Abstract
Increasing evidence points to the functional importance of alternative splice variations in cancer pathophysiology. Two splice variants are derived from the CASP9 gene via the inclusion (Casp9a) or exclusion (Casp9b) of a four exon cassette. Here we show that alternative splicing of Casp9 is dysregulated in non-small cell lung cancers (NSCLC) regardless of their pathological classification. Based on these findings we hypothesized that survival pathways activated by oncogenic mutation regulated this mechanism. In contrast to K-RasV12 expression, EGFR overexpression or mutation dramatically lowered the Casp9a/9b splice isoform ratio. Moreover, Casp9b downregulation blocked the ability of EGFR mutations to induce anchorage-independent growth. Furthermore, Casp9b expression blocked inhibition of clonogenic colony formation by erlotinib. Interrogation of oncogenic signaling pathways showed that inhibition of PI3K or Akt dramatically increased the Casp9a/9b ratio in NSCLC cells. Finally, Akt was found to mediate exclusion of the exon 3,4,5,6 cassette of Casp9 via the phosphorylation state of the RNA splicing factor SRp30a via serines 199, 201, 227 and 234. Taken together, our findings demonstrate that oncogenic factors activating the PI3Kinase/Akt pathway can regulate alternative splicing of Casp9 via a coordinated mechanism involving the phosphorylation of SRp30a.
Keywords: SRp30a, alternative splicing, erlotinib, Akt
Introduction
NSCLC comprises over 80% of all lung cancers diagnosed, thus there is an urgent need for further understanding of the molecular mechanisms of NSCLC, thereby defining new molecular targets for the treatment of this disease(1). In NSCLC, inappropriate activation of the PI3 Kinase pathway by direct oncogenic mutation or from a K-Ras or EGFR mutation has been associated with the development of NSCLC(2–4). A critical component of the PI3 Kinase nexus is the proto-oncogene Akt, a serine/threonine kinase. Akt is activated by recruitment to membranes through direct contact of its PH-domain with PtdIns(3,4,5)P3 and subsequent phosphorylation on two key residues, threonine 308 and serine 473. Previously, several laboratories reported that Akt is constitutively active in NSCLC, and experimental models have shown that Akt activation contributes to the resistance of NSCLC to chemotherapy.
An important player in the contrasting signaling cascade of apoptosis, with roles in both the sensitivity of NSCLC to chemotherapy and radiation, is Casp9. Casp9 is a member of the caspase family of proteases and is activated upon the formation of trimeric complex with cytochrome c and APAF-1, termed the apoptosome. Casp9 has roles in both the response of cells to undergo chemotherapy-induced cell death and the suppression of oncogenic transformation(5–15). The CASP9 gene produces two antagonistic isoforms, the pro-apoptotic Casp9a and the pro-survival Casp9b, via the inclusion/exclusion of an exon 3, 4, 5, 6 cassette(7, 12). The Casp9b isoform (exon exclusion) lacks catalytic activity while retaining key interacting domains (e.g. CARD)(7, 12). Casp9b acts as an endogenous inhibitor of Casp9a by competing with the full-length Casp9a for binding to the apoptosome (7, 12). Casp9b has also been surmised to directly interact with Casp9a blocking the auto-proteolysis of the enzyme(7). In this study, Casp9 splicing was shown to be dysregulated in NSCLC tumors and cell lines, and regulated by the PI3K/Akt pathway. Furthermore, this study demonstrates that Akt exerts its effects via the phospho-status the RNA trans-factor SRp30a.
Materials and Methods
Cell culture
A549, H2347, H358, H226, H2170, H596, H460, H1792, H1299, H520, H1703, H2030, H838, HCC827, and H292 cells were obtained from ATCC and grown in 50% RPMI 1640 and 50% DMEM supplemented with L-glutamine, 100 units/ml penicillin G sodium and 100 μg/ml streptomycin sulfate. NHBE cells, from Clonetics (Cambrex Bio Science), were maintained in basal bronchial epithelial growth media (BEGM) supplemented with SingleQuot Kits-growth factors, cytokines and supplements (Cambrex). HBEC-3KTs, HBEC-3KT K-RasV12, wild-type EGFR and mutant EGFR HBEC-3KT cell lines were previously created and characterized(16). HBEC3-KT (E746-A750 del EGFR, K-RasV12) cell lines stably expressing control or Casp9b shRNA were created using lentivirus followed by selection with 20ug/mL hygromycin (Lentigen). To generate an A549, H838, and HCC827 cell line stably expressing Casp9b cDNA, 1.5 × 105 cells were transfected with pcDNA3.1(−)/Casp9b using Effectene followed by selection with zeocin. For all comparison studies between cell lines, cells were plated in keratinocyte serum-free medium without supplements overnight prior to analysis.
Quantitative RT-PCR
Competitive
Total RNA (1 μg) was reverse-transcribed using Superscript III and oligo (dT) and analyzed for Casp9, Bcl-x, caspase 8, and caspase 2 splice variants as previously described(17). Real-time: Total RNA was used for real-time PCR for Casp9a, Casp9b and 18s using TaqMan PCR master mix and the Applied Biosystems 7500 Real-Time PCR System. Casp9a and 18s Q-PCR primers were ID numbers Hs00154261_m1 and Hs99999901_s1. The Q-PCR primers for Casp9b were 5′-TTTGGTGATGTCGGAGCAGA-3′ (forward) and 5′-GATTTGGTGATGTCGGAGCA-3′ (reverse) and 5′-TTCCCCTGAAGACGAGTCCCCTGG-3′ (probe).
Inhibitor studies/adenoviral infection
Cells (1.2 × 105) were infected 2hrs with constitutively active Akt2, PTEN, or Null adenovirus (50MOI). After 48hr, total RNA and protein was isolated.
Clonogenic and soft agar assays
For clonogenic assays, 150 viable cells were seeded into 6-well tissue culture dishes with complete growth media. 2hrs post-plating, cells were treated with erlonitib. After 24hrs, cells were rinsed and placed in complete media for 12 days. Colonies were counted following fixation with methanol and staining (0.1% crystal violet). The soft agar assays, cells (103) were accomplished as previously described(16).
Phosphorylation assays
24hrs after transfection, cells were scraped into 150 μL of NP-40 lysis buffer (50mM Tris-HCL [pH 8.0], 150mM NaCl, 1% NP-40,100ug of phenylmethyl-sulfonyl fluoride/mL, and protease inhibitor cocktail. The lysates were incubated with 500 U of alkaline phosphatase for 1 hr. at 37°C. Wild-type and mutant SRp30a proteins were detected using anti-T7 tag antibody.
Western immunoblotting
Immunoblotting was accomplished as previously described(18) using primary antibodies, anti-Casp9 (Assay Designs), anti-Akt1 (Santa Cruz), anti-Akt2 (Santa Cruz), anti-Akt phospho-S473 (Cell Signaling), anti-Akt pan (Cell Signaling), anti-ERK1/2 (Santa Cruz), anti-ERK1/2 phospho-Thr202/204 (Cell Signaling), anti-PDK1 (Santa Cruz), anti-T7 tag (Novagen), and anti-β-actin (Sigma-Aldrich). Secondary antibodies were HRP-conjugated goat anti-mouse or anti-rabbit (Sigma-Aldrich).
Transfections
The mammalian expression vectors pCGT7-SRp30a-WT and SRp30a-RD are previously described(19). To generate SRp30a phospho-mutants, site-directed mutagenesis was performed using QuikChange XL Site-Directed Mutagenesis protocol. To analyze Casp9 splicing, 1.5 × 104 cells were transfected with 0.5 μg of pCGT7-SRp30a +/− 0.2 μg of the Casp9 minigene using Effectene as previously described(17). After 24hrs, RNA and protein were isolated. For downregulation of Akt1, Akt2, and PDK1, cell lines were transfected with either Akt1, Akt2, or PDK1 SMARTpool multiplex siRNA or control siRNA (Dharmacon) using Dharmafect 1 as previously described(18). After 48hrs, RNA and protein were isolated.
