Summary
The p53 tumor suppressor and its paralogs p63 and p73 are at the crux of a network modulating cellular responses against potentially tumorigenic events. p53 acts primarily as a transcription factor, regulating the expression of both coding and non-coding RNAs, as well as the activity of RNA processing complexes. In line with their anti-tumorigenic function, p53 and p63 have recently been implicated in restricting tumor cell invasion. In parallel, a growing number of non-canonical target genes have been added to the p53 repertoire. These include genes encoding for proteins that impinge on a broad spectrum of cellular functions, from cell metabolism to stem cell renewal. The p53 story is still far from being fully told.
Introduction
Humans with germline p53 mutations are affected by the Li-Fraumeni syndrome, characterized by very high cancer susceptibility[1–2]. p53 knockout mice develop tumors with short latency and 100% penetrance [3]. In approximately 50% of human cancers p53 is mutated; in many of the remaining 50%, the function of the retained wild type (wt) p53 protein is compromised by deregulation of upstream or downstream components of the p53 pathway [4]. En masse, these observations demonstrate the critical role of p53 in tumor prevention.
In unstressed cells, p53 is constitutively restrained by Mdm2, an E3 ubiquitin ligase that promotes p53 degradation; the Mdm2 gene is positively regulated by p53, defining a negative feedback loop that controls p53 activity. Cellular stress relieves Mdm2’s inhibitory effects, triggering p53 stabilization and activation. Once activated, p53 facilitates DNA repair and inhibits the proliferation of potentially tumorigenic cells, chiefly through instigating cell cycle arrest, senescence or apoptosis.
The p53 response is elicited by a wide variety of stress signals conducive to or associated with malignant transformation, such as DNA damage, oncogene activation, abnormal mitosis, loss of cell-cell contact and hypoxia [5]. Although seemingly dissimilar, many of these signals may actually converge on one another. Biochemically, p53 is a potent transcriptional regulator capable of controlling the expression of hundreds of genes [4–5]. Within this context, it interacts with numerous cofactors and binding partners that modulate its transcriptional output. The p53 gene family includes two additional members, p63 and p73, also acting as transcriptional modulators.
The great interest in p53 has spawned numerous excellent reviews. Therefore, we will focus only on a limited set of recent studies, pertaining particularly to new functions of p53 and family.
p53 and metabolism
Recent years have seen a renaissance of interest in the links between cancer and metabolism; p53 research is no exception.
p53 engages in an intricate interplay with reactive oxygen species (ROS). Under conditions of mild, physiological oxidative stress, p53 preferentially induces expression of antioxidant genes; when ROS production is aberrantly high, p53 instead activates pro-oxidant genes that may facilitate apoptosis, along with overt proapoptotic genes such as PUMA, Bax and Pig3 [6]*. Antioxidant genes upregulated by p53 include glutathione peroxidase 1 (GPX1), mitochondrial superoxide dismutase 2 (SOD2), aldehyde dehydrogenase 4 family member A1 (ALDH4A1) and sestrin 1 and 2 (SESN1 and SESN2). The induction of antioxidant genes by p53 is likely aimed to minimize the genotoxic danger that even basal levels of ROS pose to DNA. Strikingly, dietary supplementation with the antioxidant N-acetylcysteine (NAC) completely abolished the incidence of lymphoma in p53−/− mice [6], implicating p53’s antioxidant function as essential for its tumor suppressing action.
Even under normal physiological conditions, p53 may participate in homeostatic regulation of ROS formation and metabolic processes by maintaining the optimal mode of glucose metabolism and energy boost in response to dips in ATP levels. Reliance on aerobic glycolysis (the Warburg Effect) is a trademark of tumor cells.
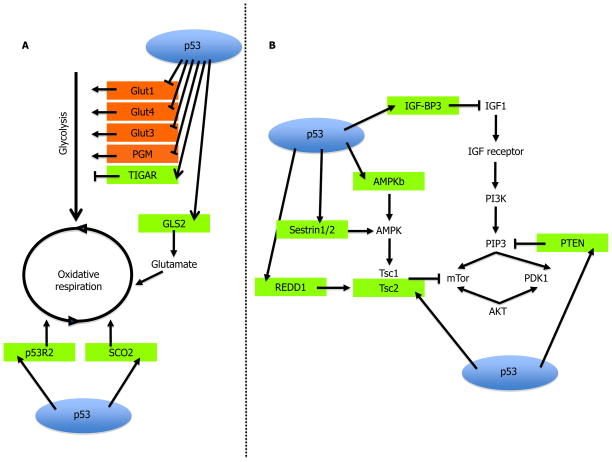
The reprogramming of metabolic pathways endows cancer cells with multiple growth advantages. These include better growth in low oxygen conditions and mobilization of pathways that promote nucleotide biosynthesis and production of fatty acids for lipid biosynthesis, necessary for intensive proliferation [7]. p53 antagonizes the Warburg Effect and inhibits glycolysis by decreasing glucose uptake [8], inducing glycolysis-repressing genes [9] and enhancing mitochondrial respiration [10] (Figure 1A).
