|---|---|---|---|
| Gaps | Breaks | Isochromatid breaks | Exchanges | Triradials | Rings | Complexes | ||
| +/+ | 134 | 1 | 1 | |||||
| +/– | 103 | 1 | ||||||
| +/– | 101 | 1 | 1 | |||||
| –/– | 77 | 3 | 38 | 14 | 82 | 14 | 2 | 11 |
| –/– | 64 | 2 | 20 | 11 | 69 | 14 | 7 | |
Aberrations are classified according to Savage (1976).
A role for Xrcc2 in the prevention of apoptosis, especially in the developing nervous system
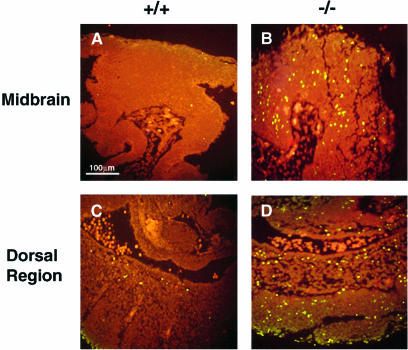
To investigate the cellular pathology linked to the morphological defects and lethality associated with Xrcc2 disruption, we used the TUNEL (terminal deoxytransferase-mediated dUTP-fluorescein nick end labelling) assay as an indicator of apoptotic death. At E9.5, there was a marked elevation of apoptotic cells throughout Xrcc2–/– embryonic tissues when compared with littermate or stage-matched wild-type embryos. Cell death in Xrcc2–/– embryos was highest in caudal and dorsal regions, and developing areas of the brain as shown in Figure 4.
Fig. 4. Extensive cell death in Xrcc2–/– E9.5 embryos (sagittal sections) as shown by TUNEL staining (green). (A and B) midbrain and (C and D) dorsal region at the level of the forelimb of stage-matched Xrcc2+/+ (A and C) and Xrcc2–/– embryos (B and D).
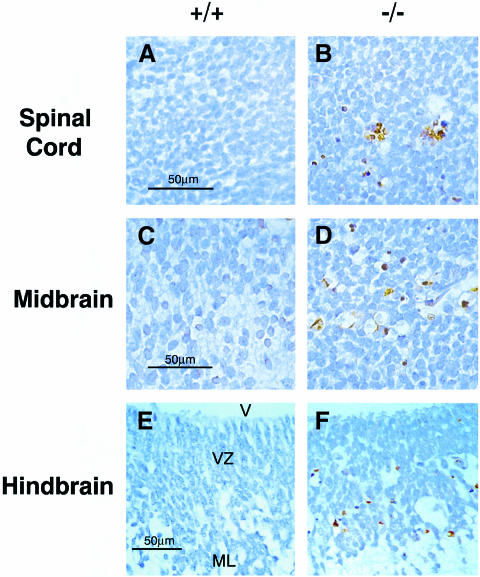
At E11.5 and later stages, extensive cell death was detected in Xrcc2–/– embryos, although this was largely confined to the developing central and peripheral nervous system. TUNEL-positive cells were observed in the spinal cord, midbrain, hindbrain and forebrain in Xrcc2–/– embryos, with TUNEL staining localized to pyknotic nuclei as expected (Figures 5 and 6). In other organs, including the lungs, heart and liver, abnormal numbers of apoptotic cells were not observed (data not shown).
Fig. 5. Extensive cell death in the developing CNS of Xrcc2–/– E11.5–12.5 embryos. TUNEL staining (DAB) in (A and B) the spinal cord at E12.5, (C and D) the midbrain at E12.5, and (E and F) the hindbrain at E11.5 in sagittal sections of Xrcc2+/+ (A, C and E) and Xrcc2–/– (B, D and F) littermates. Nuclei are counterstained with haematoxylin. V, fourth ventricle; VZ, ventricular zone; ML, mantle layer.
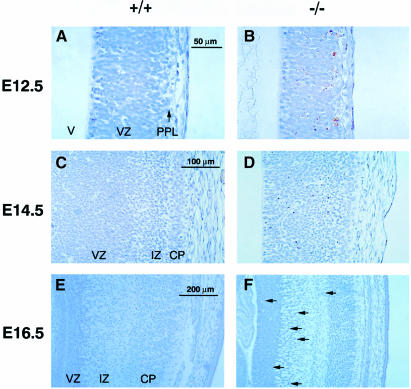
Fig. 6. Extensive apoptosis in Xrcc2–/– embryos reflects the spatial and temporal pattern of neurogenesis. TUNEL staining for sagittal sections of the developing cortex of the forebrain at (A and B) E12.5, (C and D) E14.5 and (E and F) E16.5 for wild-type (A, C and E) and Xrcc2–/– (B, D and F) embryos. At E16.5, arrows indicate TUNEL-positive nuclei. V, lateral ventricle; VZ, ventricular zone; PPL, primordial plexiform layer; IZ, intermediate zone; CP, cortical plate.
