Abstract
Purpose
The purpose of this study was to develop a population pharmacokinetic (PK) model for 3-AP pharmacokinetics and to evaluate the effect of ABCB1 polymorphisms on the pharmacokinetic profile of 3-AP and to assess the relationship between 3AP disposition and patient covariates.
Methods
A total of 40 patients with advanced cancer from two phase 1 studies were included in the population PK model building. Patients received 3-AP 25–105 mg/m2 IV on day 1. 3-AP plasma and erythrocyte levels were sampled at 10 timepoints over a 24-hour period and measured by a validated HPLC method. Data were analyzed by a nonlinear mixed-effects modeling approach using the NONMEM system.
Results
3AP pharmacokinetics were described as a 3-compartment model with first-order elimination. One compartment representing the plasma and another representing erythrocyte concentrations. Gender was associated with volume of distribution, in which women had a lower V2. The number of cycles administered was associated with clearance; those with decreased clearance were more likely to receive less than 2 cycles before going off study.
Conclusion
This study suggests that monitoring 3-AP plasma concentrations in the first cycle and dose adjustment in those with decreased clearance may be helpful in decreasing toxicity associated with the 3-AP.
Keywords: Triapine®, 3-aminopyridine-2-carboxaldehyde thiosemicarbazone, population pharmacokinetics, phase 1
INTRODUCTION
3-Aminopyridine-2-carboxaldehyde thiosemicarbazone (3-AP, Triapine®, Vion Pharmaceuticals, Inc., New Haven, CT) is a novel small molecule inhibitor of the M2 metal binding site of ribonucletoide reductatse (RR) in development as an anti-cancer agent (1,2). 3-AP has been evaluated in a number of phase I and II clinical trials for a variety of malignancies as both a single agent and in combination with other cytotoxic agents (3–9) demonstrating promising activity in hematological malignancies (8, 9) as well as melanoma and prostate cancer (4), however its development as an anticancer agent has been limited by toxicity. Currently, little is known about the pharmacokinetics of 3-AP and a better understanding may help guide future drug development.
Toxicities of 3-AP are similar to those seen with other cytotoxic chemotherapy agents and include neutropenia, thrombocytopenia, anemia, fatigue, nausea, and diarrhea and appear dose related (3–9). 3-AP may also cause methemoglobinemia, which can be severe in subjects with G6PD deficiency (10,11), however even in trials of 3-AP where G6PD deficient patients were excluded, methemoglobinemia has been reported (4,12,13).
p-glycoprotein (pgp), is a 170–180 kDa plasma membrane-associated protein and the product of the multidrug resistance (ABCB1) gene (14). Rappa and colleagues (15) demonstrated in vitro that the presence of the ABCB1 gene makes cells 2–3 fold more resistant to 3-AP, suggesting that 3-AP is a substrate for pgp. Several common polymorphisms in the ABCB1 gene are associated with decreased pgp activity and our hypothesis was that individuals with ABCB1 variants associated with decreased pgp activity may have altered pharmacokinetics of 3-AP.
The purpose of this study was to estimate the population pharmacokinetic parameters of 3-AP and to describe the relationship between patient specific covariates, including age, gender, weight, performance status, body surface area, concurrent chemotherapy, ABCB1 genotype cycles of chemotherapy received and toxicity.
METHODS
Patients
A total of 40 patients with advanced stage primary or metastatic tumors from two phase 1 studies were included in the population PK model building.
Study Design
The phase 1 studies have been described in detail (16, 17). Patients received escalating doses of 3-AP and concurrent chemotherapy with either irinotecan or doxorubicin, which is described in Table 1 For both studies, 3-AP was supplied by Vion Pharmaceuticals, Inc., and distributed by the Cancer Therapy Evaluation Program, the Division of Cancer Treatment and Diagnosis, National Cancer Institute. Blood samples were collected on Day 1 of Cycle 1 at pre-infusion, 1–2 minutes just before the end of the infusion, and at 10, 20, 30, 45 minutes and 1, 2, 4.5, 6, 8, 10 and 22 hours after the end of the infusion.
Table 1.