Figure 1. Metabolic regulation by p53.
(A) p53 transcriptionally induces numerous inhibitors of mTOR, thus negatively affecting cell growth; (B) p53 inhibits glycolysis and facilitates oxidative respiration via transcriptional regulation of relevant genes. p53-induced genes are colored green; p53-repressed genes are red.
p53 also indirectly impinges on metabolism via the mTOR pathway (Figure 1B). Besides responding to glucose levels, mTOR senses changes in availability of amino acids, ATP/AMP and growth factors [11]. AMP-activated kinase (AMPK) is one of the major upstream inhibitors of mTOR activity. Perhaps not surprisingly, AMPK both activates and is activated by p53 in response to energetic stress [12–13]. Other p53 response genes negatively affecting the mTOR pathway include IGF-BP3, PTEN, TSC2, Sestrin1/2 and REDD1 (reviewed in [11]). Thus, p53 leads a multifaceted campaign against the Warburg Effect, both under normal and metabolically challenged conditions. The involvement of p53 in maintaining metabolic homeostasis raises the intriguing possibility that loss of p53 might make cancers more susceptible to drugs that target metabolic pathways.
mTOR is also a negative regulator of autophagy, a process affording cell survival during nutrient starvation by catabolic breakdown of cellular components. Autophagy also contributes to genome stability by destroying potentially harmful cytoplasmic organelles, such as defective mitochondria which otherwise would emit genotoxic ROS. Accordingly, p53 can positively affect autophagy, both by inhibiting mTOR activity and by transactivating pro-autophagic genes such as DRAM [14]*[15]*. Yet, p53 was also reported to inhibit autophagy, particularly under conditions where p53 is cytoplasmic [16]*. To make the picture even more complex, autophagy can promote tumor cell survival under stress, including chemotherapy. Indeed, tumor cells retaining wtp53 may reap a survival advantage from the improved autophagic response endowed by their p53, thereby ingeniously distorting the anti-cancer apparatus into a pro-cancer machinery. Additional forays into the links between p53 and autophagy will likely be rewarding.
p53 and non-coding RNAs
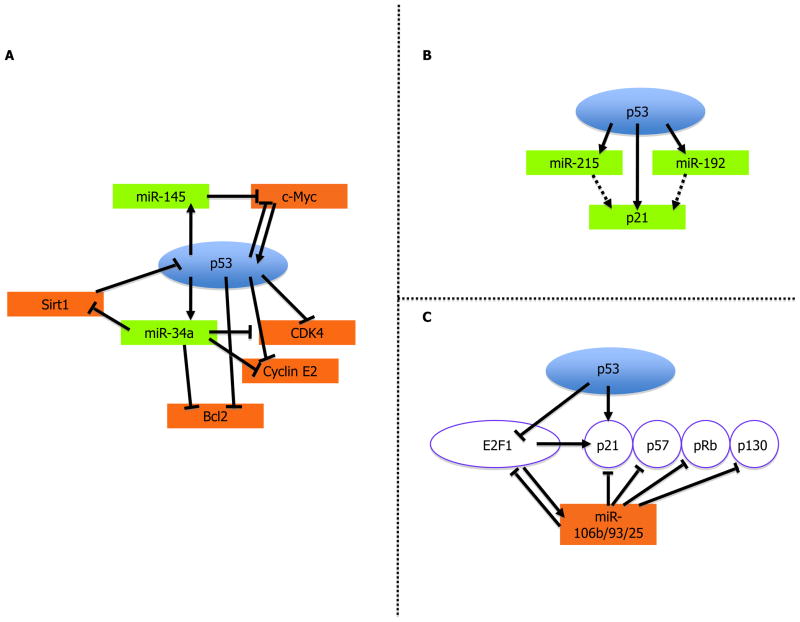
MicroRNAs (miRNAs) are short non-coding RNA molecules that regulate protein levels by binding to specific mRNAs, inhibiting their translation and often also accelerating their degradation. As is the case for protein-coding mRNAs, miRNA expression patterns are also grossly altered in cancer. p53 modulates the expression of numerous miRNA species, including miR-34a,b and c [17–18]. It may not be coincidental that some of the mRNA species targeted by p53-responsive miRNAs are also directly transcriptionally modulated by p53. In this way, p53 governs either the amplification or fine-tuning of signals that impinge on cell fate. For example, miR-34a contributes to p53-dependent apoptosis, as well as cell cycle arrest and senescence [17–19]. Validated targets of miR-34a include CDK4, Cyclin E2, Bcl2 and c-Met, all of which are also transcriptionally repressed by p53 (Figure 2A). An additional interesting target of miR-34a is the deacetylase Sirt1. Since Sirt1 is a negative regulator of p53, its downregulation by miR-34a defines a positive feedback loop that amplifies p53 activity [20].