Neurogenesis in normal embryos is a spatially and temporally regulated process: in normal cortex it commences at E11–12.0, peaks at around E12.5 and E13.5, and then decreases to lower levels by E18 (Bayer and Altman, 1991). This pattern was reflected in the TUNEL staining of developing lateral cortex of the forebrain of Xrcc2–/– embryos (Figure 6). Neurons arise from mitotic progenitor cells in the ventricular zone (VZ), and newly formed post-mitotic neurons initially migrate to form the primordial plexiform layer (PPL); at later stages this migration occurs through an intermediate zone (IZ) to form the cortical plate (CP), the future cortical grey matter. Extensive cell death was first observed in the migrating cells at E12.5 (Figure 6B) and increased until E13.5–14.5, with large numbers of apoptotic cells apparent in the VZ and IZ (Figure 6D). By E16.5, cell death was substantially reduced (Figure 6F), continuing at gradually lower levels to E18.5 (data not shown). Few TUNEL-staining cells were detected in littermate controls over the same period (Figure 6A, C and E). Neurogenesis in the hindbrain precedes that of the forebrain, commencing at E11.0 and proceeding to completion by E14.0 (Nornes and Carry, 1978). Correspondingly, extensive cell death was detected in the hindbrain of Xrcc2–/– embryos at E11.5–12.5, but had ceased by E14.5 (data not shown). No increased TUNEL staining was seen in mature neurons or in the cerebellum, where development extends into the post-natal period.
To confirm the identity of apoptotic cells, we stained E12.0 Xrcc2–/– sections of forebrain precortex with TUNEL reagents and a TUJ1 antibody specific to β3-tubulin, an early marker of post-mitotic neurons (Lee et al., 1990). In the frontal cortex (Figure 7A and B), TUNEL staining occurs within the VZ and at the boundary with the PPL where the majority of TUJ1-staining neurons are found. In the septal area of the developing cortex (Figure 7C and D), neurogenesis is more advanced and TUNEL-positive cells localize predominantly within the PPL and to a lesser extent in the VZ. Similar data were obtained for the hindbrain (not shown). Therefore, the extensive apoptosis in Xrcc2–/– embryos occurred within the early post-mitotic neuronal cell population, during or shortly following the completion of migration to the PPL layer.
Fig. 7. Apoptosis in early post-mitotic neurons of Xrcc2–/– embryos. Sagittal section of (A and B) the frontal cortex and (C and D) the septal area of the developing forebrain in an E12.0 Xrcc2-null embryo, stained with propidium iodide (A and C), and double stained with TUNEL reagents (green) and the anti-β3-tubulin antibody, TUJ1 (red) (B and D). TUNEL-positive nuclei are localized predominantly within the newly generated, TUJ1-staining neuronal cell population of the PPL layer. V, lateral ventricle; VZ, ventricular zone; PPL, primordial plexiform layer.
At E14.5–16.5 (Figure 6) and E18.5 (data not shown), the thickness of the cortical plate in Xrcc2–/– embryos is reduced when compared with wild-type littermates. Therefore, it is evident that the extensive cell death results in a severely abnormal cortical structure.
Discussion
Early embryonic death, apoptosis and genetic instability
Disruption of the Xrcc2 gene in mice leads to embryonic lethality, but the mutant embryos appeared normal up to E8.5. Thereafter, retardation in growth and development occurred, and by E12.5 about three-quarters of the mutant embryos were dead. The majority of Xrcc2–/– embryos that survived this period remained viable until respiratory failure at birth.