Dose Escalation Scheme
| 3-AP Irinotecan | ||
|---|---|---|
| Level (n) | Irinotecan | 3-AP |
| −2 | 75 mg/m2 | 45 mg/m2 |
| −1 | 100 mg/m2 | 60 mg/m2 |
| 1 | 150 mg/m2 | 85 mg/m2 |
| 2 | 150 mg/m2 | 60 mg/m2 |
| 3 | 200 mg/m2 | 60 mg/m2 |
| 4 | 250 mg/m2 | 60 mg/m2 |
| 5 | 300 mg/m2 | 60 mg/m2 |
| 6 | 300 mg/m2 | 85 mg/m2 |
| 3-AP Doxorubicin | ||
| Level (n) | Doxorubicin | 3-AP |
| 1a | 45 mg/m2 | 25 mg/m2 |
| 1 | 60 mg/m2 | 25 mg/m2 |
| 2a | 45 mg/m2 | 45 mg/m2 |
| 2 | 60 mg/m2 | 45 mg/m2 |
All patients were required to have Eastern Cooperative Oncology Group performance status of 0–2; adequate bone marrow (WBC > 3,000/µl, absolute neutrophil count > 1,500/µl, platelet > 100,000/µl); adequate hepatic function (total bilirubin within institutional normal limit and alanine aminotransferase, ≤ 2.5 × the institutional upper limit of normal); adequate renal function (creatinine ≤ 1.5 mg/dl or measured creatinine clearance ≥ 60 ml/min/1.73m2 for patients with creatinine levels about institutional normal); an LVEF > 45%; no G6PD deficiency and life expectancy greater than 12 weeks. Patients receiving irinotecan were required to be heterozygous or wild-type for the UGT1A1. Since all patients had essentially normal hepatic, renal, cardiac and hematologic function at study entry, these variables were not modeled as covariates. Adverse events were evaluated using the National Cancer Institute Common Toxicity Criteria, version 3.0 guidelines. The determination of antitumor efficacy was based on objective tumor assessments made according to RECIST. Baseline imaging-based tumor assessments were performed at baseline and all were repeated after two cycles. The dose escalation scheme is shown in Table 1 and baseline demographics are in Table 2.
Table 2.
Baseline Patient Characteristics
| 3-AP Irinotecan N=20 |
3-AP Doxorubicin N=20 |
|
|---|---|---|
| Age (years) | 59 (29–72) | 61 (34–84) |
| Weight (kg) | 79±13 | 80±15 |
| BSA (m2) | 1.92±.02 | 1.95±.02 |
| ECOG Performance Status (0/1) | 3/17 | 2/18 |
| Gender (male/female) | 9/11 | 13/7 |
| Mean Cycle Number (range) | 2.6 (1–8) | 2.6 (1–8) |
| Grade III–IV Neutropenia | 8/20 | 17/20 |
| Grade III–IV GI toxicity | 5/20 | 1/20 |
| Grade III–IV Hypoxia | 3/20 | 0/20 |
| ABCB1 C1236T (CC, CT, TT) | 0.28, 0.28, 0.44 | 0.32, 0.63, 0.05 |
| ABCB1 G2677T (GG, GT, TT) | 0.26, 0.26, 0.47 | 0.35, 0.50, 0.15 |
| ABCB1 C3435T (CC, CT, TT) | 0.21, 0.37, 0.42 | 0.15, 0.50, 0.35 |
Analytical Procedure
HPLC with UV detection was used to analyze the serum and erythrocyte samples for 3-AP concentrations as previously described by Murren (18). A Spectra Physics P2000 HPLC system was used. Chromatographic separation was achieved using a Supelco Discovery C18 column (5 µM, 250 mm × 4.6 mm; Supelco, St. Louis, MO) with detection at 400 nm. Plasma or erythrocyte samples (0.5 ml) were extracted with 1.0 ml of methanol (containing 4 mM EDTA). After centrifugation, the extract was concentrated to dryness and was reconstituted with 0.25 ml of a solvent consisting of 10% acetonitrile and 90% mobile Phase A [20 mM potassium phosphate buffer, 15 mM 1-heptanesulfonic acid, and 1 mM EDTA (pH 3.0)]. The reconstituted solution sample (30 µl) was then injected into the HPLC system. External calibration standards were prepared in pooled control human plasma and were processed identically to test samples. The validated assay is linear over 0.02–10 µg/ml for plasma (r2=0.99), with an intraday variability ranging from a coefficient of variation (CV) of 0.41%–3.4% and an interday variability ranging from a CV of 2.60%–5.5%. The lower limit of quantitation was 0.078 ug/ml and an absolute recovery from plasma of 92%.
Genotyping Procedure
Pyrosequencing assays for the common ABCB1 polymorphisms C1236T, G2677T/A and C3435T (19) were developed. Samples were collected at baseline in a DNA Paxgene tube (Quiagen,Valencia, CA) and DNA was extracted as recommended as by the manufacturer.
PCR Amplification
Specific oligonucleotide primers for amplification by PCR of ABCB1 gene fragments from genomic DNA were derived from known sequences [GenBank accession no: AC005068] using Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). PCR for pyrosequencing was performed in 40 µl reactions containing 20 µl PCR Master Mix (Promega, Madison, WI), 10 pmol forward and reverse primer (IDT Technology, Coraville, IA), 14 µl of nuclease free water and 10–100 ng of genomic DNA. PCR amplification was performed under the following conditions: initial denaturation at 95°C for 5 minutes, 50 cycles of denaturation at 95°C for 30 seconds, annealing at 52°C for 30 seconds, and extension at 72°C for 30 seconds, followed by a final extension step at 72°C for 5 minutes.