Figure 2. miRNAs in the service of p53.
(A) p53 induces the expression of miRNAs that target p53-repressed genes; (B) p53 induces the expression of miRNAs that enhance p53 activity output by augmenting p21 levels; (C) p53 represses miRNAs that target E2F1-induced antiproliferative genes, thereby restricting cell proliferation. See text for further details.
Other miRs transactivated by p53 include miR-192, miR-215 [21] and miR-145 [22]. MiRs-192 and -215 both upregulate p21, a canonical p53 target gene product, defining a feed-forward cycle that restricts cell proliferation (Figure 2B). On the other hand, miR-145 downregulates c-Myc [22]*, a proto-oncogene that is also transcriptionally repressed by p53 (Figure 2A).
A cluster of cancer-associated miRNAs, including miR-106b/93/25 and others, is repressed by p53 in an E2F1-mediated manner [23]*. These miRNAs target antiproliferative genes that are themselves E2F1 targets (Figure 2C); accordingly, overexpression of these miRNAs promotes cell proliferation [23]*. By repressing them, p53 tilts the balance towards growth arrest and senescence.
Other classes of non-coding RNA such as large intervening non-coding RNA (lincRNA) are also regulated by p53 [24]. lincRNAs may guide chromatin remodeling factors to target loci or else act together with transcription factors (perhaps also p53?) to modulate pre-existing transcriptional programs. For instance, the p53-induced lincRNA TUG1 facilitates repression of cell cycle-related genes through binding the polycomb repression complex PRC2 [25].
A new twist in the regulation of miRNA expression by p53 was revealed by showing that the DNA-binding domain of p53 binds to the Drosha complex in response to DNA damage. Drosha cleaves primary miRNA transcripts into hairpin structures (pre-miRs) that are subsequently processed into mature, functional miRNAs by another endonuclease complex, Dicer. p53 binding enhances recruitment of Drosha to target precursor miRNA and its processing activity towards a subset of miRNAs [26]**. By modulating Drosha activity, p53 might alter the inventory of pre-miRs available for Dicer operation. Interestingly, one of the miRNAs whose processing is altered in such manner is miR-145, a transcriptional target of p53.
The p53 family and tumor cell invasion
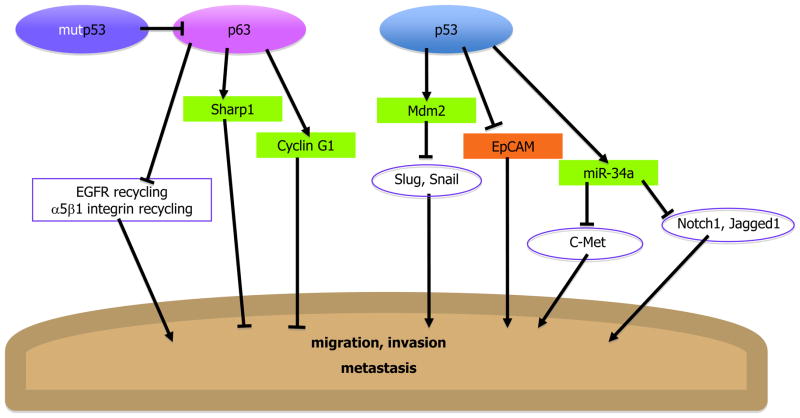
Increased invasiveness of cancer cells is a major driver of metastasis and malignancy. This has not escaped the attention of p53 and its family member p63 (Figure 3). Loss of p53 augments cancer cell invasion [27]; conversely, p53 activation suppresses migration and invasion [28]. This inhibitory effect of p53 is partly mediated by Mdm2, which promotes the ubiquitination and degradation of Slug and Snail, pivotal transcription factors that drive tumor cell invasiveness [29–30]. The anti-invasive role of Mdm2, a well-established oncogene, is rather intriguing. Or perhaps not; hyperproliferation and invasiveness are emerging as two uncoupled and often opposing properties of cancer cells, underpinning distinct stages in tumor progression. In addition, p53 inhibits invasion by modulation of cell adhesion proteins such as EpCAM [31]. Furthermore, p53-regulated miRNAs such as miR-34a, which downregulates c-Met, Notch1 and Jagged1, also contribute to p53’s ability to repress migration and invasion [32–33]. The critical importance of p53’s anti-invasive action for cancer suppression is reinforced by recent experiments employing a mouse model of Wnt-driven intestinal carcinogenesis (Y. Ben-Neriah, personal communication).