Statistical analysis
When appropriate, the data are presented as the mean ± S.E. Data points were compared using a two-tailed, unpaired Student’s t test, and the P-values calculated. P-values less than 0.05 were considered significant.
Results and Discussion
Casp9 RNA splicing is dysregulated in NSCLC tumors and cell lines
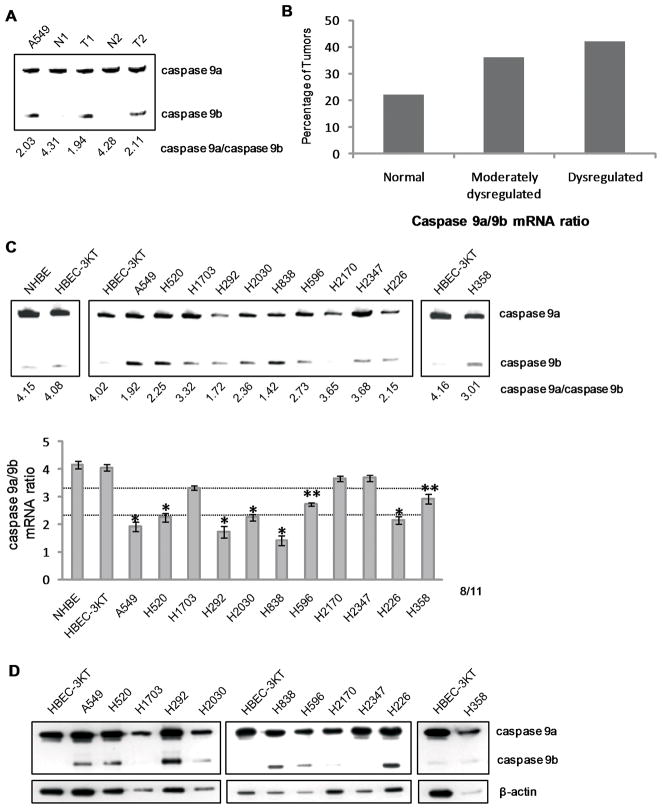
In this study, we examined the hypothesis that Casp9 RNA splicing was dysregulated in all pathologies of NSCLC. Utilizing total RNA from pathologist-verified human NSCLC samples, quantitative/competitive RT-PCR analysis was performed to determine the degree of dysregulation in the Casp9a/9b ratio as compared to matched, normal lung tissue controls (Supplemental Table I)(17). Tumor samples were categorized into three groups respectively: normal, a Casp9a/9b mRNA ratio of >3.3; moderately dysregulated, a Casp9a/9b mRNA ratio of 2.3–3.3; and highly dysregulated, a Casp9a/9b mRNA ratio <2.3 (Figure 1A and B). The normal group corresponds to the normal ratio of Casp9a/9b mRNA observed in non-transformed cells: the moderately dysregulated group corresponds to a ratio of Casp9a/9b reported to have a significant, but minor effect on Casp9 activity(15, 20); and the highly dysregulated group corresponds to a ratio of Casp9a/9b reported to significantly reduce Casp9 activity and inhibit the association of Casp9a with APAF-1(15, 17, 20). Analysis(18, 21–23) of Casp9 splice variants demonstrated that 36% of NSCLCs examined presented a moderately dysregulated Casp9a/9b mRNA ratio (N=149). Importantly, 42% of tumors demonstrated a > 50% decrease in the Casp9a/9b ratio. Thus, the ratio of Casp9a/9b mRNA is significantly lower in a high percentage of NSCLC tumors irrespective of NSCLC subtype.
Figure 1. The Casp9a/9b mRNA ratio is dysregulated in NSCLC tumors and transformed lung epithelial cells.
cDNAs from pathologist-verified lung adeno-, squamous, and large cell carcinomas (Origene) underwent quantitative/competitive PCR for expression of Casp9 splice variants. A) Representation of matched pair analysis used for the degree of Casp9a/9b dysregulation in NSCLC tumors. N= normal, T=tumor. B) RT-PCR analysis of Casp9 splice variants showing 36% of NSCLC tumors are moderately dysregulated (C9a/9b ratio of 2.2–3.3) and 42% of NSCLC tumors are highly dysregulated (C9a/9b ratio <2.2) (N=149). C) RT-PCR analysis of Casp9 splice variants from NSCLC and HBEC-3KT cells. The “*” indicates a dysregulated Casp9a/9b mRNA ratio and “**” indicates a moderately dysregulated Casp9a/9b mRNA ratio. D) In parallel, protein expression of Casp9 splice variants.
We then examined a pure population of non-transformed lung epithelial cells, specifically primary human bronchial epithelial cells (NHBE) and immortalized HBEC-3KT cells, for the ratio of Casp9a/9b in comparison to the transformed lung epithelial cell lines, A549, H838, H2347, H358, H2030, H226, H2170, H596, H1792, H1299, H520, H1703, and H292 cells (Supplemental Table II). HBEC-3KT cells present with a normal Casp9a/9b ratio of 4.02±0.15 as do NHBE cells (4.15±0.23) (Figure 1C). In contrast, 8 of 11 transformed lung epithelial cell lines grown under the exact same culture conditions demonstrated a significant decrease in the Casp9a/9b mRNA ratio. Importantly, the disproportionate ratio of Casp9a/9b mRNA observed in the transformed lung epithelial cell lines translated to a disproportionate ratio of Casp9a/9b protein expression (Figure 1D). We further validated the decrease in the Casp9a/9b mRNA ratio of A549s in comparison to HBEC-3KTs via Q-PCR (Supplemental Figure 1), reconfirming the quantitative nature of the assay as also previously shown by ribonuclease protection assay(21). These data indicate that a significant portion of NSCLC tumors and transformed lung epithelial cells demonstrate severe dysregulation of the alternative splicing of Casp9 to favor a pro-survival/pro-oncogenic phenotype.