Pyrosequencing
The Pyrosequencing primers were designed using SNP Primer Design Software Version 1.01 (http://www.pyrosequencing.com). Briefly, 35 µl of biotinylated PCR product was immobilized on strep-tavidin-coated Sepharose beads (Amersham Biosciences, Piscataway, NJ) with binding buffer (10 mmol/L Tris-HCl, 2 mol/L NaCl, 1 mmol/L EDTA, and 0.1% Tween 20, pH 7.6). After room temperature incubation with constant agitation for 10 minutes, the strands were separated and treated with 70% ethanol, denaturation solution (0.2 mol/L NaOH) and washing buffer (10 mmol/L Tris-Acetate, pH 7.6). The beads, containing the biotinylated template, were released into wells with a 40 µl mixture of anneal-ing buffer (20 mmol/L Tris-Acetate, 2 mmol/L Magnesium Acetate Tetrahydrate, pH 7.6) and 21 pmol of sequencing primer (IDT Technology, Coraville, IA). Incubation was carried out at 80°C for 2 minutes. Genotyping was subsequently performed using a PSQ 96 SNP Reagent Kit and PSQ 96MA system (Bio-tage AB, Uppsala, Sweden). Genotypes were resolved on the basis of peak height measurements using PSQ96 SNP Software, version 1.2 AQ. Subjects were categorized as wild-type, heterozygote or variant.
Population Pharmacokinetic Model Development
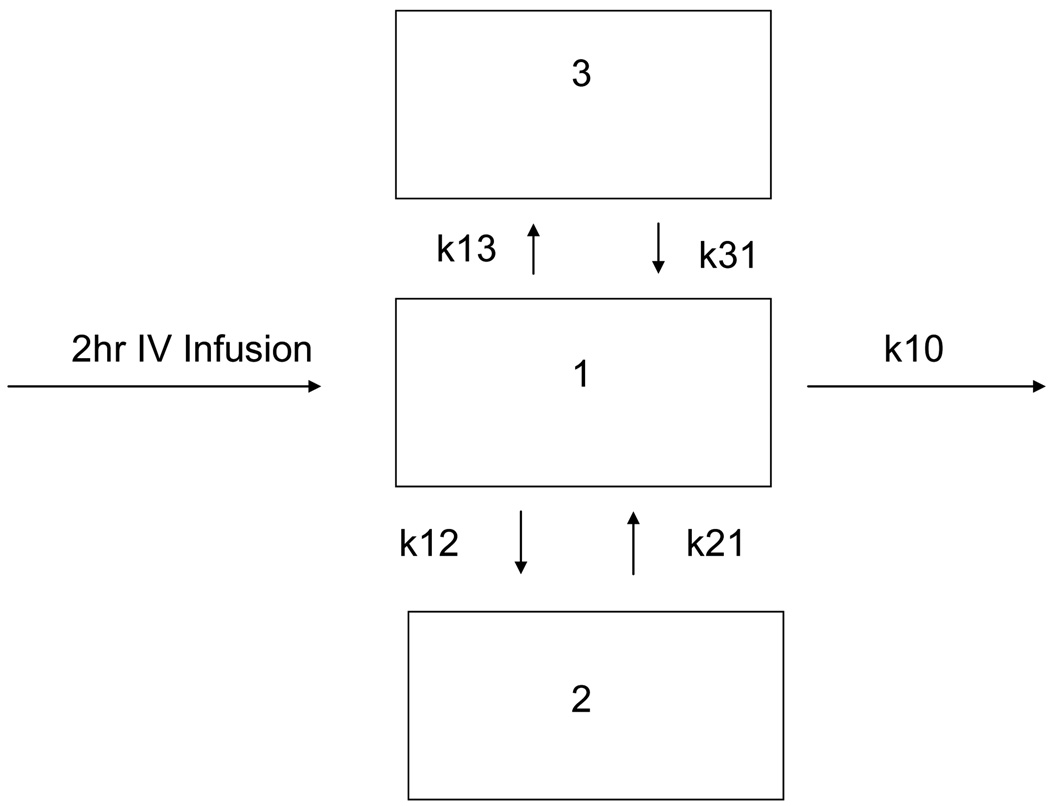
Data were analyzed by a nonlinear mixed-effects modeling approach using the NONMEM system (Version VI, NONMEM Project Group, UCSF/Globomax and PDx-Pop Version 3.1). Xpose4 and S-PLUS (Insightful Corp, Seattle, Washington) were used for goodness-of-fit assessment and model evaluation. A 3-compartment model with first-order elimination was used (subroutine ADVAN11 TRANS 4) as described in Figure 1. The central compartment was associated with plasma concentrations and one of the peripheral compartments was associated with erythrocyte concentrations. Both plasma and erythrocyte concentrations were simultaneously analyzed in the single model.
Figure 1.
Base pharmacokinetic model for 3-AP in plasma and erythrocytes. Compartment 1 is plasma sampling, compartment 2 is sampling in erythrocytes.
The pharmacokinetic parameters obtained from the model were clearance from compartment 1(CL1), clearance from compartment 2 (CL2), clearance from compartment 3 (CL3), volume of distribution for compartment 1, plasma (V1), volume of distribution for compartment 2, erythrocytes (V2) volume of distribution for compartment 3 (V3).