Figure 3. p53 and p63 inhibit cell migration, invasion and metastasis.
p53 and p63 transcriptionally activate inhibitors of invasion and repress enhancers of invasion. Mutp53 inhibits p63, thereby abrogating its anti-invasion activity.
Like its cousin p53, p63 also modulates adhesion, migration and invasion [34–37]. The picture is confounded by the existence of multiple p63 isoforms, either possessing or lacking the N-terminal transactivation domain (TAp63 and Delta-Np63, respectively), whose transcriptional effects and biological impact are often opposite. It remains to be firmly established how each distinct p63 isoform impinges on tumor cell invasion. Yet, recent work has highlighted a new interesting aspect of this story. About half of all human tumors harbor p53 mutations, often with excessive accumulation of mutant p53 (mutp53) protein. Mounting evidence indicates that such mutp53 proteins acquire cancer-promoting gain of function activities, including promotion of metastasis [38]. It now emerges that the latter may be largely due to inhibition of anti-invasive and anti-migratory effects of p63 by mutp53 [39–40]*. The outcome of this activity of mutp53 is repression of anti-invasion genes [39], enhancement of integrin and EGF receptor (EGFR) recycling [40] driving activation of the EGFR pathway [41], and eventual promotion of metastasis. This might offer an appealing explanation to the observation that p53 mutations often correlate with advanced, invasive stages of tumor progression.
p53 in stem cells and aging- two sides of the same coin?
Recently, there has been a flourish of publications demonstrating that p53 deficiency facilitates reprogramming of differentiated cells into induced pluripotent stem (iPS) cells, closely resembling embryonic stem (ES) cells [42–46]*. The exact nature of the antagonism between p53 and reprogramming pathways is still debated. One possibility is that the iPS procedure indirectly causes DNA damage, driving p53 to activate a barrier of anti-proliferative senescence [47]. Alternatively, explicit pro-differentiation effects of p53 [48] might actively inhibit the iPS program. Indeed, in frog development, p53 interacts with Smad transcriptional regulators to direct embryonic germ layer specification [49]. Interestingly, p53 was reported to protect mouse ES cells from DNA damage not by exerting cell cycle arrest or apoptosis, but by inducing differentiation via suppression of the stem cell (SC)-specific genes Nanog [50] and Oct4 [51].
Adult SCs share with ES cells pluripotency and capacity to self-renew. Yet, these long-lived renewable reservoirs may provide a cellular compartment with increased neoplastic potential. Germline deletion of p53 in mice with critically short telomeres spares damaged SCs from apoptosis and protracts their survival [52–53]. The skin of such p53-deleted mice displays improved wound healing and hair growth, apparently due to increased numbers of epidermal SCs [54]. Similarly, elegant in vivo competition experiments show that p53-deficient mouse hematopoietic stem cells (HSCs) have improved repopulation capacity in transplantation assays, while the outcompeted p53-proficient SCs acquire senescence-like features [55]. Surprisingly, recent analysis of human HSCs [56]* reveals a strikingly different picture. As expected, p53-deficient HSCs better resist radiation-induced apoptosis. However, upon repeated in vivo expansion without acute genotoxic insult, they actually display reduced self-renewal capacity, apparently due to persistent accumulation of unrepaired DNA damage. Thus, in human HSCs, p53 serves as a positive regulator of self-renewal, by maintaining rigorous genome-integrity quality-control. Beyond illustrating the complexity of the links between p53 and SCs, these findings also raise the alarming possibility that mechanisms of p53-mediated tumor suppression may differ between mouse and human.