The EGF pathway regulates Casp9 RNA splicing in a pro-oncogenic fashion
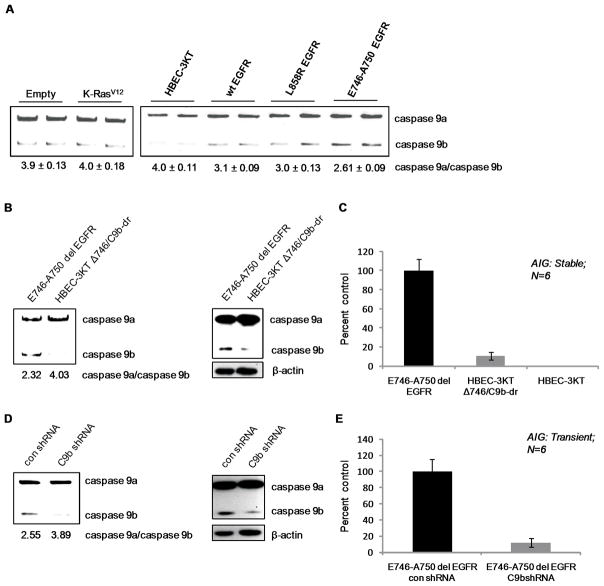
In essentially all epithelial cancers, including NSCLC, one or more members of the family of epidermal growth factor receptor (EGFR) genes are either overexpressed or mutated(4). As a large percentage of NSCLC tumors and cell lines demonstrated a dysregulated ratio of Casp9a/9b, we next examined whether this common oncogene in NSCLC affected Casp9 RNA splicing in HBEC-3KT cells. Whereas low expression of K-RasV12 in HBEC-3KT cells(16) had no discernable effect on the ratio of Casp9a/9b mRNA, the overexpression of wild-type EGFR, the expression of L858R mutation in EGFR, and the expression of the del E746-A750 EGFR mutant induced a significant reduction in the Casp9a/9b ratio (Figure 2A). These HBEC-3KT cell lines expressing either wild-type or mutant EGFR have previously been reported to exhibit enhanced phosphorylation of EGFR in the absence of EGF, in contrast to HBEC-3KTs expressing K-RasV12(16). We also reconfirmed these previous findings showing increased phosphorylation of AKT in HBEC-3KTs expressing wild-type or mutant EGFR, as well as demonstrate an increase in the phosphorylation of ERK1/2 in HBEC-3KTs expressing K-RasV12 (Supplemental Figure 2A,B). Importantly, the del E746-A750 mutation of EGFR induced the most significant decrease in the Casp9a/9b ratio correlating with the ability of this EGFR mutant to induce anchorage-independent growth (AIG)(16). Therefore, we hypothesized that increased expression of Casp9b was a major mechanism in conferring AIG of HBEC-3KT ΔE746-A750 del cells. To test this hypothesis, an E746-A750 del EGFR clonal cell line (HBEC-3KT Δ746/C9b-dr) stably expressing Casp9b shRNA was produced. The expression of Casp9b in the HBEC-3KT Δ746/C9b-dr clonal cell line was reduced to normal immunoreactive levels, and presented with a normal Casp9a/Casp9b ratio (Figure 2B). Importantly, the HBEC-3KT Δ746/C9b-dr cells demonstrated a complete loss of AIG compared to E746-A750 del EGFR cells stably expressing control shRNA (Figure 2C). These effects did not require stable expression and were not due to integration artifacts as short term/transient downregulation of Casp9b also inhibited the AIG of E746-A750 del EGFR cells (Figure 2D,E). Furthermore, K-RasV12 expressing HBEC-3KTs with Casp9b downregulated exhibited no difference in their AIG capacity, demonstrating specificity for oncogenic EGFR (Supplemental Figure 3). Thus, the distal mechanism of Casp9 splicing plays a major and specific role in the ability of EGFR signaling to confer AIG. The significance of this finding stems from the knowledge that EGFR mutation occurs in a large percentage of NSCLCs, roughly 5 to 10%, and overexpressed EGFR along with its ligands occur in approximately 70% of NSCLCs. Interestingly, EGFR mutation/overexpression is also considered an early event in NSCLC since the mutation is also found in normal epithelial cells prior to metaplasia and adenoma formation rationalizing the hypothesis that the alternative splicing of Casp9 plays a role in early events leading to the formation of NSCLC.
Figure 2. EGFR regulates Casp9 RNA splicing.
A) RT-PCR analysis of Casp9 splice variants from vector control-, K-RasV12-expressing HBEC-3KT cells, wild-type EGFR-, L858R EGFR-, and E746-A750 del EGFR-expressing HBEC-3KT cells(16). B, D) Characterization of HBEC3-KT Δ746 cells B) stably and D) transiently expressing Casp9b or control shRNA by RT-PCR and western blot analysis. C, E) Colony formation assay in soft agar of HBEC-3KT Δ746 cells C) stably and E) transiently expressing Casp9b or control shRNA. Data are depicted as mean ± S.E. represented as percent control.
Modulation of Casp9 RNA splicing regulates the ability of erlotinib to inhibit the colony formation of NSCLC cells
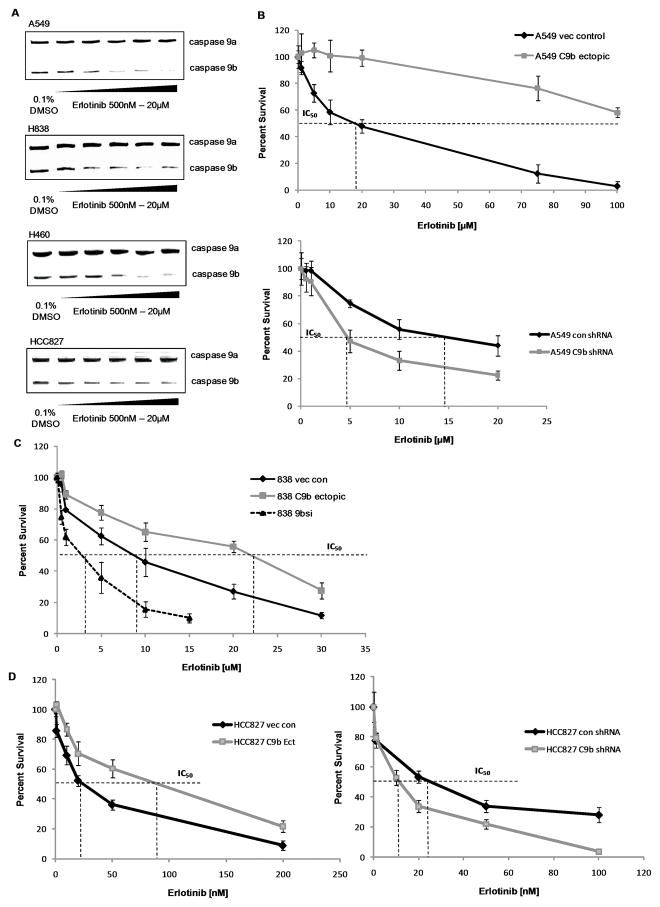
Based on the above data, we were prompted to investigate the effect of erlotinib, a clinically relevant inhibitor of the human EGFR, on the Casp9a/9b ratio. Treatment of A549, H838, H460, and HCC827 cells with erlotinib led to a dose-dependent increase in the Casp9a/9b ratio (Figure 3A). We further validated the increase in the Casp9a/9b ratio via Q-PCR (Supplemental Figure 4A,B). Additionally, the effectiveness of erlotinib treatment was shown by loss of Akt phosphorylation (Supplemental Figure 4C). Next, the effect of lowering the Casp9a/9b ratio on the sensitivity of A549 cells to erlotinib was examined. Low ectopic expression of Casp9b dramatically inhibited the ability of erlotinib to suppress cell survival (e.g. induced a resistant phenotype) (Figure 3B). In contrast, downregulation of Casp9b sensitized A549 cells to erlotinib as shown by a dramatic reduction in the IC50 (14.9 μM for control shRNA to 4.8 μM for C9b shRNA) (Figure 3B). To determine translatability, the effect of lowering the Casp9a/9b ratio on the sensitivity of H838 and HCC827 cells to erlotinib was also examined. H838 cells ectopically expressing Casp9b cDNA significantly inhibited the ability of erlotinib to suppress cell survival increasing the IC50 (8.5 μM for vector control to 22.2 μM for C9b cDNA) (Figure 3C). Conversely, downregulation of Casp9b sensitized H838s to erlotinib reducing the IC50 to 2.9 μM for C9b shRNA cells (Figure 3C). Additionally, these same effects translated to HCC827 cells, a cell line has been reported to be highly sensitive to erlotinib-induced growth inhibition(24). Specifically, HCC827 cells ectopically expressing Casp9b cDNA demonstrated an increase in the IC50 of erlotinib (22.2 nM for vector control to 89.9 nM for C9b cDNA (Figure 3D). Conversely, downregulation of Casp9b reduced the IC50 to 11.7 nM for C9b shRNA cells (Figure 3D). Thus, Casp9 RNA splicing plays a major role in the sensitivity of NSCLC cells to erlotinib. As erlotinib has shown promise in clinical trials significantly improving the survival rate of NSCLC patients (25, 26), examining the alternative splicing of Casp9 may have future predictive/prognostic value for a subset of patients or allow for determination of erlotinib responsiveness in NSCLC tumors.