Early embryonic death is found for some other gene disruptions affecting HRR. Lethality arising from the disruption of Rad51, a central gene in the homologous recombination–repair pathway, occurs even earlier than for Xrcc2–/– embryos (before E8.5) (Lim and Hasty, 1996; Tsuzuki et al., 1996). Disruption of the Brca1 and Brca2 genes, which have been associated with Rad51 and HRR activity (Venkitaraman, 1999), generally leads to death before E9.5 (Hakem et al., 1998). These findings show that some HRR genes are critical for survival at early stages of development, probably because they perform crucial roles in combating DNA damage, particularly in the rapidly proliferating cells of the early embryo. Severe impairment of embryonic cell proliferation is characteristic of the Rad51 and Brca gene disruptions, whereas the loss of Xrcc2 does not appear to compromise proliferation significantly in isolated blastocysts (Table II) or embryos up to E8.5. Xrcc2 disruption may have less impact than Rad51 or Brca gene disruptions because its role in HRR is to facilitate the process (see Introduction). However, at mid-gestation, corresponding to the onset of organogenesis, sufficient cellular loss (or loss in specific tissues) may have occurred in many Xrcc2–/– embryos to compromise their viability. Those surviving this phase have morphological defects (truncation, exencephaly) attributable to significant but not critical cell loss sustained in mid-gestation. Interestingly, the recent disruption of another Rad51-like gene, Rad51L3, which has been found to physically associate with Xrcc2 (see Introduction), similarly leads to mid-gestation lethality and truncation of the caudal tail region (Pittman and Schimenti, 2000).
In accordance with the timing of lethality, we observed apoptosis throughout Xrcc2–/– embryos at E9.5, although by about E12.5 it was largely confined to post-mitotic cells in neurological tissue. Given the role of Xrcc2 in DNA repair, it is likely that an accumulation of unrepaired or misrepaired damage triggers apoptosis. It is important to note that, unlike some other repair pathways, HRR is a mechanism for high-fidelity repair of damage such as DNA double-strand breaks (see Introduction). Consistent with loss of this pathway, there is a highly elevated frequency of chromosomal aberrations in Xrcc2–/– embryonic cells (Table III). The occurrence of chromatid aberrations in Xrcc2–/– cells is indicative of persistent genetic instability, and isochromatid breaks may signify failed recombination between sister chromatids in post-replicative cells. In agreement with our earlier findings with an established line of Xrcc2-deficient hamster cells, irs1 (Jones et al., 1987), we found a moderate increase in the sensitivity of Xrcc2–/– embryonic cells to ionizing radiation (Table II). Thus, exposure to a DNA strand break-inducing agent augments the response to endogenous damage. However, while the Xrcc2-deficient irs1 line is viable, we found that mouse embryonic cells senesced prematurely, and that mouse ES cells with both alleles of Xrcc2 disrupted could not be recovered. Similar findings have been reported for disruptions of both Rad51 (Lim and Hasty, 1996) and Brca2 (Sharan et al., 1997). These failures in mouse primary cells are likely to be due to the presence of effective checkpoint genes acting as a barrier to cell cycle progression in the presence of persistent DNA damage (Lim and Hasty, 1996; Hakem et al., 1997). In this context, it will be interesting to assess the effects of Xrcc2 disruption on a checkpoint-defective genetic background (e.g. Tp53–/–); this may allow the birth of viable offspring, and an assessment of the influence of concomitant genetic instability on cancer incidence.
Defective neurogenesis and perinatal lethality
The elimination of a few per cent of neuronal cells normally accompanies cellular proliferation in mammalian neurogenesis (Thomaidou et al., 1997), but the amount of apoptosis in Xrcc2–/– embryos considerably exceeds the normal level. In recent studies of mouse embryos in which evolutionarily conserved genes of the NHEJ repair pathway were disrupted, it has also been found that elevated levels of neuronal apoptosis occur at E12.5 onwards. Importantly, the extent of apoptosis depends on which of the NHEJ genes is disrupted: a massive increase in apoptosis of post-mitotic neurons occurs in ligase IV- and XRCC4-deficient embryos, but more subtle increases occur in Ku70- and Ku80-deficient embryos (Barnes et al., 1998; Frank et al., 1998; Gao et al., 1998; Gu et al., 2000). Ligase IV- and XRCC4-deficient embryos have genotypes in Mendelian ratios up until about E15 but do not survive to birth, while Ku70 and Ku80 deficiencies allow survival to give viable, although growth-retarded mice. Adult Ku70-deficient mice have also been reported to suffer from aganglionosis in the gut, similar to that found in the congenital disorder of the enteric nervous system known as Hirschsprung syndrome (Li et al., 1998). Severe apoptotic damage to the nervous system may therefore be a cause of embryonic lethality (Copp et al., 1990; Gu et al., 2000), while less severe damage is compatible with life. In support of this link, mice doubly deficient in p53 and either ligase IV or XRCC4 show abrogation of the neurological defects and increased survival (Frank et al., 2000; Gao et al., 2000).