Tumors represent rare perturbed clonal outgrowths, whose continuous propagation may rely on a subset of tumor-initiating cells with SC-like properties. In a mouse model of ErbB2-driven breast cancer, cultured p53−/− mammospheres were found enriched for self-renewing “SCs” due to loss of p53 control over asymmetric cell division [57]**. In the hematopoietic system, K-Ras activation instigates a burst of hyperproliferation, but subsequent p53-driven terminal differentiation of stem and progenitor cells provides a p53-dependent barrier against limitless proliferation of undifferentiated leukemia-initiating cells [58]. Prevention of expansion of the cancer-initiating cell pool thus emerges as an important tumor suppressor activity of p53.
The anti-proliferative effect of p53 might not be all advantageous. In fact, mice with hyperactive p53 amass fewer HSCs due to decreased self-renewal capacity, probably accounting for their accelerated aging phenotype [59]; the same may hold for SCs of other tissues. That being said, other studies suggest an anti-aging effect of p53: transgenic mice with only a mild increase in p53 exhibit a reduced rate of age-related oxidative damage, more efficient clearing of DNA-damaged cells and enhanced longevity [60]. Interestingly, in mice and humans, p53 activity declines with age [61], probably contributing to the concomitant increase in cancer frequency. It remains to be determined whether the diminished p53 function promotes aging or, contrarily, the reduced p53 function is a consequence of the aging process.
Conclusion
Since its discovery more than 30 years ago, a massive amount of data has accumulated that attests to the tumor suppressing role of p53. However, as we keep exploring the intricacies of p53 activity, more and more of its diverse functions are cropping up. The field of tumor suppression is experiencing a growing interest in « esoteric » subjects such as metabolism and stem cells. Perhaps, with knowledge from this broader picture, we will also better understand the workings of cancer cells.
Acknowledgments
Work in the authors’ laboratories is supported by grants from the National Cancer Institute (R37 CA40099), the Flight Attendant Medical Research Institute, the European Commission (OncomiRs, FP7 Contract 201102 and INFLACARE, FP7 Contract 223151), the Robert Bosch Foundation, and the M.D. Moross Cancer Institute. M.O. is the incumbent of the Andre Lwoff Professorial Chair in Molecular Biology at the Weizmann institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Malkin D. p53 and the Li-Fraumeni syndrome. Cancer Genet Cytogenet. 1993;66:83–92. doi: 10.1016/0165-4608(93)90233-c. [DOI] [PubMed] [Google Scholar]
- 2.Chang F, Syrjanen S, Tervahauta A, Syrjanen K. Tumourigenesis associated with the p53 tumour suppressor gene. Br J Cancer. 1993;68:653–661. doi: 10.1038/bjc.1993.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 4.Vousden KH, Prives C. Blinded by the Light. The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 5.Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6*.Sablina AA, Budanov AV, Ilyinskaya GV, Agapova LS, Kravchenko JE, Chumakov PM. The antioxidant function of the p53 tumor suppressor. Nat Med. 2005;11:1306–1313. doi: 10.1038/nm1320. This study reports that p53 modulates a pro- or anti-oxidant transcriptional output depending on the extent of ROS damage. Most remarkably, it shows that intracellular increases of ROS are pivotal contributors to the cancer prone phenotype of p53-null mice, highlighting the importance of p53 antioxidant action for tumor suppression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev. 2008;18:54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawauchi K, Araki K, Tobiume K, Tanaka N. p53 regulates glucose metabolism through an IKK-NF-kappaB pathway and inhibits cell transformation. Nat Cell Biol. 2008;10:611–618. doi: 10.1038/ncb1724. [DOI] [PubMed] [Google Scholar]
- 9.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 10.Ma W, Sung HJ, Park JY, Matoba S, Hwang PM. A pivotal role for p53: balancing aerobic respiration and glycolysis. J Bioenerg Biomembr. 2007;39:243–246. doi: 10.1007/s10863-007-9083-0. [DOI] [PubMed] [Google Scholar]