Figure 3. Casp9 RNA splicing regulates the ability of erlotinib to inhibit colony formation.
A) RT-PCR analysis of Casp9 splice variants from A549, H838, and H460 cells treated with erlotinib. Clonogenic plate assays of B) A549 cells, C) H838 cells and D) HCC827 cells expressing ectopic Casp9b cDNA, vector control, Casp9b shRNA or control shRNA after 24hr in the presence of erlotinib. Data are mean ± S.E. represented as percent control.
The PI3K/PDK1/Akt pathway regulates Casp9 RNA splicing
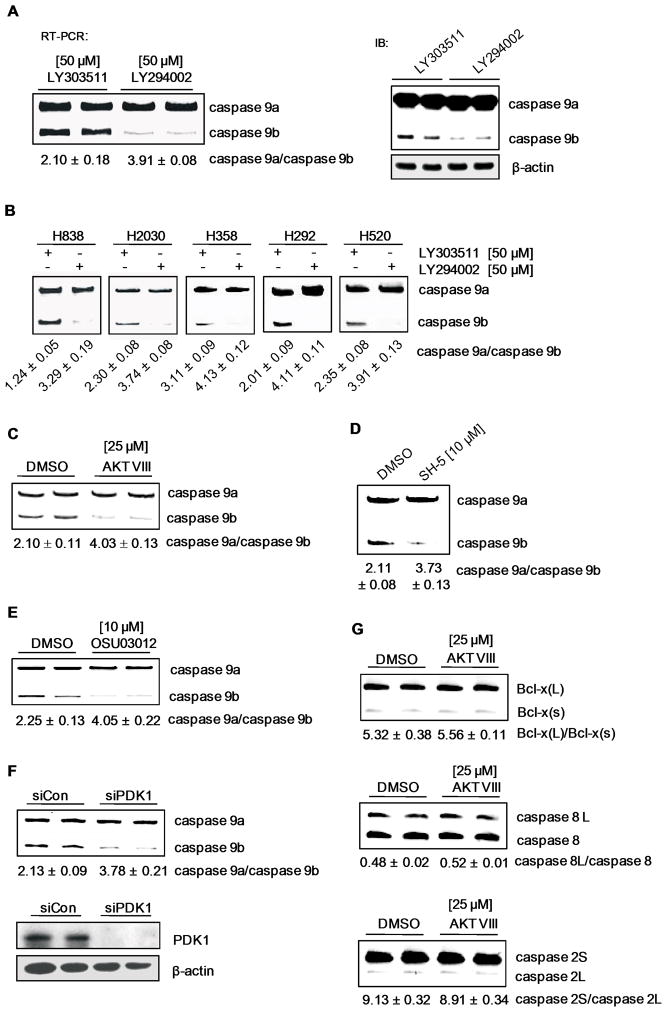
We next hypothesized that a major mitogenic signaling pathway activated by EGFR regulates Casp9 RNA splicing to favor the production of Casp9b. To investigate this hypothesis, mitogenic pathways were examined for effects on the ratio of Casp9a/9b utilizing small-molecule inhibitors at doses/times reported in the scientific literature for A549 cells(27–33). Only treatment with the PI3K inhibitor, LY 294002 [50 μM], resulted in a significant increase in the ratio of Casp9a/9b, compared to the inactive, structurally-related compound, LY303511 (Figure 4A) (Supplementary Table III). Specifically, the Casp9a/9b ratio increased from 2.10 ± 0.18 for control samples to 3.91 ± 0.08 for inhibitor-treated samples (p < 0.0008 (n=6)). The inhibition of PI3K effectively returned the Casp9a/9b ratio to the ratio observed in non-transformed lung epithelial cells. Importantly, this effect on the Casp9a/9b ratio translated to the protein level (Figure 4A).
Figure 4. The PI3K/PDK1/Akt pathway regulates Casp9 RNA splicing.
RT-PCR analysis and western immunoblot of Casp9 splice variants from A) A549s and B) H838s, H2030s, H358s, H292s, and H520s treated with either LY294002 [50 μM] or LY303511 [50 μM]. C) RT-PCR analysis of Casp9 splice variants and the corresponding Casp9a/9b mRNA ratios from A549 cells treated with 0.1% DMSO control, Akt inhibitor VIII [25 μM], D) SH-5 [25 μM], or E) OSU03012 [10 μM]. F) RT-PCR analysis of Casp9 splice variants and western immunoblot analysis of PDK1 from A549s transfected with scrambled siRNA [100 nM] or PDK1 siRNA [100 nM]. G) A549 cells were treated with Akt inhibitor VIII [25 μM] and analyzed by RT-PCR for Bcl-x, caspase 8, and caspase 2 splice variants. Data are expressed as means ± S.E.
To determine translatability, H838, H2030, H358, H292, and H520 cells were also treated with LY294002 (Figure 4B). Again, the Casp9a/9b ratio was dramatically increased (H838 cells from 1.24 ± 0.05 to 3.29 ± 0.19; H2030 cells from 2.30 ± 0.08 to 3.74 ± 0.08; H358 cells from 3.11 ± 0.09 to 4.13 ± 0.12; H292 cells from 2.01 ± 0.09 to 4.11 ± 0.11; and H520 cells from 2.35 ± 0.08 to 3.91 ± 0.13). Therefore, the ability of the PI3K pathway to regulate the alternative splicing of Casp9 translates to multiple NSCLC cell lines.
Akt/PKB, SGK, PKCξ, and PKCδ are downstream of PI3K(34), and to investigate the downstream effector of PI3K responsible for regulating Casp9 RNA splicing, small-molecule inhibitors in conjunction with RNAi technology was employed. Treatment of A549 cells with the PKC inhibitors GÖ6976 [10 μM] and GÖ6983 [10 μM] (Supplemental Figure 5A,B; Supplemental Table III) resulted in no significant change in the Casp9a/b ratio. Conversely, treatment of A549 cells with the Akt inhibitor, Akt VIII [25 μM], and the phosphatidylinositol analog, SH-5 [10 μM], resulted in an increased Casp9a/9b ratio to the same extent as PI3K inhibition (from 2.10 ± 0.11 for DMSO to 4.03 ± 0.13 for Akt VIII (n=6) p<0.01; and from 2.10 ± 0.08 for DMSO to 3.73 ± 0.13 for SH-5 (n=4) p<0.01) (Figure 4C,D). In addition, treatment of cells with the PDK1 inhibitor, OSU03012 [10 μM], or siRNA downregulation of PDK1 (Figure 4E and F) resulted in a significantly increased Casp9a/9b ratio (from 2.25 ± 0.13 for control to 4.05 ± 0.22 for OSU03012 (n=4), p<0.01; and from 2.13 ± 0.09 for siRNA controls to 3.78 ± 0.21 for siPDK1 (n=4) p<0.01). To verify appropriate inhibition of downstream signaling pathways, the phosphorylation status of Akt was examined via western immunoblotting (Supplemental Figure 6A–C).
These data demonstrate that Akt regulates Casp9 RNA splicing in an anti-apoptotic/pro-survival manner. This effect was specific for Casp9 RNA splicing, and not a generalized effect on constitutive RNA splicing, as no effect on the alternative splicing of caspase 8, caspase 2, and Bcl-x pre-mRNA was observed with Akt VIII treatment (Figure 4G). Furthermore, co-treatment of A549 cells with both Akt VIII and LY294002 could not further increase the Casp9a/9b ratio demonstrating a linear pathway with Akt as the downstream effector of PI3K (Supplemental Figure 7).