The finding of cell lethality in the developing nervous system of embryos with disrupted NHEJ genes led to the suggestion that the NHEJ pathway may be particularly important for neurogenesis, perhaps because there is no back-up from other repair pathways such as HRR (Gu et al., 2000). However, our data now show that HRR as exemplified by the function of Xrcc2 is important in ensuring correct neuronal development, since the loss of this pathway leads to defects similar to the loss of components of NHEJ. Neurological defects were not observed in Rad51 and Brca gene disruptions, presumably because the embryos died before such assessments could be made. However, there is one report of Brca1 disruption where the embryos survived to E11–13 and develop mental abnormalities including exencephaly were reported (Gowen et al., 1996).
It is unclear at present why post-mitotic neurons in particular are adversely affected. This cell type may be especially sensitive to apoptotic stimuli (Gao et al., 1998), or there may be increased levels of oxidative damage in these cells leading to the formation of both single- and double-strand breaks in DNA (Barnes et al., 1998). The idea that there is an increased level of DNA breakage in certain parts of the developing brain has led to the further speculation that the NHEJ pathway has a specific role in creating genetic diversity in neurons: it has been proposed that NHEJ can rejoin DNA breaks at defined genomic sites, in a similar way to the involvement of this pathway in the generation of antigen-recognition diversity in the immune system (Chun and Schatz, 1999; Gilmore et al., 2000). This speculation is made less attractive by our findings, at least in terms of a specific role for NHEJ, since another repair pathway (HRR) seems to be equally important in the repair of this damage. This conclusion is supported by a very recent study on the effects of disruption of the gene encoding DNA polymerase β (polβ), a component of the DNA base-excision repair pathway. Null polβ embryos suffer defective neurogenesis and perinatal lethality associated with respiratory failure (Sugo et al., 2000). The similarity of findings for null polβ and Xrcc2–/– embryos also supports the idea that unrepaired/misrepaired DNA damage can affect neurological development in ways that predict later failures in specific organs, such as the lungs.
Taken with other recent work, our findings indicate that unrepaired or misrepaired DNA damage arising from the loss of individual components of several different repair pathways can lead to the apoptosis observed in the developing nervous system and the failure to survive at birth. Further, the severity of the phenotype in mice may relate to the specific role of the disrupted gene, although functional redundancy among genes may play a role in mitigating severity. In this context it will be interesting to look for subtle alterations of the nervous system in mice with other HRR gene disruptions, such as Rad52 (Rijkers et al., 1998) and Rad54 (Essers et al., 1997), where these survive to birth.
Materials and methods
Construction of the targeting vector
A 17 kb genomic fragment containing exons II and III of murine Xrcc2 was isolated from a 129/Sv mouse genomic library (Lambda FixII; Stratagene) by screening with a human Xrcc2 cDNA probe. Following restriction mapping and the determination of intron–exon boundaries, fragments were subcloned into pBluescript SK(+) (Stratagene). To generate a targeting vector, 1.6 kb HindIII–EcoRI and 4.5 kb EcoRI–XbaI fragments, from 5′ and 3′ to exon III, respectively, were inserted into pBTII SK(+) (Stratagene) flanking either side of a PGK-neo cassette, with a PGK-HSVtk gene positioned upstream of the short arm. Neo and tk markers were from pPNT (Tybulewicz et al., 1991).
Targeting of ES cells and generation of germ-line chimeras
MAC3 ES cells (M.Maconochie, unpublished) derived from mouse strain 129/Sv/Ev were cultured as described previously (Ramirez-Solis et al., 1993). NotI-linearized targeting vector was introduced into ES cells by electroporation, and colonies surviving G418 (180 µg/ml active ingredient) and gancyclovir (1 µM) selection were screened by Southern blot analysis of BamHI-digested genomic DNA; a 1.8 kb XhoI–BstXI fragment 5′ to the region of targeting was used as a probe. To generate chimeras, a correctly targeted ES clone was injected into C57Bl/6 blastocysts followed by transfer to pseudopregnant foster mothers (C57Bl/6 × CBA F1 hybrid). Resultant high-grade agouti-marked chimeric offspring were mated to C57Bl/6 females, and germ-line transmission was demonstrated by the presence of agouti coat colour in the F1 offspring. The presence of a disrupted Xrcc2 allele was confirmed by Southern blot analysis. The animal studies described were carried out under the guidance issued by the Medical Research Council in ‘Responsibility in the Use of Animals for Medical Research’ (July, 1993) and Home Office Project Licence 30/1544.