- 11.Feng Z, Levine AJ. The regulation of energy metabolism and the IGF-1/mTOR pathways by the p53 protein. Trends Cell Biol. 2010;20:427–434. doi: 10.1016/j.tcb.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, Birnbaum MJ, Thompson CB. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 13.Feng Z, Hu W, de Stanchina E, Teresky AK, Jin S, Lowe S, Levine AJ. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res. 2007;67:3043–3053. doi: 10.1158/0008-5472.CAN-06-4149. [DOI] [PubMed] [Google Scholar]
- 14*.Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci U S A. 2005;102:8204–8209. doi: 10.1073/pnas.0502857102. This is the first demonstration that p53 is a regulator of autophagy. Specifically, it shows that activation of p53 upon glucose deprivation leads to autophagy by inhibiting mTOR via AMPK and TSC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15*.Crighton D, Wilkinson S, O’Prey J, Syed N, Smith P, Harrison PR, Gasco M, Garrone O, Crook T, Ryan KM. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. This study describes a novel regulator of autophagy that is transcriptionally induced by p53, thus providing a new molecular link between p53 and autophagy. [DOI] [PubMed] [Google Scholar]
- 16*.Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D’Amelio M, Criollo A, Morselli E, Zhu C, Harper F, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–687. doi: 10.1038/ncb1730. This study reports that, opposite to the transcriptional effects of p53 that promote autophagy, cytoplasmic p53 can actually inhibit autophagy, illustrating the importance of p53 subcellular localization in determination of cell fate. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 18.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christoffersen NR, Shalgi R, Frankel LB, Leucci E, Lees M, Klausen M, Pilpel Y, Nielsen FC, Oren M, Lund AH. p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death Differ. 2010;17:236–245. doi: 10.1038/cdd.2009.109. [DOI] [PubMed] [Google Scholar]
- 20.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci U S A. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Braun CJ, Zhang X, Savelyeva I, Wolff S, Moll UM, Schepeler T, Orntoft TF, Andersen CL, Dobbelstein M. p53-Responsive micrornas 192 and 215 are capable of inducing cell cycle arrest. Cancer Res. 2008;68:10094–10104. doi: 10.1158/0008-5472.CAN-08-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22*.Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, Elble R, Watabe K, Mo YY. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci U S A. 2009;106:3207–3212. doi: 10.1073/pnas.0808042106. This study reveals a new facet of the importance of p53-regulated miRNAs, namely inhibition of cell proliferation by miRNA-mediated c-Myc repression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23*.Brosh R, Shalgi R, Liran A, Landan G, Korotayev K, Nguyen GH, Enerly E, Johnsen H, Buganim Y, Solomon H, et al. p53-Repressed miRNAs are involved with E2F in a feed-forward loop promoting proliferation. Mol Syst Biol. 2008;4:229. doi: 10.1038/msb.2008.65. This study highlights the ability of p53 to coordinately modulate an “orchestra” of miRNAs, which are both targets and regulators of E2F. It provides a network view of p53 action. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26**.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. This study reveals a novel surprising biochemical and regulatory feature of p53. Specifically, it shows that p53 not only works as a transcription factor to modulate the expression of specific miRNAs, but also changes the cellular miRNA landscape by a very different mechanism, namely binding Drosha and facilitating selective miRNA processing. [DOI] [PubMed] [Google Scholar]
- 27.Gadea G, de Toledo M, Anguille C, Roux P. Loss of p53 promotes RhoA-ROCK-dependent cell migration and invasion in 3D matrices. J Cell Biol. 2007;178:23–30. doi: 10.1083/jcb.200701120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moran DM, Maki CG. Nutlin-3a induces cytoskeletal rearrangement and inhibits the migration and invasion capacity of p53 wild-type cancer cells. Mol Cancer Ther. 2010;9:895–905. doi: 10.1158/1535-7163.MCT-09-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang SP, Wang WL, Chang YL, Wu CT, Chao YC, Kao SH, Yuan A, Lin CW, Yang SC, Chan WK, et al. p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nat Cell Biol. 2009;11:694–704. doi: 10.1038/ncb1875. [DOI] [PubMed] [Google Scholar]