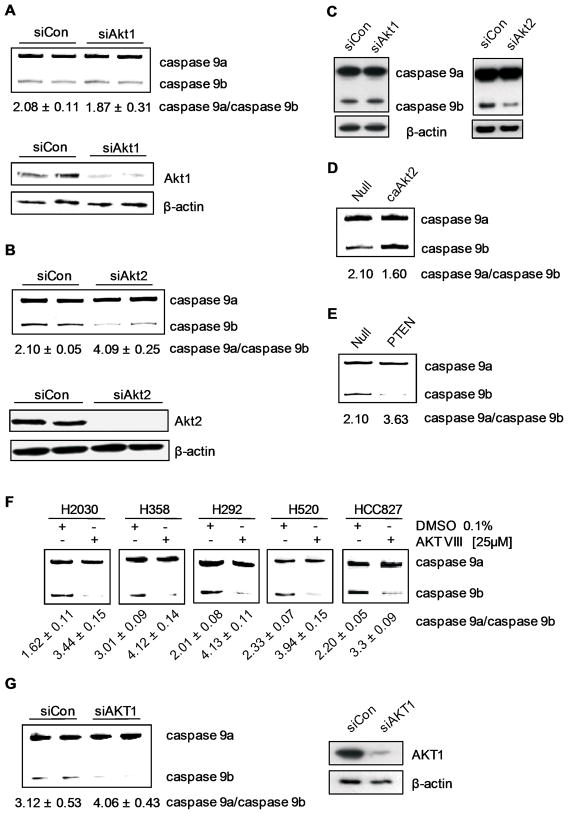
To examine the Akt isoform responsible for regulating the alternative splicing, siRNA was again utilized. Downregulation of Akt1 using multiplex siRNA resulted in no significant change in the Casp9a/b ratio (p > 0.34) (Figure 5A). In contrast, downregulation of Akt2 resulted in a dramatic increase in the Casp9a/b ratio from 2.10 ± 0.05 to 4.09 ± 0.25, p < 0.01 (n=5) (Figure 5B). Again, this effect translated to the protein level (Figure 5C).
Figure 5. Akt regulates Casp9 RNA splicing via PIP3 membrane interactions.
A) RT-PCR analysis of Casp9 splice variants from A549s transfected with control siRNA, Akt1 siRNA, or B) Akt2 siRNA. C) Western blot analysis for Casp9. D) RT-PCR analysis of Casp9 splice variants from A549s infected with null, constitutively active Akt2, or E) PTEN adenovirus. F) H2030s, H358s, H292s, H520s, and HCC827s were treated with 0.1% DMSO or Akt inhibitor VIII [25 μM] for 24 hrs. Total RNA was isolated and RT-PCR performed for Casp9 splice variants. G) RT-PCR analysis of Casp9 splice variants from H358s transfected with control siRNA or Akt1 siRNA. Data are expressed as means ± S.E.
To further validate a role for Akt2, expression of constitutively-active Akt2 (always membrane bound via myristoylation) induced a significant decrease in the Casp9a/9b ratio (Figure 5D). Furthermore, ectopic expression of PTEN induced the contrasting effect (Figure 5E). Therefore, Akt2 regulates Casp9 RNA splicing via PIP3 membrane interactions in A549 cells, and also requires PDK1 phosphorylation.
To determine translatability, H2030, H292, H358, H520, and HCC827 cells were treated with Akt VIII. Indeed, all cell lines also demonstrated a dramatic increase in the Casp9a/9b ratio (Figure 5F). Additionally, Akt1 and Akt2 were downregulated in H358 cells to determine the Akt isoform responsible for regulating Casp9 RNA splicing. In contrast to A549s, downregulation of Akt1, but not Akt2 induced a significant increase in the Casp9a/9b ratio (Figure 5G). Therefore, multiple Akt isoforms have the potential to regulate Casp9 RNA splicing in NSCLC cells.
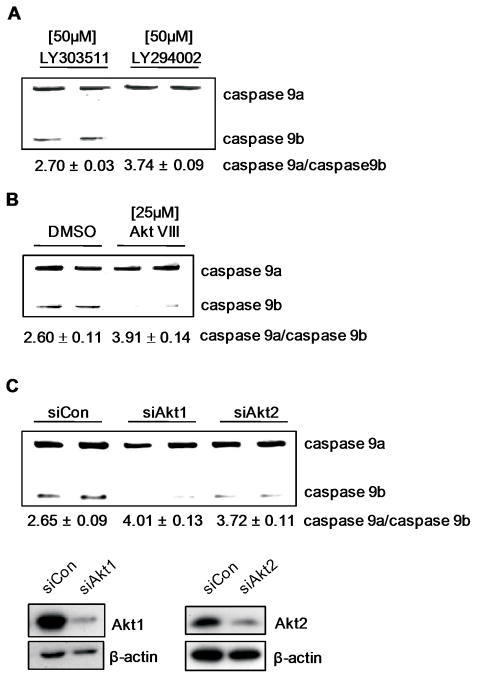
Lastly, as with the NSCLC cell lines, the ability of del 746–750 EGFR mutation to induce a lowering of the Casp9a/9b ratio in HBEC-3KT cells was dependent on PI3K and Akt as treatment with LY294002 and Akt VIII inhibitor increased the ratio of Casp9a/9b (Figure 6A,B). These data suggest that EGFR overexpression/mutation correlates with Akt activation affecting the alternative splicing of Casp9. We further extended these results by examining the Akt isoform responsible for regulating Casp9 RNA splicing in HBEC-3KT (del E746-A750 EGFR) cells. Interestingly, siRNA against both Akt1 and Akt2 (Figure 6C) resulted in a significantly increase Casp9a/Casp9b ratio as compared to control siRNA (from 2.65 ± 0.05 for siRNA control to 4.01 ± 0.13 for siAkt1 (n=3), p<0.01; and to 3.72 ± 0.11 for siAkt2 (n=3) p<0.01). Therefore, multiple Akt isoforms are responsible for regulating the alternative splicing of Casp9. The finding that the Akt pathway regulates this splicing mechanism important for EGFR conferring AIG makes logical sense in relation to cellular transformation, as this pathway is found constitutively active in approximately 58% of NSCLC cell lines and tumors(35, 36). This pathway is also linked to constitutive EGFR activity, NF-κB activation, and the ability of oncogenic Ras to transform various cell types(37–39). Thus, the Akt pathway activated by the EGFR receptor is extremely important in a therapeutic sense, and this study suggests that the alternative splicing of Casp9 is a key distal mechanism in the biological role of this pathway in NSCLC development/maintenance.
Figure 6. Akt regulates Casp9 RNA splicing in E746-A750 del EGFR-expressing HBEC-3KT cells.
A) RT-PCR analysis of Casp9 splice variants from E746-A750 EGFR-expressing HBEC3-KTs treated with LY294002 [50M], or LY303511 [50M], and B) Akt inhibitor VIII [25M] or 0.1% DMSO. C) RT-PCR analysis of Casp9 splice variants from E746-A750 EGFR-expressing HBEC3-KTs transfected with control siRNA, Akt1 siRNA, or Akt2 siRNA. Data are expressed as means ± S.E.