The base pharmacokinetic model was developed without including patient-specific covariates. Covariates were then selected based on their possible effects on the pharmacokinetics of 3-AP. Fifteen covariates—including weight, body surface area [BSA], gender, performance status, age, ABCB1[ABCB1 C1236T, G2667T, C3436T] genotypes, toxicity [grade III–IV myelosuppression, gastrointestinal toxicity, hypoxia] concurrent chemotherapy [irinotecan, doxorubicin], response and number of cycles administered were available for testing. During stepwise covariate model building, covariates were only tested on those PK parameters that might make an important difference in dosing 3-AP, namely CL, V1 and V2.
Patient-specific covariates were tested by improving the goodness of fit assessed by the likelihood ratio test and visual inspection of diagnostic plots. The likelihood ratio test was used to assess the significance of a covariate in the model. A decrease in the objective function value (OFV) of at least 3.8 (χ2, P ≤ 0.05, df = 1) was considered significant for adding a single covariate into the model. The standard backward elimination step was ultimately not applied in this analysis due to only identifying two covariates that influenced the parameters.
The first-order conditional estimation method with interaction (FOCE-I) was used for all analyses. Between-subject variabilities were modeled under the assumption of log-normally distributed parameters. Residual unexplained variability was explored using combinations of additive and proportional models, and allowing for distinct error structures for plasma and erythrocyte concentrations.
A model qualification step was implemented using a visual predictive check for the plasma and erythrocyte compartments separately. Using the parameters of the final model, 100 individuals were simulated using a parametric bootstrap approach. The plasma and erythrocyte concentrations for each were simulated and the median, 5th and 95th quantiles were calculated.
RESULTS
3-AP Pharmacokinetics
A total of 398 plasma concentrations and 392 erythrocytes concentrations from 40 subjects were available for analysis. Baseline demographic characteristics and genotyping results for patients included in this analysis are described in Table 2. Only variables considered as potential covariates for developing the population pharmacokinetic model are included.
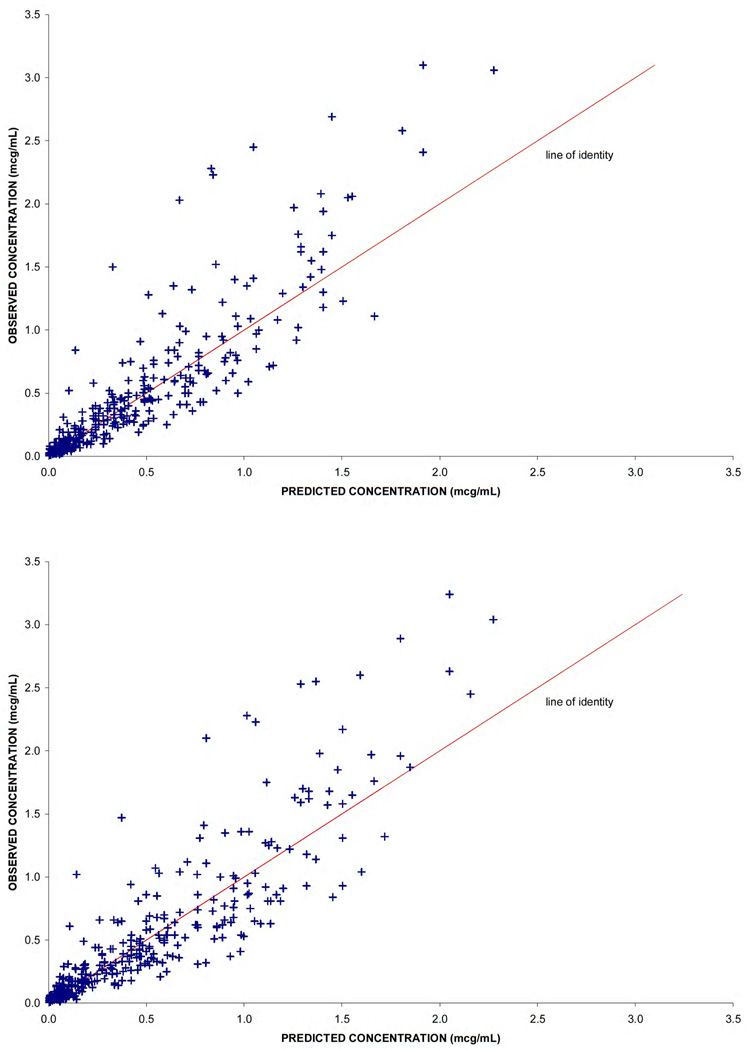
All model parameters estimates and their relative standard errors are described in Table 3. Plots of the observed concentration versus time for plasma and erythrocyte pools are presented in Figure 2. Observed versus predicted concentrations for plasma and erythrocyte compartments are presented in Figure 3. These figures demonstrate the model was adequate to describe the data. Other diagnostic plots including weighted residuals versus time and versus prediction were examined and evaluated to be acceptable (figures not shown).
Table 3.