- 30.Lim SO, Kim H, Jung G. p53 inhibits tumor cell invasion via the degradation of snail protein in hepatocellular carcinoma. FEBS Lett. 2010;584:2231–2236. doi: 10.1016/j.febslet.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 31.Sankpal NV, Willman MW, Fleming TP, Mayfield JD, Gillanders WE. Transcriptional repression of epithelial cell adhesion molecule contributes to p53 control of breast cancer invasion. Cancer Res. 2009;69:753–757. doi: 10.1158/0008-5472.CAN-08-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pang RT, Leung CO, Ye TM, Liu W, Chiu PC, Lam KK, Lee KF, Yeung WS. MicroRNA-34a suppresses invasion through downregulation of Notch1 and Jagged1 in cervical carcinoma and choriocarcinoma cells. Carcinogenesis. 2010;31:1037–1044. doi: 10.1093/carcin/bgq066. [DOI] [PubMed] [Google Scholar]
- 33.Li N, Fu H, Tie Y, Hu Z, Kong W, Wu Y, Zheng X. miR-34a inhibits migration and invasion by down-regulation of c-Met expression in human hepatocellular carcinoma cells. Cancer Lett. 2009;275:44–53. doi: 10.1016/j.canlet.2008.09.035. [DOI] [PubMed] [Google Scholar]
- 34.Barbieri CE, Tang LJ, Brown KA, Pietenpol JA. Loss of p63 leads to increased cell migration and up-regulation of genes involved in invasion and metastasis. Cancer Res. 2006;66:7589–7597. doi: 10.1158/0008-5472.CAN-06-2020. [DOI] [PubMed] [Google Scholar]
- 35.Carroll DK, Carroll JS, Leong CO, Cheng F, Brown M, Mills AA, Brugge JS, Ellisen LW. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat Cell Biol. 2006;8:551–561. doi: 10.1038/ncb1420. [DOI] [PubMed] [Google Scholar]
- 36.Gu X, Coates PJ, Boldrup L, Nylander K. p63 contributes to cell invasion and migration in squamous cell carcinoma of the head and neck. Cancer Lett. 2008;263:26–34. doi: 10.1016/j.canlet.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 37.Kommagani R, Leonard MK, Lewis S, Romano RA, Sinha S, Kadakia MP. Regulation of VDR by deltaNp63alpha is associated with inhibition of cell invasion. J Cell Sci. 2009;122:2828–2835. doi: 10.1242/jcs.049619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol. 2010;2:a001107. doi: 10.1101/cshperspect.a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39*.Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, Solari A, Bobisse S, Rondina MB, Guzzardo V, et al. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137:87–98. doi: 10.1016/j.cell.2009.01.039. This study describes a novel mechanism for promotion of metastasis by cancer-associated mutant p53 proteins (mutp53). Specifically, it describes a concerted action of oncogenic Ras, mutp53 and TGF-beta, which leads to p63 inactivation and abrogation of its anti-metastatic effects. [DOI] [PubMed] [Google Scholar]
- 40*.Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S, Lukashchuk N, Gillespie DA, Ludwig RL, Gosselin P, et al. Mutant p53 drives invasion by promoting integrin recycling. Cell. 2009;139:1327–1341. doi: 10.1016/j.cell.2009.11.026. Complementing #39, this study provides an additional mechanism whereby mutp53 promotes metastasis by p63 inhibition. Sepcifically, mutp53 is shown to drive increased invasion by enhancing integrin and EGFR trafficking. [DOI] [PubMed] [Google Scholar]
- 41.Dong P, Xu Z, Jia N, Li D, Feng Y. Elevated expression of p53 gain-of-function mutation R175H in endometrial cancer cells can increase the invasive phenotypes by activation of the EGFR/PI3K/AKT pathway. Mol Cancer. 2009;8:103. doi: 10.1186/1476-4598-8-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Belmonte JC. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46*.Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, Pinho S, Silva JC, Azuara V, Walsh M, et al. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23:2134–2139. doi: 10.1101/gad.1811609. The above five articles (#42–45) demonstate the inhibitory role of p53 on iPS reprogramming, and suggest that p53 may act as an important barrier against cancer by restraining the emergence of cells with “stem-like” properties. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Banito A, Gil J. Induced pluripotent stem cells and senescence: learning the biology to improve the technology. EMBO Rep. 2010;11:353–359. doi: 10.1038/embor.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Molchadsky A, Rivlin N, Brosh R, Rotter V, Sarig R. p53 is balancing development, differentiation and de-differentiation to assure cancer prevention. Carcinogenesis. 2010 doi: 10.1093/carcin/bgq101. [DOI] [PubMed] [Google Scholar]