How Casp9b is acting to drive AIG is more of an enigma. As we demonstrated, removal of Casp9b blocked the ability of EGFR (del E746-A750) to induce AIG. This is likely not attributed to the blockade of an initiator caspase such as Casp9a, and suggests a role in cell signaling for Casp9b. In this regard, the possibility that Casp9b acts as a signaling molecule has been reported by Latchman and co-workers(5). Specifically, this laboratory group showed that ectopic expression of Casp9b induced the activation of NF-κB irrespective of caspase activation(5). Activation of NF-κB by Casp9b expression also “fits” well with cooperation with K-Ras mutations for the induction of cellular transformation based on the findings of Ma and Baldwin(40, 41). These laboratory groups showed in several different ways that NF-κB activation enhances the ability of oncogenic ras to induce cellular transformation. Coupled with the knowledge that EGFR overexpression/mutation leads to both cooperation with oncogenic ras in cellular transformation and NF-κB activation, a role for Casp9b in these pathways important for cellular transformation is logical. Thus, Casp9b may act as a scaffolding protein to elicit downstream signaling events with roles outside the simple inactivation of Casp9a. This possibility is far from inconceivable as the initiator caspase, caspase 8, is reported to recruit cell survival factors such as PI3 Kinase subunits(42–44). Although this caspase is an initiator of extrinsic pathways of apoptosis, Casp9b may be playing an analogous role in survival signaling as an initiator of the intrinsic pathway of apoptosis.
The phospho-status of SRp30a modulates the effect of Akt signaling on Casp9 RNA splicing
Our laboratory reported that SRp30a was a required enhancer factor for the inclusion of the exon 3,4,5,6 cassette of Casp9(18). Furthermore, SRp30a has been demonstrated to be a specific target of Akt in vitro(45, 46). Therefore, we hypothesize that the phospho-status of SRp30a regulates the inclusion of the exon 3,4,5,6 cassette of Casp9, downstream of Akt activation. To investigate this hypothesis, we employed an established phospho-mimic of SRp30a, SRp30a-RD, in which the majority of serine residues in the RS domain were mutated to aspartic acid(19). Co-expression of SRp30a-RD with a functional Casp9 minigene induced a significant decrease in the Casp9a/9b ratio compared to wild-type SRp30a and empty vector controls (Figure 7A). Importantly, expression of SRp30a-RD also induced a significant decrease in the endogenous Casp9a/9b ratio as compared to wild-type SRp30a and empty vector controls (Figure 7B).
Figure 7. The phospho-status of SRp30a regulates Casp9 RNA splicing.
A,B,C) RT-PCR analysis of A, C) Casp9 minigene-derived transcripts or B, D) endogenous Casp9 splice variants from A549s transfected with the A) Casp9 minigene and SRp30a-WT or SRp30a-RD, B) SRp30a-WT or SRp30a-RD, C,D) the indicated SRp30a phospho-mutants. E,F,G,H) Western blot analysis of the phosphorylation state of SRp30a. Lysates were analyzed for the expression/migration of T7-tagged SRp30a. E) A549s were transfected with the indicated T7-tagged SRp30a constructs and total cell lysates were incubated in the presence of either active or denatured alkaline phosphatase (AP). F) HBEC-3KT and A549 cells transfected with the indicated T7-tagged SRp30a constructs. G) Endogenous SRp30a from A549 cells treated with 0.1% DMSO or erlotinib [50μM] for 48hr. Total protein lysates were incubated in the presence of either active or denatured alkaline phosphatase (AP). H) Endogenous SRp30a from A549 cells treated with 0.1% DMSO or Akt VIII [50μM]. I) RT-PCR analysis of endogenous Casp9 splice variants from A549s transfected with SRp30a-WT or SRp30a-QD ± Akt inhibitor VIII [25μM]. Data are expressed as means ± S.E.
To determine the serine residue/residues of SRp30a required for regulating the alternative splicing of Casp9, site-directed replacement mutagenesis was utilized. Multiple serine residues in the RS domain of SRp30a contain recognized motifs for Akt phosphorylation and numerous residues have been validated by mass spectrometric analysis (Supplemental Table IV)(45). These serine residues were individually mutated into aspartic acid to produce phospho-mimics (Supplemental Table IV). Co-expression of only the SRp30a-S199D, SRp30a-S201D, SRp30a-S227D, and SRp30a-S234D mutants with a functional Casp9 minigene decreased the Casp9a/9b ratio compared to wild type SRp30a control (Figure 7C).
We extended these results to produce a SRp30a double phospho-mutant (SRp30a-S199,201D), a SRp30a triple phospho-mutant (SRp30a-S199,201,234D), and a SRp30a quadruple (SRp30a-QD) mutant harboring serine to aspartic acid mutations at residues 199, 201, 227 and 234. Expression of SRp30a double and triple phospho-mutants with the Casp9 minigene further decreased the Casp9a/9b ratio (Supplemental Figure 8). Ectopic expression of SRp30a-QD induced a reduction in the Casp9a/9b ratio comparable to the SRp30a-RD mutant. Conversely, a quadruple dephospho-mimic of SRp30a (SRp30a-QA) induced the opposite effect in A549 cells (Figure 7D). These effects could not be due to localization issues as both SRp30a quadruple mutants are localized in the nucleus and were expressed in equivalent amounts (Supplemental Figure 9).
To demonstrate that these phospho-sites are hyper-phosphorylated in NSCLC, the phosphorylation status of the transiently expressed SRp30a was analyzed by comparing the electrophoretic migration profiles in the presence of either alkaline phosphatase (AP) or denatured AP. A dramatic increase in the migration of SRp30a-WT was observed after treatment with AP, indicating that this protein is phosphorylated in A549 cells. In contrast, the migration of SRp30a-QD and SRp30a-QA after treatment with alkaline phosphatase was significantly decreased in comparison to the migration of SRp30a-WT, indicating that these proteins are phosphorylated to a significantly lesser extent (Figure 7E). These data show that serine199, 201, 227, and 234 exist in a phosphorylated state in A549 cells. Moreover, the electrophoretic migration profiles of A549s and HBEC3-KT cells after transfection with SRp30a-WT and SRp30a-QA indicate that SRp30a exists in a decreased phosphorylated state in non-transformed cells versus NSCLC cells (Figure 7F).
We extended these results to determine the effect of inhibiting the EGFR/Akt pathway on the phosphorylation status of SRp30a in A549 cells. As predicted, an increase in the migration of endogenous SRp30a was observed after treatment with erlotinib (Figure 7G), as well as after treatment with the Akt inhibitor, Akt VIII (Figure 7H). To determine whether the PI3K/Akt pathway regulates Casp9 RNA splicing in a phospho-SRp30a-dependent manner, SRp30a-QD was expressed in the presence or absence of Akt VIII inhibitor. In the presence of SRp30a-QD, Akt VIII inhibitor was unable to increase the ratio of the Casp9a/9b to the same extent as compared to wild-type SRp30a (Figure 7I). Therefore, the Akt pathway regulates the alternative splicing of Casp9 at least partially via the phospho-state of SRp30a on serine199, 201, 227, and 234.
These data solidify a role for phosphorylation of SRp30a in regulating the alternative splicing of Casp9, but also suggest additional regulating mechanisms. In this regard, our laboratory recently has found that the RNA trans-factor, hnRNP L, acts as a repressor for the inclusion of the exon 3,4,5,6 cassette of Casp9, and its repressor activity is regulated by the phosphorylation status of serine52 (17). Thus, we hypothesize that the EGFR/PI3K/Akt pathway may also regulate the phospho-status of hnRNP L at serine52, suggesting a coordinated interplay between these two trans-factors in regulating the alternative splicing of Casp9. This possibility is logical as Lynch and coworkers showed the ability of SRp30a and hnRNP L to directly compete for binding to the exon 5 regulatory sequence of CD45; and that this interplay between SRp30a and hnRNPL influences the extent of exon inclusion(47).
The phospho-state of SRp30a regulating the inclusion of the exon 3,4,5,6 cassette also “fits” with our previous findings that ceramide induced both the dephosphorylation of SRp30a and the inclusion of the Casp9 exon cassette. SRp30a was also required for ceramide effects on the inclusion of the exonic cassette of Casp9. Thus, the regulation of SRp30a phosphorylation and the alternative splicing of Casp9 may be a key distal point by which ceramide acts as a tumor suppressing/cell senescence agent as the ceramide signaling and PI3 kinase/Akt pathway are well established to antagonize each other (Supplemental Figure 10).