Double targeting of Xrcc2 in ES cells
HM-1 ES cells (kindly supplied by Drs D.Melton and J.McWhir, Edinburgh University) were initially targeted with a vector based on that shown in Figure 1A, with the neo selection cassette replaced by the Hprt mini-gene (containing an internal XhoI restriction site) (Selfridge et al., 1992). Selection following electroporation was in medium containing hypoxanthine (100 µM), aminopterin (0.4 µM) and thymidine (16 µM) (Life Technologies) and gancyclovir (2 µM). An Xrcc2+/– cell line was re-electroporated in a second step with the neo-based vector (Figure 1A). Selection was with G418 (250 µg/ml) and gancyclovir (2 µM). Gene disruption was determined by Southern blot analysis of XhoI-digested genomic DNA in each case.
In vitro culture and irradiation of blastocysts
Ionizing radiation sensitivity of embryos was determined as described previously (Sharan et al., 1997). Blastocysts from Xrcc2+/– intercrosses were isolated at E3.5 and washed in M16 medium (Sigma) before exposure to γ-radiation (60Co, 4 Gy at 1.4 Gy/min). Blastocysts were cultured for 6 days in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 20% fetal bovine serum, 0.1 mM sodium pyruvate, 100 µM β-mercaptoethanol, 2 mM glutamine, 100 µM non-essential amino acids, and antibiotics. Inner cell mass area was determined by confocal microscopy after cell staining with 3,3′-dihexyloxacarbo-cyanine iodide.
PCR genotyping of blastocysts and embryos
Genotype determination of Xrcc2 was performed on cell lysates prepared by digestion of either blastocysts, whole embryos or yolk sacs in lysis buffer (50 mM Tris–HCl pH 8.0, 1 mM EDTA, 0.5% Tween-20, 0.5 µg/ml Proteinase K) for 0.5–2 h at 55°C followed by Proteinase K inactivation at 95°C for 5 min. Three primers were used in the PCR reaction. A single 5′ sense primer to the Xrcc2 intron upstream of exon III (5′-GGTTCTATCTTGTGCTTTTGTGTGTTTA-3′), and 3′ antisense primers to exon III (5′-TCTGTTTTCCCCGTTCCTTCTG-3′) and the Pgk-1 promoter of the neo cassette (5′-GCATGCTCCAGACTGCCTTGG-3′) were used to detect wild-type and mutant alleles, respectively. Amplification for 30–35 cycles at 94°C for 35 s, 58°C for 1 min and 72°C for 45 s gave fragments of 545 bp (wild type) and 280 bp (mutant), separated by electrophoresis through a 1.5% agarose gel. Wild-type (Wt) mouse genomic DNA (129Sv) and a plasmid (pX2) containing neo and Xrcc2 sequences mimicking a targeted locus were used as controls.
Chromosome analysis
Tissue from E15.5 embryos was macerated and cells disaggregated by incubation in trypsin. Cells were incubated overnight in DMEM–15% fetal calf serum, then exposed to colcemid (0.05 µg/ml) for 2 h, trypsinized and incubated in hypotonic solution (1:1, 0.075 M KCl:0.034 M trisodium citrate) for 15 min at 37°C. Cells were fixed in 3:1 ethanol:acetic acid and dropped onto slides, before staining with Giemsa for cytogenetic analysis. FISH used chromosome-specific probes for chromosomes 2 [fluorescein isothiocyanate (FITC)-labelled and biotinylated in a ratio of 3:1], 4 (FITC-labelled) and 11 (biotinylated), together with a cy3-labelled pan-centromeric probe (Cambio, UK).
Histological analysis and TUNEL assay
Embryos were fixed in 4% paraformaldehyde, paraffin embedded and serially sectioned (4 µm). Sections for TUNEL assay were dewaxed and permeabilized according to standard procedures, and an in situ Cell Death Detection Kit used as per the manufacturer’s instructions (Boehringer Mannheim). Samples were analysed by fluorescent microscopy or following immunostaining with alkaline phosphatase-conjugated anti-fluorescein antibody (Boehringer Mannheim) and reaction with 3,3′- diaminobenzidine (DAB). Nuclei were counterstained with propidium iodide or haematoxylin. Immunostaining of post-mitotic neurons was performed using monoclonal anti-β3-tubulin antibody TUJ1 (BAbCO) followed by Texas-Red-conjugated horse anti-mouse IgG (Vector Laboratories).