- 49.Piccolo S. p53 regulation orchestrates the TGF-beta response. Cell. 2008;133:767–769. doi: 10.1016/j.cell.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 50.Lin T, Chao C, Saito S, Mazur SJ, Murphy ME, Appella E, Xu Y. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol. 2005;7:165–171. doi: 10.1038/ncb1211. [DOI] [PubMed] [Google Scholar]
- 51.Qin H, Yu T, Qing T, Liu Y, Zhao Y, Cai J, Li J, Song Z, Qu X, Zhou P, et al. Regulation of apoptosis and differentiation by p53 in human embryonic stem cells. J Biol Chem. 2007;282:5842–5852. doi: 10.1074/jbc.M610464200. [DOI] [PubMed] [Google Scholar]
- 52.Begus-Nahrmann Y, Lechel A, Obenauf AC, Nalapareddy K, Peit E, Hoffmann E, Schlaudraff F, Liss B, Schirmacher P, Kestler H, et al. p53 deletion impairs clearance of chromosomal-instable stem cells in aging telomere-dysfunctional mice. Nat Genet. 2009;41:1138–1143. doi: 10.1038/ng.426. [DOI] [PubMed] [Google Scholar]
- 53.Chin L, Artandi SE, Shen Q, Tam A, Lee SL, Gottlieb GJ, Greider CW, DePinho RA. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97:527–538. doi: 10.1016/s0092-8674(00)80762-x. [DOI] [PubMed] [Google Scholar]
- 54.Flores I, Blasco MA. A p53-dependent response limits epidermal stem cell functionality and organismal size in mice with short telomeres. PLoS One. 2009;4:e4934. doi: 10.1371/journal.pone.0004934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bondar T, Medzhitov R. p53-mediated hematopoietic stem and progenitor cell competition. Cell Stem Cell. 2010;6:309–322. doi: 10.1016/j.stem.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56*.Milyavsky M, Gan OI, Trottier M, Komosa M, Tabach O, Notta F, Lechman E, Hermans KG, Eppert K, Konovalova Z, et al. A Distinctive DNA Damage Response in Human Hematopoietic Stem Cells Reveals an Apoptosis-Independent Role for p53 in Self-Renewal. Cell Stem Cell. 2010 doi: 10.1016/j.stem.2010.05.016. This study reports that, contrary to what has been found in mouse models, p53 actually enhances self-renewal capability of adult human stem cells by assuring the intactness of their genomes. It raises the intriguing possibility that p53-mediated tumor suppression mechanisms differ between mouse and human. [DOI] [PubMed] [Google Scholar]
- 57**.Cicalese A, Bonizzi G, Pasi CE, Faretta M, Ronzoni S, Giulini B, Brisken C, Minucci S, Di Fiore PP, Pelicci PG. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell. 2009;138:1083–1095. doi: 10.1016/j.cell.2009.06.048. This study highlights a novel cancer-suppressor role of p53. Specifically, it shows that p53 facilitates asymetrical stem cell division, which serves to limit the stem cell pool. In p53’s absence, cancer-initiating “stem” cells divide symmertrically, such that one “cancer stem cell” gives rise to two of its kind, constantly increasing the number of such tumor-driving cells. [DOI] [PubMed] [Google Scholar]
- 58.Zhao Z, Zuber J, Diaz-Flores E, Lintault L, Kogan SC, Shannon K, Lowe SW. p53 loss promotes acute myeloid leukemia by enabling aberrant self-renewal. Genes Dev. 2010;24:1389–1402. doi: 10.1101/gad.1940710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Y, Elf SE, Miyata Y, Sashida G, Huang G, Di Giandomenico S, Lee JM, Deblasio A, Menendez S, Antipin J, et al. p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell. 2009;4:37–48. doi: 10.1016/j.stem.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matheu A, Maraver A, Klatt P, Flores I, Garcia-Cao I, Borras C, Flores JM, Vina J, Blasco MA, Serrano M. Delayed ageing through damage protection by the Arf/p53 pathway. Nature. 2007;448:375–379. doi: 10.1038/nature05949. [DOI] [PubMed] [Google Scholar]
- 61.Feng Z, Hu W, Teresky AK, Hernando E, Cordon-Cardo C, Levine AJ. Declining p53 function in the aging process: a possible mechanism for the increased tumor incidence in older populations. Proc Natl Acad Sci U S A. 2007;104:16633–16638. doi: 10.1073/pnas.0708043104. [DOI] [PMC free article] [PubMed] [Google Scholar]