Final Parameter Estimates and 95% Confidence Intervals from NONMEM
| Parameter | Estimate | Relative SE (%) |
|---|---|---|
| Cl (L/h) | 25.0 | 5.96 |
| In subjects receiving only one cycle | 17.0 | 14.6* |
| V1 (L) | 5.68 | 12.4 |
| Cl2 (L/h) | 99.6 | 15.0 |
| V2 (L) (female) | 19.0 | 11.6 |
| V2 (L) (male) | 34.8 | 15.5* |
| Cl3 (L/h) | 8.76 | 10.8 |
| V3 (L) | 40.4 | 19.0 |
| Variability in Cl (%CV) | 34.8 | 25.1 |
| Variability in V1 (%CV) | 60.6 | 38.4 |
| Variability in V2 (%CV) | 41.5 | 28.7 |
| RUV (%CV) | 17.5 | 18.3 |
| RUV (SD) | 0.0168 | 35.4 |
Cl, total body clearance; V apparent volume of distribution; CV, coefficient of variation; RUV residual unexplained variability; relative SE, measure of precision computed by dividing the standard error (SE) by the value of the estimate × 100.
Relative SE expressed in terms of the multiplicative term, for the covariate
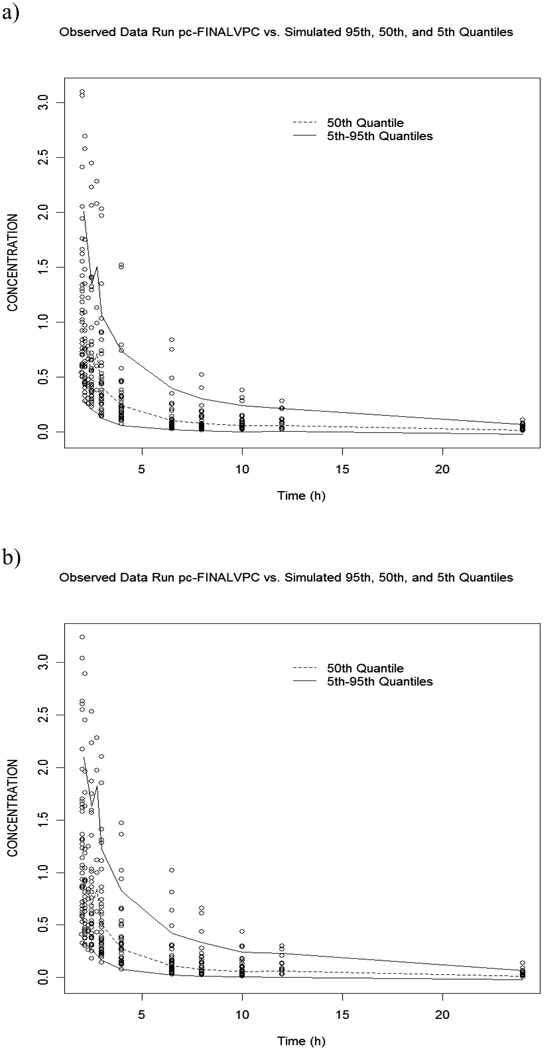
Figure 2.
Symbols are the observed versus model predicted 3-AP concentrations in a) plasma and b) erythrocytes following administration as a 2 hour infusion in 40 subjects. The dashed line is the median from the visual predictive check, and the solid lines represent the 5th and 95th quantiles.
Figure 3.
Diagnostic plots of the observed versus model predicted 3-AP concentrations and weighted residuals in a) plasma and b) erythrocytes.
Covariate Model
Fifteen covariates were evaluated in step-wise model building. V2 was significantly smaller in women, 19 L compared to men 34.8 L. A significantly lower CL was associated with patients who received only one cycle of chemotherapy, compared to patients who were able to tolerate more than one cycle. Patients receiving only one cycle of chemotherapy had a CL of 17 L/h compared to patients receiving more than one cycle of chemotherapy who had a CL of 25 L/h. Concurrent chemotherapy, BSA and ABCB1 genotypes did not influence pharmacokinetic parameters.
Variance Model
As is commonly the case in data sets with relatively small numbers of subjects, between-subject variabilities could not be determined on all PK parameters. That does not mean variability does not exist in all parameters, but that the data are simply insufficient to uniquely identify all the sources of between-subject variability. The identified between-subject variability in CL, V1 and V2 were all moderate (see Table 3) and within a range that would be expected from a drug with these PK characteristics. A combined additive and proportional error model was found to be appropriate to describe the observed concentrations. Additionally, it was not necessary to model residual variability separately for observations in the plasma and erythrocyte compartments.
The visual predictive checks of the plasma (upper panel) and erythrocyte (lower panel) compartments, presented in Figure 2, are adequate to support the final model and parameter estimates. The dashed line represents the median of the simulated data and the solid lines define the 5th and 95th quantiles of the simulated data. Ideally, the 50th quantile should fall in the center of the data, with approximately 5% of the observed data falling above the 95th quantile and 5% below. For the plasma data, 9.9% falls outside of the 5th to 95th quantiles, and 12.6% of the erythrocyte concentrations are outside of the range. While both of these values are reasonable and the median is centered within the data, there is a slight tendency noted to underpredict concentrations relative to the observed values. This could result from and inadequate PK model, a slight bias in a PK parameter or a variance structure that is not most appropriately defined as a log-normal distribution. Given the very small deviations that are observed in the predictive check, it was not considered worth pursuing alternative model specifications.