RT–PCR analysis
Total RNA was extracted from embryonic tissue (E15.5) using TRIzol reagent (Life Technologies). The mRNA in 5 µg total RNA was reverse transcribed using oligo(dT)12–18 primers and Superscript II reverse transcriptase (Life Technologies), according to the supplier’s protocol. PCR to determine the presence of Xrcc2 mRNA was performed using a 5′ sense, exon II-specific primer (5′-GAAGGCAGAAGCTCGTTGAAAGAA-3′) and 3′ antisense primer to exon III (5′-CAGGGCTTCCAGGGAGTGC-3′). PCR conditions were 94°C for 45 s, 53°C for 1 min and 72°C for 1 min, for 31 cycles. As a control for reverse transcription, the following primers were used for amplification of glyceraldehyde-3-phosphate dehydrogenase (Gapdh) cDNA: 5′ sense primer, 5′-GAGAAACCTGCCAAGTATGATGAC-3′; 3′ antisense primer, 5′-TGATGGTATTCAAGAGAGTAGGGAG-3′. The products of amplification, 354 and 421 bp for Xrcc2 and Gapdh cDNAs, respectively, were separated by electrophoresis on a 1.5% agarose gel.
Acknowledgments
Acknowledgements
We thank Terry Hacker, Nicola Powles, Stuart Townsend and the Harwell Transgenic Facility for technical support; David Papworth for statistical analysis; Ruth Arkell for advice; and the European Commission for partial financial support (contract F14P-CT950010).
References
- Barnes D.E., Stamp,G., Rosewell,I., Denzel,A. and Lindahl,T. (1998) Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr. Biol., 8, 1395–1398. [DOI] [PubMed] [Google Scholar]
- Baumann P. and West,S.C. (1998) Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem. Sci., 23, 247–251. [DOI] [PubMed] [Google Scholar]
- Bayer S.A. and Altman,J. (1991) Neocortical Development. Raven Press, New York, NY. [Google Scholar]
- Braybrooke J.P., Spink,K.G., Thacker,J. and Hickson,I.D. (2000) The RAD51 family member, RAD51L3, is a DNA-stimulated ATPase that forms a complex with XRCC2. J. Biol. Chem., 275, 29100–29106. [DOI] [PubMed] [Google Scholar]
- Cartwright R., Tambini,C.E., Simpson,P.J. and Thacker,J. (1998) The XRCC2 DNA repair gene from human and mouse encodes a novel member of the recA/RAD51 family. Nucleic Acids Res., 26, 3084–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun J. and Schatz,D.G. (1999) Developmental neurobiology: alterna tive ends for a familiar story? Curr. Biol., 9, R251–R253. [DOI] [PubMed] [Google Scholar]
- Copp A.J., Brook,F.A., Estibeiro,J.P., Shum,A.S. and Cockroft,D.L. (1990) The embryonic development of mammalian neural tube defects. Prog. Neurobiol., 34, 363–403. [DOI] [PubMed] [Google Scholar]
- Essers J., Hendriks,R.W., Swagemakers,S.M., Troelstra,C., de Wit,J., Bootsma,D., Hoeijmakers,J.H. and Kanaar,R. (1997) Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell, 89, 195–204. [DOI] [PubMed] [Google Scholar]
- Frank K.M., Sekiguchi,J.M., Seidl,K.J., Swat,W., Rathbun,G.A., Cheng,H.L., Davidson,L., Kangaloo,L. and Alt,F.W. (1998) Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature, 396, 173–177. [DOI] [PubMed] [Google Scholar]
- Frank K.M. et al. (2000) DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Mol. Cell, 5, 993–1002. [DOI] [PubMed] [Google Scholar]
- Game J.C. (1993) DNA double-strand breaks and the RAD50–RAD57 genes in Saccharomyces. Semin. Cancer Biol., 4, 73–83. [PubMed] [Google Scholar]
- Gao Y. et al. (1998) A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell, 95, 891–902. [DOI] [PubMed] [Google Scholar]
- Gao Y. et al. (2000) Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature, 404, 897–900. [DOI] [PubMed] [Google Scholar]
- Gilmore E.C., Nowakowski,R.S., Caviness,V.S.,Jr and Herrup,K. (2000) Cell birth, cell death, cell diversity and DNA breaks: how do they all fit together? Trends Neurosci., 23, 100–105. [DOI] [PubMed] [Google Scholar]