In conclusion, the presented study reports several major findings taking a comprehensive approach. First, the dysregulation of the alternative splicing of Casp9 toward a pro-survival phenotype was demonstrated in NSCLC. Second, a survival/mitogenic/oncogenic pathway involving EGFR, PI3K and Akt was shown to regulate this splicing mechanism. Lastly, the phospho-state of SRp30a was shown to regulate this distal mechanism via Akt signaling. Therefore, the presented study demonstrates a novel and key distal mechanism in NSCLC and provides new target mechanisms for the development of therapeutics to combat the cancer with the highest mortality rate.
Supplementary Material
Acknowledgments
This work was supported by grants from the Veteran’s Administration (VA Merit Review I and a Research Career Scientist Award to C.E.C.), from the National Institutes of Health (HL072925 (C.E.C), CA117950 (C.E.C), NH1C06-RR17393 (VCU), NCI Lung Cancer SPORE P50CA70907 (J.D.M.), NRSA-T32 Fellowship (DSW), and from NASA, NNJO5HD36G (J.D.M).
The Abbreviations
- CARD
caspase recruitment domains
- HBEC
human bronchial epithelial cells
- Casp9
Caspase 9
References
- 1.Walker S. Updates in non-small cell lung cancer. Clinical journal of oncology nursing. 2008;12(4):587–96. doi: 10.1188/08.CJON.587-596. [DOI] [PubMed] [Google Scholar]
- 2.Hirsch FR, Varella-Garcia M, Bunn PA, Jr, et al. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol. 2003;21(20):3798–807. doi: 10.1200/JCO.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 3.Li AR, Chitale D, Riely GJ, et al. EGFR mutations in lung adenocarcinomas: clinical testing experience and relationship to EGFR gene copy number and immunohistochemical expression. J Mol Diagn. 2008;10(3):242–8. doi: 10.2353/jmoldx.2008.070178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rowinsky EK. The erbB family: targets for therapeutic development against cancer and therapeutic strategies using monoclonal antibodies and tyrosine kinase inhibitors. Annual review of medicine. 2004;55:433–57. doi: 10.1146/annurev.med.55.091902.104433. [DOI] [PubMed] [Google Scholar]
- 5.Stephanou A, Scarabelli TM, Knight RA, Latchman DS. Antiapoptotic activity of the free caspase recruitment domain of procaspase-9: a novel endogenous rescue pathway in cell death. J Biol Chem. 2002;277(16):13693–9. doi: 10.1074/jbc.M108530200. [DOI] [PubMed] [Google Scholar]
- 6.Hajra KM, Liu JR. Apoptosome dysfunction in human cancer. Apoptosis. 2004;9(6):691–704. doi: 10.1023/B:APPT.0000045786.98031.1d. [DOI] [PubMed] [Google Scholar]
- 7.Seol DW, Billiar TR. A caspase-9 variant missing the catalytic site is an endogenous inhibitor of apoptosis. The Journal of biological chemistry. 1999;274(4):2072–6. doi: 10.1074/jbc.274.4.2072. [DOI] [PubMed] [Google Scholar]
- 8.Wu GS, Ding Z. Caspase 9 is required for p53-dependent apoptosis and chemosensitivity in a human ovarian cancer cell line. Oncogene. 2002;21(1):1–8. doi: 10.1038/sj.onc.1205020. [DOI] [PubMed] [Google Scholar]
- 9.Philchenkov A, Zavelevich M, Kroczak TJ, Los M. Caspases and cancer: mechanisms of inactivation and new treatment modalities. Exp Oncol. 2004;26(2):82–97. [PubMed] [Google Scholar]
- 10.Giaccia AJ, Kastan MB. The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev. 1998;12(19):2973–83. doi: 10.1101/gad.12.19.2973. [DOI] [PubMed] [Google Scholar]
- 11.Liang Q, Liou AK, Ding Y, et al. 6-Hydroxydopamine induces dopaminergic cell degeneration via a caspase-9-mediated apoptotic pathway that is attenuated by caspase-9dn expression. J Neurosci Res. 2004;77(5):747–61. doi: 10.1002/jnr.20198. [DOI] [PubMed] [Google Scholar]
- 12.Srinivasula SM, Ahmad M, Guo Y, et al. Identification of an endogenous dominant-negative short isoform of caspase-9 that can regulate apoptosis. Cancer research. 1999;59(5):999–1002. [PubMed] [Google Scholar]
- 13.Yacoub A, Mitchell C, Hong Y, et al. MDA-7 regulates cell growth and radiosensitivity in vitro of primary (non-established) human glioma cells. Cancer Biol Ther. 2004;3(8):739–51. doi: 10.4161/cbt.3.8.968. [DOI] [PubMed] [Google Scholar]
- 14.Kominsky DJ, Bickel RJ, Tyler KL. Reovirus-induced apoptosis requires mitochondrial release of Smac/DIABLO and involves reduction of cellular inhibitor of apoptosis protein levels. J Virol. 2002;76(22):11414–24. doi: 10.1128/JVI.76.22.11414-11424.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ebert AD, Chen F, He X, Cryns VL, Bohn MC. A tetracycline-regulated adenovirus encoding dominant-negative caspase-9 is regulated in rat brain and protects against neurotoxin-induced cell death in vitro, but not in vivo. Exp Neurol. 2005;191 (Suppl 1):S80–94. doi: 10.1016/j.expneurol.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 16.Sato M, Vaughan MB, Girard L, et al. Multiple oncogenic changes (K-RAS(V12), p53 knockdown, mutant EGFRs, p16 bypass, telomerase) are not sufficient to confer a full malignant phenotype on human bronchial epithelial cells. Cancer research. 2006;66(4):2116–28. doi: 10.1158/0008-5472.CAN-05-2521. [DOI] [PubMed] [Google Scholar]
- 17.Goehe R, Shultz JC, Murudkar C, et al. hnRNP L regulates the tumorigenic capacity of lung cancer xenografts in mice via caspase 9 pre-mRNA processing. Journal of Clinical Investigation. 2010 doi: 10.1172/JCI43552. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Massiello A, Chalfant CE. SRp30a (ASF/SF2) regulates the alternative splicing of caspase-9 pre-mRNA and is required for ceramide-responsiveness. Journal of lipid research. 2006;47(5):892–7. doi: 10.1194/jlr.C600003-JLR200. [DOI] [PubMed] [Google Scholar]
- 19.Cazalla D, Zhu J, Manche L, Huber E, Krainer AR, Caceres JF. Nuclear export and retention signals in the RS domain of SR proteins. Molecular and cellular biology. 2002;22(19):6871–82. doi: 10.1128/MCB.22.19.6871-6882.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hakem R, Hakem A, Duncan GS, et al. Differential requirement for caspase 9 in apoptotic pathways in vivo. Cell. 1998;94(3):339–52. doi: 10.1016/s0092-8674(00)81477-4. [DOI] [PubMed] [Google Scholar]
- 21.Chalfant CE, Rathman K, Pinkerman RL, et al. De novo ceramide regulates the alternative splicing of caspase 9 and Bcl-x in A549 lung adenocarcinoma cells. Dependence on protein phosphatase-1. The Journal of biological chemistry. 2002;277(15):12587–95. doi: 10.1074/jbc.M112010200. [DOI] [PubMed] [Google Scholar]
- 22.Massiello A, Roesser JR, Chalfant CE. SAP155 Binds to ceramide-responsive RNA cis-element 1 and regulates the alternative 5′ splice site selection of Bcl-x pre-mRNA. Faseb J. 2006;20(10):1680–2. doi: 10.1096/fj.05-5021fje. [DOI] [PubMed] [Google Scholar]