DISCUSSION
As the development of 3-AP has been limited by toxicity, our primary purpose was to determine if toxicity was associated with pharmacokinetic parameters. We identified a relationship between CL and the number cycles the patient received, where a significantly reduced clearance was associated with patients who stopped therapy after receiving only one cycle of chemotherapy. This 32% decrease in CL is a population-based finding and was found irrespective of which starting dose to which a patient was randomized. Overall, eight patients went off study after only one cycle of therapy. Of these eight patients, four experienced dose limiting toxicities and three others discontinued therapy because of toxicity that was not defined as dose limiting. Only one patient discontinued therapy for progressive disease, which was based on physician and patient judgement and did not meet RECIST criteria. It is important to point out that this evaluation provides an association and does not determine a cause-and-effect relationship. It is unlikely that stopping therapy after one cycle causes a decrease in clearance. Nonetheless, if a patient had to terminate therapy after one cycle, they tended to have lower 3-AP clearance. The mechanism of reduced clearance is unknown. A polymorphism in ABCB1 is not likely the explanation as these polymorphisms did not contribute to pharmacokinetic variability. However, a larger controlled clinical trial powered to detect an appropriate difference would be necessary to demonstrate that finding. Reduced renal and hepatic function are also not explanations, as normal and renal and hepatic function were required for study entry. A potential explanation may be variable intracellular binding of 3-AP to ribonucleotide reductase and changes in intracellular clearance (22).
Prior investigations have demonstrated that ABCB1 genotype was predictive of both outcome (20) and toxicity (20, 21), in patients treated with 3-AP and an in vitro evaluation suggested 3-AP was a substrate for p-glycoprotein (15). We therefore hypothesized individuals with variant genotypes had altered pharmacokinetics and evaluated this relationship in the current study, showing the ABCB1 genotype did not influence 3-AP pharmacokinetic parameters. While some studies have shown a pharmacokinetic relationship between ABCB1 genotype and plasma concentrations, a number of studies have failed to demonstrate a relationship (23). This may be explained by the wide and variable distribution of ABCB1 including the blood brain barrier, GI tract, liver and tumor cells, all of which may distribute 3-AP back to a central compartment in a variable manner, resulting in no consistent PK relationship between ABCB1 genotype and 3-AP plasma concentrations.
BSA was not identified as a significant covariate for any 3-AP PK parameter. While BSA based dosing is widely used in oncology and generally believed to reduce inter-individual pharmacokinetic variability, comparisons between flat dosing strategies and BSA based dosing have generally shown them to be equivalently ineffective in decreasing inter-individual variability (24). Since gender did influence V2, a gender specific, flat dose could be considered in future clinical trials.
The relationship between toxicity and exposure was further evaluated by assessing grade III–IV myelosuppression and gastrointestinal toxicity. However, nearly universal myelosuppression was observed in both clinical studies, even with lower doses (25–45mg/m2) of 3-AP used in combination with doxorubicin. In addition, only five patients experienced gastrointestinal toxicity, all of which were observed in the study with concurrent irinotecan. Since gastrointestinal toxicity with irinotecan is a well known dose limiting toxicity of irinotecan and no toxicities were noted in the doxorubicin arm, this finding suggests 3-AP contributed little to the observed GI toxicity.
When considering grade III–IV hypoxia, only three patients experienced episodes of hypoxia. All of these occurred at the highest dose levels of 3-AP administered in this study of 60–85mg/m2. This is consistent with other trials of 3-AP where G6PD deficient patients were excluded, methemoglobinemia has still been reported at doses ranging from 96–105mg/m2 in 5–25% of subjects. A potential explanation for this toxicity may be preferential distribution or accumulation of 3-AP into erythrocytes. Our data is best fit by a three compartment model, with V2, sampled from the erythrocyte compartment, having a 3–6 fold increased volume when compared to V1, sampled from the plasma compartment. This suggests extensive intracellular distribution of 3-AP in the erythrocyte compartment. We also noted a significantly decreased V2 in female subjects, compared to males. This may have a protective effect, as all hypoxia episodes occurred in male subjects, even though 50% (8 male, 8 female) of the subjects treated at the highest doses of 3-AP were female.
CONCLUSION
This paper describes a population pharmacokinetic model for 3-AP in cancer patients enrolled in phase I clinical trials. A three compartment model is the best fit for the data, and 3-AP appears to accumulate in V2, or the erythrocyte sampled compartment. Reduced CL by an unknown mechanism is associated with receiving less than 2 cycles of chemotherapy, but not individual toxicity. This study suggests that monitoring 3-AP plasma concentrations in the first cycle and dose adjustment in those with decreased clearance may be helpful in decreasing toxicity associated with the 3-AP. Additionally, a gender specific, flat dosing schedule may be considered in future clinical trials.