- Gowen L.C., Johnson,B.L., Latour,A.M., Sulik,K.K. and Koller,B.H. (1996) Brca1 deficiency results in early embryonic lethality characterized by neuroepithelial abnormalities. Nature Genet., 12, 191–194. [DOI] [PubMed] [Google Scholar]
- Griffin C.S., Simpson,P.J., Wilson,C.R. and Thacker,J. (2000) Mammalian recombination–repair genes XRCC2 and XRCC3 promote correct chromosome segregation. Nature Cell Biol., 2, 757–761. [DOI] [PubMed] [Google Scholar]
- Gu Y., Sekiguchi,J., Gao,Y., Dikkes,P., Frank,K., Ferguson,D., Hasty,P., Chun,J. and Alt,F.W. (2000) Defective embryonic neurogenesis in Ku-deficient but not DNA-dependent protein kinase catalytic subunit-deficient mice. Proc. Natl Acad. Sci. USA, 97, 2668–2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakem R., de la Pompa,J.L., Elia,A., Potter,J. and Mak,T.W. (1997) Partial rescue of Brca1 (5-6) early embryonic lethality by p53 or p21 null mutation. Nature Genet., 16, 298–302. [DOI] [PubMed] [Google Scholar]
- Hakem R., de la Pompa,J.L. and Mak,T.W. (1998) Developmental studies of Brca1 and Brca2 knock-out mice. J. Mammary Gland Biol. Neoplasia, 3, 431–445. [DOI] [PubMed] [Google Scholar]
- Jeggo P.A. (1998) DNA breakage and repair. Adv. Genet., 38, 185–218. [DOI] [PubMed] [Google Scholar]
- Johnson R.D., Liu,N. and Jasin,M. (1999) Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature, 401, 397–399. [DOI] [PubMed] [Google Scholar]
- Jones N.J., Cox,R. and Thacker,J. (1987) Isolation and cross-sensitivity of X-ray-sensitive mutants of V79-4 hamster cells. Mutat. Res., 183, 279–286. [DOI] [PubMed] [Google Scholar]
- Kanaar R. et al. (1996) Human and mouse homologs of the Saccharomyces cerevisiae RAD54 DNA repair gene: evidence for functional conservation. Curr. Biol., 6, 828–838. [DOI] [PubMed] [Google Scholar]
- Kanaar R., Hoeijmakers,J.H. and van Gent,D.C. (1998) Molecular mechanisms of DNA double strand break repair. Trends Cell Biol., 8, 483–489. [DOI] [PubMed] [Google Scholar]
- Lee M.K., Tuttle,J.B., Rebhun,L.I., Cleveland,D.W. and Frankfurter,A. (1990) The expression and posttranslational modification of a neuron-specific β-tubulin isotype during chick embryogenesis. Cell Motil. Cytoskel., 17, 118–132. [DOI] [PubMed] [Google Scholar]
- Li G.C., Ouyang,H., Li,X., Nagasawa,H., Little,J.B., Chen,D.J., Ling,C.C., Fuks,Z. and Cordon-Cardo,C. (1998) Ku70: a candidate tumor suppressor gene for murine T cell lymphoma. Mol. Cell, 2, 1–8. [DOI] [PubMed] [Google Scholar]
- Liang F., Han,M., Romanienko,P.J. and Jasin,M. (1998) Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl Acad. Sci. USA, 95, 5172–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim D.S. and Hasty,P. (1996) A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol., 16, 7133–7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N. et al. (1998) XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol. Cell, 1, 783–793. [DOI] [PubMed] [Google Scholar]
- Muris D.F. et al. (1994) Cloning of human and mouse genes homologous to RAD52, a yeast gene involved in DNA repair and recombination. Mutat. Res., 315, 295–305. [DOI] [PubMed] [Google Scholar]
- Nornes H.O. and Carry,M. (1978) Neurogenesis in spinal cord of mouse: an autoradiographic analysis. Brain Res., 159, 1–6. [DOI] [PubMed] [Google Scholar]
- Pittman D.L. and Schimenti,J.C. (2000) Midgestation lethality in mice deficient for the RecA-related gene, Rad51d/Rad51l3. Genesis, 26, 167–173. [DOI] [PubMed] [Google Scholar]
- Ramirez-Solis R., Davis,A.C. and Bradley,A. (1993) Gene targeting in embryonic stem cells. Methods Enzymol., 225, 855–878. [DOI] [PubMed] [Google Scholar]