- 23.Massiello A, Salas A, Pinkerman RL, Roddy P, Roesser JR, Chalfant CE. Identification of two RNA cis-elements that function to regulate the 5′ splice site selection of Bcl-x pre-mRNA in response to ceramide. The Journal of biological chemistry. 2004;279(16):15799–804. doi: 10.1074/jbc.M313950200. [DOI] [PubMed] [Google Scholar]
- 24.Amann J, Kalyankrishna S, Massion PP, et al. Aberrant epidermal growth factor receptor signaling and enhanced sensitivity to EGFR inhibitors in lung cancer. Cancer research. 2005;65(1):226–35. [PubMed] [Google Scholar]
- 25.Perez-Soler R, Chachoua A, Hammond LA, et al. Determinants of tumor response and survival with erlotinib in patients with non--small-cell lung cancer. J Clin Oncol. 2004;22(16):3238–47. doi: 10.1200/JCO.2004.11.057. [DOI] [PubMed] [Google Scholar]
- 26.Cappuzzo F, Ciuleanu T, Stelmakh L, et al. SATURN: A double-blind, randomized, phase III study of maintenance erlotinib versus placebo following nonprogression with first-line platinum-based chemotherapy in patients with advanced NSCLC. J Clin Oncol. 2009;27:15s. [Google Scholar]
- 27.Barnett SF, Defeo-Jones D, Fu S, et al. Identification and characterization of pleckstrin-homology-domain-dependent and isoenzyme-specific Akt inhibitors. The Biochemical journal. 2005;385(Pt 2):399–408. doi: 10.1042/BJ20041140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen YL, Lin PC, Chen SP, et al. Activation of nonsteroidal anti-inflammatory drug-activated gene-1 via extracellular signal-regulated kinase 1/2 mitogen-activated protein kinase revealed a isochaihulactone-triggered apoptotic pathway in human lung cancer A549 cells. The Journal of pharmacology and experimental therapeutics. 2007;323(2):746–56. doi: 10.1124/jpet.107.126193. [DOI] [PubMed] [Google Scholar]
- 29.Clarke CJ, Guthrie JM, Hannun YA. Regulation of neutral sphingomyelinase-2 (nSMase2) by tumor necrosis factor-alpha involves protein kinase C-delta in lung epithelial cells. Molecular pharmacology. 2008;74(4):1022–32. doi: 10.1124/mol.108.046250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyamoto-Yamasaki Y, Yamasaki M, Tachibana H, Yamada K. Induction of endoreduplication by a JNK inhibitor SP600125 in human lung carcinoma A 549 cells. Cell biology international. 2007;31(12):1501–6. doi: 10.1016/j.cellbi.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 31.Monick MM, Cameron K, Powers LS, et al. Sphingosine kinase mediates activation of extracellular signal-related kinase and Akt by respiratory syncytial virus. American journal of respiratory cell and molecular biology. 2004;30(6):844–52. doi: 10.1165/rcmb.2003-0424OC. [DOI] [PubMed] [Google Scholar]
- 32.Sandquist JC, Means AR. The C-terminal tail region of nonmuscle myosin II directs isoform-specific distribution in migrating cells. Molecular biology of the cell. 2008;19(12):5156–67. doi: 10.1091/mbc.E08-05-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas KW, Monick MM, Staber JM, Yarovinsky T, Carter AB, Hunninghake GW. Respiratory syncytial virus inhibits apoptosis and induces NF-kappa B activity through a phosphatidylinositol 3-kinase-dependent pathway. The Journal of biological chemistry. 2002;277(1):492–501. doi: 10.1074/jbc.M108107200. [DOI] [PubMed] [Google Scholar]
- 34.Cooper GM. The Cell: A Molecular Approach. 2. Sunderland, Missachusetts: Sinauer Associates, Inc; 2000. [Google Scholar]
- 35.Balsara BR, Pei J, Mitsuuchi Y, et al. Frequent activation of AKT in non-small cell lung carcinomas and preneoplastic bronchial lesions. Carcinogenesis. 2004;25(11):2053–9. doi: 10.1093/carcin/bgh226. [DOI] [PubMed] [Google Scholar]
- 36.Brognard J, Clark AS, Ni Y, Dennis PA. Akt/protein kinase B is constitutively active in non-small cell lung cancer cells and promotes cellular survival and resistance to chemotherapy and radiation. Cancer research. 2001;61(10):3986–97. [PubMed] [Google Scholar]
- 37.Moscatello DK, Holgado-Madruga M, Emlet DR, Montgomery RB, Wong AJ. Constitutive activation of phosphatidylinositol 3-kinase by a naturally occurring mutant epidermal growth factor receptor. The Journal of biological chemistry. 1998;273(1):200–6. doi: 10.1074/jbc.273.1.200. [DOI] [PubMed] [Google Scholar]
- 38.Grandage VL, Gale RE, Linch DC, Khwaja A. PI3-kinase/Akt is constitutively active in primary acute myeloid leukaemia cells and regulates survival and chemoresistance via NF-kappaB, Mapkinase and p53 pathways. Leukemia. 2005;19(4):586–94. doi: 10.1038/sj.leu.2403653. [DOI] [PubMed] [Google Scholar]
- 39.Rodriguez-Viciana P, Warne PH, Khwaja A, et al. Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell. 1997;89(3):457–67. doi: 10.1016/s0092-8674(00)80226-3. [DOI] [PubMed] [Google Scholar]
- 40.Wang P, Shi T, Ma D. Cloning of a novel human caspase-9 splice variant containing only the CARD domain. Life sciences. 2006;79(10):934–40. doi: 10.1016/j.lfs.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 41.Mayo MW, Wang CY, Cogswell PC, et al. Requirement of NF-kappaB activation to suppress p53-independent apoptosis induced by oncogenic Ras. Science (New York, NY) 1997;278(5344):1812–5. doi: 10.1126/science.278.5344.1812. [DOI] [PubMed] [Google Scholar]
- 42.Frisch SM. Caspase-8: fly or die. Cancer research. 2008;68(12):4491–3. doi: 10.1158/0008-5472.CAN-08-0952. [DOI] [PubMed] [Google Scholar]
- 43.Krumschnabel G, Manzl C, Villunger A. Caspase-2: killer, savior and safeguard--emerging versatile roles for an ill-defined caspase. Oncogene. 2009;28(35):3093–6. doi: 10.1038/onc.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krumschnabel G, Sohm B, Bock F, Manzl C, Villunger A. The enigma of caspase-2: the laymen’s view. Cell death and differentiation. 2009;16(2):195–207. doi: 10.1038/cdd.2008.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blaustein M, Pelisch F, Tanos T, et al. Concerted regulation of nuclear and cytoplasmic activities of SR proteins by AKT. Nature structural & molecular biology. 2005;12(12):1037–44. doi: 10.1038/nsmb1020. [DOI] [PubMed] [Google Scholar]
- 46.Patel NA, Kaneko S, Apostolatos HS, et al. Molecular and genetic studies imply Akt-mediated signaling promotes protein kinase CbetaII alternative splicing via phosphorylation of serine/arginine-rich splicing factor SRp40. The Journal of biological chemistry. 2005;280(14):14302–9. doi: 10.1074/jbc.M411485200. [DOI] [PubMed] [Google Scholar]
- 47.Motta-Mena L, Heyd F, Lynch K. Context-dependent regulatory mechanism of the splicing factor hnRNPL. Molecular cell. doi: 10.1016/j.molcel.2009.12.027. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.