Acknowledgments
Supported by: U01CA062491 “Early Clinical Trials of Anti-Cancer Agents with Phase I Emphasis” NCI; CTEP Translational Research Initiative Funding 24XS090, and 1ULRR025011 Clinical and Translational Science Award of the National Center for Research Resources, NIH and the American College of Clinical Pharmacy
References
- 1.Finch RA, Liu MC, Cory AH, Cory JG, Sartorelli AC. Triapine (3-aminopyridine-2-carboxaldehyde thiosemicarbazone; 3-AP): an inhibitor of ribonucleotide reductase with antineoplastic activity. Adv Enzyme Regul. 1999;39:3–12. doi: 10.1016/s0065-2571(98)00017-x. [DOI] [PubMed] [Google Scholar]
- 2.Cory JG, Cory AH, Rappa G, Lorico A, Liu MC, Lin TS, Sartorelli AC. Inhibitors of ribonucleotide reductase. Comparative effects of amino- and hydroxy-substituted pyridine-2-carboxaldehyde thiosemicarbazones. Biochem Pharmacol. 1994;48:335–344. doi: 10.1016/0006-2952(94)90105-8. [DOI] [PubMed] [Google Scholar]
- 3.Schelman WR, Holen K, Mulkerin D, Kolesar J, Thomas J, Kruse M, Oliver K, Marnocha R, Eickhoff J, Wilding G. Journal of Clinical Oncology, 2006 ASCO Annual Meeting Proceedings Part I. Vol 24, No. 18S (June 20 Supplement); 2006. p. 12011. [Google Scholar]
- 4.Chang JE, Morgan Meadows S, Traynor A, Kolesar J, Marnocha R, Lee F, Eickoff J, Beth E, Binger K, Wilding G. Journal of Clinical Oncology, 2006 ASCO Annual Meeting Proceedings Part I. Vol 24, No. 18S (June 20 Supplement); 2006. p. 13168. [Google Scholar]
- 5.Attia S, Kolesar J, Mahoney MR, Pitot HC, Laheru D, Heun J, Huang W, Eickhoff J, Erlichman C, Holen KD. A phase 2 consortium (P2C) trial of 3-aminopyridine-2-carboxaldehyde thiosemicarbazone (3-AP) for advanced adenocarcinoma of the pancreas. Invest New Drugs. 2008 Aug;26(4):369–379. doi: 10.1007/s10637-008-9123-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mackenzie MJ, Saltman D, Hirte H, Low J, Johnson C, Pond G, Moore MJ. A Phase II study of 3-aminopyridine-2-carboxaldehyde thiosemicarbazone (3-AP) and gemcitabine in advanced pancreatic carcinoma. A trial of the Princess Margaret hospital Phase II consortium. Invest New Drugs. 2007 Dec;25(6):553–558. doi: 10.1007/s10637-007-9066-3. [DOI] [PubMed] [Google Scholar]
- 7.Knox JJ, Hotte SJ, Kollmannsberger C, Winquist E, Fisher B, Eisenhauer EA. Phase II study of Triapine in patients with metastatic renal cell carcinoma: a trial of the National Cancer Institute of Canada Clinical Trials Group (NCIC IND.161) Invest New Drugs. 2007 Oct;25(5):471–477. doi: 10.1007/s10637-007-9044-9. [DOI] [PubMed] [Google Scholar]
- 8.Karp JE, Giles FJ, Gojo I, Morris L, Greer J, Johnson B, Thein M, Sznol M, Low J. A phase I study of the novel ribonucleotide reductase inhibitor 3-aminopyridine-2-carboxaldehyde thiosemicarbazone (3-AP, Triapine) in combination with the nucleoside analog fludarabine for patients with refractory acute leukemias and aggressive myeloproliferative disorders. Leuk Res. 2008 Jan;32(1):71–77. doi: 10.1016/j.leukres.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gojo I, Tidwell ML, Greer J, Takebe N, Seiter K, Pochron MF, Johnson B, Sznol M, Karp JE. Phase I and pharmacokinetic study of Triapine, a potent ribonucleotide reductase inhibitor, in adults with advanced hematologic malignancies. Leuk Res. 2007 Sep;31(9):1165–1173. doi: 10.1016/j.leukres.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 10.Dalal BI, Kollmannsberger C. Drug-induced haemolysis and methaemoglobinaemia in glucose 6-phosphate dehydrogenase deficiency. Br J Haematol. 2005 May;129(3):291. doi: 10.1111/j.1365-2141.2005.05404.x. [DOI] [PubMed] [Google Scholar]
- 11.Foltz LM, Dalal BI, Wadsworth LD, Broady R, Chi K, Eisenhauer E, Kobayashi K, Kollmannsburger C. Recognition and management of methemoglobinemia and hemolysis in a G6PD-deficient patient on experimental anticancer drug Triapine. Am J Hematol. 2006 Mar;81(3):210–211. doi: 10.1002/ajh.20547. [DOI] [PubMed] [Google Scholar]