- Rijkers T., Van Den Ouweland,J., Morolli,B., Rolink,A.G., Baarends,W.M., Van Sloun,P.P., Lohman,P.H. and Pastink,A. (1998) Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol. Cell. Biol., 18, 6423–6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage J.R. (1976) Classification and relationships of induced chromosomal structural changes. J. Med. Genet., 13, 103–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild D., Lio,Y., Collins,D.W., Tsomondo,T. and Chen,D.J. (2000) Evidence for simultaneous protein interactions between human Rad51 paralogs. J. Biol. Chem., 275, 16443–16449. [DOI] [PubMed] [Google Scholar]
- Selfridge J., Pow,A.M., McWhir,J., Magin,T.M. and Melton,D.W. (1992) Gene targeting using a mouse HPRT minigene/HPRT-deficient embryonic stem cell system: inactivation of the mouse ERCC-1 gene. Somat. Cell Mol. Genet., 18, 325–336. [DOI] [PubMed] [Google Scholar]
- Sharan S.K. et al. (1997) Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature, 386, 804–810. [DOI] [PubMed] [Google Scholar]
- Shinohara A. and Ogawa,T. (1995) Homologous recombination and the roles of double-strand breaks. Trends Biochem. Sci., 20, 387–391. [DOI] [PubMed] [Google Scholar]
- Shinohara A., Ogawa,H. and Ogawa,T. (1992) Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell, 69, 457–470. [DOI] [PubMed] [Google Scholar]
- Shinohara A., Ogawa,H., Matsuda,Y., Ushio,N., Ikeo,K. and Ogawa,T. (1993) Cloning of human, mouse and fission yeast recombination genes homologous to RAD51 and recA. Nature Genet., 4, 239–243. [DOI] [PubMed] [Google Scholar]
- Sugo N., Aratani,Y., Nagashima,Y., Kubota,Y. and Koyama,H. (2000) Neonatal lethality with abnormal neurogenesis in mice deficient in DNA polymerase β. EMBO J., 19, 1397–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P. (1997) Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev., 11, 1111–1121. [DOI] [PubMed] [Google Scholar]
- Tambini C.E., George,A.M., Rommens,J.M., Tsui,L.C., Scherer,S.W. and Thacker,J. (1997) The XRCC2 DNA repair gene: identification of a positional candidate. Genomics, 41, 84–92. [DOI] [PubMed] [Google Scholar]
- Thacker J. (1999a) The role of homologous recombination processes in the repair of severe forms of DNA damage in mammalian cells. Biochimie, 81, 77–85. [DOI] [PubMed] [Google Scholar]
- Thacker J. (1999b) A surfeit of RAD51-like genes? Trends Genet., 15, 166–168. [DOI] [PubMed] [Google Scholar]
- Thacker J., Ganesh,A.N., Stretch,A., Benjamin,D.M., Zahalsky,A.J. and Hendrickson,E.A. (1994) Gene mutation and V(D)J recombination in the radiosensitive irs lines. Mutagenesis, 9, 163–168. [DOI] [PubMed] [Google Scholar]
- Thomaidou D., Mione,M.C., Cavanagh,J.F. and Parnavelas,J.G. (1997) Apoptosis and its relation to the cell cycle in the developing cerebral cortex. J. Neurosci., 17, 1075–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuzuki T., Fujii,Y., Sakumi,K., Tominaga,Y., Nakao,K., Sekiguchi,M., Matsushiro,A., Yoshimura,Y. and Morita,T. (1996) Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl Acad. Sci. USA, 93, 6236–6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker J.D., Jones,N.J., Allen,N.A., Minkler,J.L., Thompson,L.H. and Carrano,A.V. (1991) Cytogenetic characterization of the ionizing radiation-sensitive Chinese hamster mutant irs1. Mutat. Res., 254, 143–152. [DOI] [PubMed] [Google Scholar]
- Tybulewicz V.L., Crawford,C.E., Jackson,P.K., Bronson,R.T. and Mulligan,R.C. (1991) Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell, 65, 1153–1163. [DOI] [PubMed] [Google Scholar]
- Venkitaraman A.R. (1999) Breast cancer genes and DNA repair. Science, 286, 1100–1102. [DOI] [PubMed] [Google Scholar]