- 12.Knox JJ, Hotte SJ, Kollmannsberger C, Winquist E, Fisher B, Eisenhauer EA. hase II study of Triapine in patients with metastatic renal cell carcinoma: a trial of the National Cancer Institute of Canada Clinical Trials Group (NCIC IND.161) Invest New Drugs. 2007 Oct;25(5):471–477. doi: 10.1007/s10637-007-9044-9. [DOI] [PubMed] [Google Scholar]
- 13.Ma B, Goh BC, Tan EH, Lam KC, Soo R, Leong SS, Wang LZ, Mo F, Chan AT, Zee B, Mok T. A multicenter phase II trial of 3-aminopyridine-2-carboxaldehyde thiosemicarbazone (3-AP, Triapine) and gemcitabine in advanced non-small-cell lung cancer with pharmacokinetic evaluation using peripheral blood mononuclear cells. Invest New Drugs. 2008 Apr;26(2):169–173. doi: 10.1007/s10637-007-9085-0. [DOI] [PubMed] [Google Scholar]
- 14.Hoffmeyer S, Burk O, von Richter O, et al. Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc. Natl. Acad. Sci. USA. Mar 28;97(7):3473–3478. doi: 10.1073/pnas.050585397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rappa G, Lorico A, Liu M-C, Kruh GD, Cory AH, Cory JG, Sartorelli AC. Overexpression of the multidrug resistance genes mdr1, mdr3 and mrp in L1210 leukemia cells reisitant to inhibitors of ribonucleotide reductase. Biochemical Pharmacology. 1997;54:649–655. doi: 10.1016/s0006-2952(97)00210-4. [DOI] [PubMed] [Google Scholar]
- 16.Schelman WR, Morgan-Meadows S, Marnocha R, Lee F, Eickhoff J, Huang W, Pomplun M, Jiang Z, Alberti D, Kolesar JM, Ivy P, Wilding G, Traynor AM. A phase I study of Triapine in combination with doxorubicin in patients with advanced solid tumors. Cancer Chemother Pharmacol. 2009 May;63(6):1147–1156. doi: 10.1007/s00280-008-0890-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schelman WR, Holen K, Mulkerin D, et al. A phase I study of triapine in combination with irinotecan in refractory tumors; Journal of Clinical Oncology, 2006 ASCO Annual Meeting Proceedings Part I. Vol 24, No. 18S (June 20 Supplement); 2006. 12011”. [Google Scholar]
- 18.Murren J, Modiano M, Clairmont C, Lambert P, Savaraj N, Doyle T, Sznol M. Phase I and pharmacokinetic study of triapine, a potent ribonucleotide reductase inhibitor, administered daily for five days in patients with advanced solid tumors. Clin Cancer Res. 2003 Sep 15;9(11):4092–4100. [PubMed] [Google Scholar]
- 19.Kolesar JM, Hamidovic A, Hahn K. Clinical Significance of ABCB1 Genotyping in Oncology. J Oncol Pharm Pract. 2009 Apr 28; doi: 10.1177/1078155209104380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Traynor AM, Lee JW, Bayer GK, Tate JM, Thomas SP, Mazurczak M, Graham DL, Kolesar JM, Schiller JH. A phase II trial of Triapine(R) (NSC# 663249) and gemcitabine as second line treatment of advanced non-small cell lung cancer: Eastern Cooperative Oncology Group Study 1503. Invest New Drugs. 2009 Feb 24; doi: 10.1007/s10637-009-9230-z. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Attia S, Kolesar J, Mahoney MR, Pitot HC, Laheru D, Heun J, Huang W, Eickhoff J, Erlichman C, Holen KD. A phase 2 consortium (P2C) trial of 3-aminopyridine-2-carboxaldehyde thiosemicarbazone (3-AP) for advanced adenocarcinoma of the pancreas. Invest New Drugs. 2008 Aug;26(4):369–379. doi: 10.1007/s10637-008-9123-6. Epub 2008 Feb 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolesar JM, Schelman WR, Geiger PG, Holen KD, Traynor AM, Alberti DB, Thomas JP, Chitambar CR, Wilding G, Antholine WE. Electron paramagnetic resonance study of peripheral blood mononuclear cells from patients with refractory solid tumors treated with Triapine. J Inorg Biochem. 2008 Apr;102(4):693–698. doi: 10.1016/j.jinorgbio.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fung KL, Gottesman MM. A synonymous polymorphism in a common MDR1 (ABCB1) haplotype shapes protein function. Biochim Biophys Acta. 2009 May;1794(5):860–871. doi: 10.1016/j.bbapap.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mathijssen RH, de Jong FA, Loos WJ, van der Bol JM, Verweij J, Sparreboom A. Flat-fixed dosing versus body surface area based dosing of anticancer drugs in adults: does it make a difference? Oncologist. 2007 Aug;12(8):913–923. doi: 10.1634/theoncologist.12-8-913. [DOI] [PubMed] [Google Scholar]