Abstract
Dimerizers allowing inducible control of protein-protein interactions are powerful tools for manipulating biological processes. Here we describe genetically encoded light-inducible protein interaction modules based on Arabidopsis thaliana cryptochrome 2 and CIB1 that require no exogenous ligands and dimerize on blue light exposure with sub-second time resolution and subcellular spatial resolution. We demonstrate the general utility of this system by inducing protein translocation, transcription, and Cre-mediated DNA recombination using light.
Chemical inducers of protein dimerization1 have been extensively used to modulate protein-protein interactions in live cells, allowing inducible control of signal transduction pathways, protein translocation, transcriptional activation, and other diverse processes2. While chemical-genetic tools have opened up new avenues in experimental biology, small molecules lack tissue specificity, have temporal resolution that is limited by the time it takes for cell permeation, and can be difficult or expensive to deliver to organisms for in vivo studies.
Recently, several groups have developed genetically-encoded dimerizers based on plant photoreceptors that allow control of protein-protein interactions by light3–7. Such optical control has allowed subcellular spatial resolution and fast temporal control of protein interactions in live cells4, 5. One such system4 is based on Arabidopsis thaliana phytochrome B and PIF6, which associate upon illumination with red light and dissociate with far-red light8. When these dimerization modules are fused to protein targets, the activities of these target proteins can be controlled with light4. While this system offers rapid (sub-second) stimulation and reversibility, the interaction requires a bilin cofactor found only in plants and some light-sensing bacteria, fungi, and slime molds, making use of this system in other organisms challenging since the cofactor must be added exogenously. Another system, using the Arabidopsis LOV domain protein FKF1 and its interacting partner GIGANTEA, requires no exogenous chromophore, but the kinetics of light-triggered dimerization are slow, requiring tens of minutes5. Additionally, FKF1 and GIGANTEA remain associated for hours following light exposure, rendering the interaction effectively irreversible once induced. At present, no single optical system for induction of protein interactions has been developed that achieves fast temporal and subcellular spatial resolution without the need for exogenous cofactors.
Here we describe new dimerization modules for inducing protein interactions based on Arabidopsis CIB1, a basic helix-loop-helix (bHLH) protein, and cryptochrome 2 (CRY2), a blue light-absorbing photosensor that binds CIB1 in its photoexcited state9. These modules require no exogenous chromophore, are reversible within minutes, trigger protein translocation on a sub-second time scale, and can be activated by two-photon microscopy, allowing potential use in vivo in whole organisms.
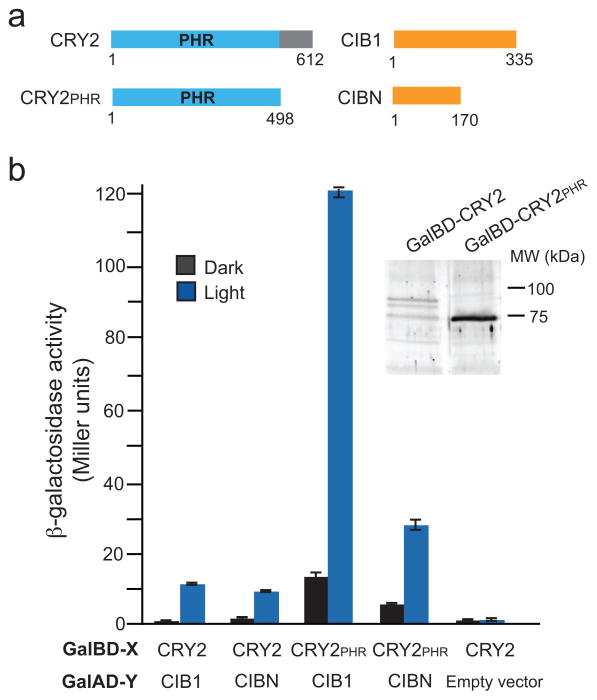
We identified minimal interaction domains for the light-induced CRY2-CIB1 interaction using the yeast two-hybrid assay (Fig. 1). Plant cryptochromes contain a conserved N-terminal photolyase homology region (PHR) that binds flavin and pterin chromophores and mediates light-responsiveness. We tested full length CRY2 and the PHR domain (CRY2PHR, aa 1-498) for interaction with full length CIB1 or a truncated version (CIBN, aa 1–170) missing the conserved bHLH domain which mediates dimerization and DNA binding9 (Fig. 1a). Reporter levels of CRY2-CIB1 and CRY2-CIBN were indistinguishable from controls in the dark, but showed clear activation upon blue light stimulation (Fig. 1b). CRY2PHR also interacted in a light-dependent manner with CIB1 and CIBN, indicating that this domain alone is sufficient to confer light-dependent specificity on the interaction. While CRY2 expression levels were very low, contributing to low levels of reporter activation, CRY2PHR expressed much better, resulting in higher levels of reporter activation in light-treated samples, but also higher basal activity with CIB1 and CIBN in dark-treated samples.
Figure 1.
Mapping of interacting domains of CRY2 and CIB1. (a) Schematic showing full-length CRY2 and CIB1 constructs used in experiments. The numbers below the proteins indicate amino acid residue position. (b)β-galactosidase activity of CRY2 and CIB1 constructs tested for interaction in the dark or in blue light (461 nm, 1.9 mW, 4 hr). The Gal4 binding domain (Gal4BD-X) and Gal4 activation domain (Gal4AD-Y) fusions used are indicated. The control vector was pGBKT7rec containing no insert. Error bars represent standard deviation (n = 3 samples). The inset panel shows immunoblot analysis of Gal4BD fusion proteins in yeast.
To test the CRY2-CIB1 interaction in mammalian cells, we coexpressed full-length CRY2 or CRY2PHR fused to the fluorescent protein mCherry (CRY2-mCh or CRY2PHR-mCh), and CIBN fused to a prenylated version of EGFP that localizes to the plasma membrane (CIBN-pmGFP) (Fig. 2a). Initial experiments revealed nuclear localization of CRY2-mCh and CIBN-pmGFP, but cytosolic localization for CRY2PHR-mCh. We mutated predicted NLS residues in CRY2-mCh and CIBN-pmGFP (Supplementary Fig. 1), which resulted in robust cytoplasmic and plasma membrane expression, respectively, when cells were maintained in the dark (Fig. 2b). We also examined a NLS-deleted version of full length CIB1 (CIB1-pmGFP), but did not use this construct further as it showed punctate perinuclear localization (data not shown).
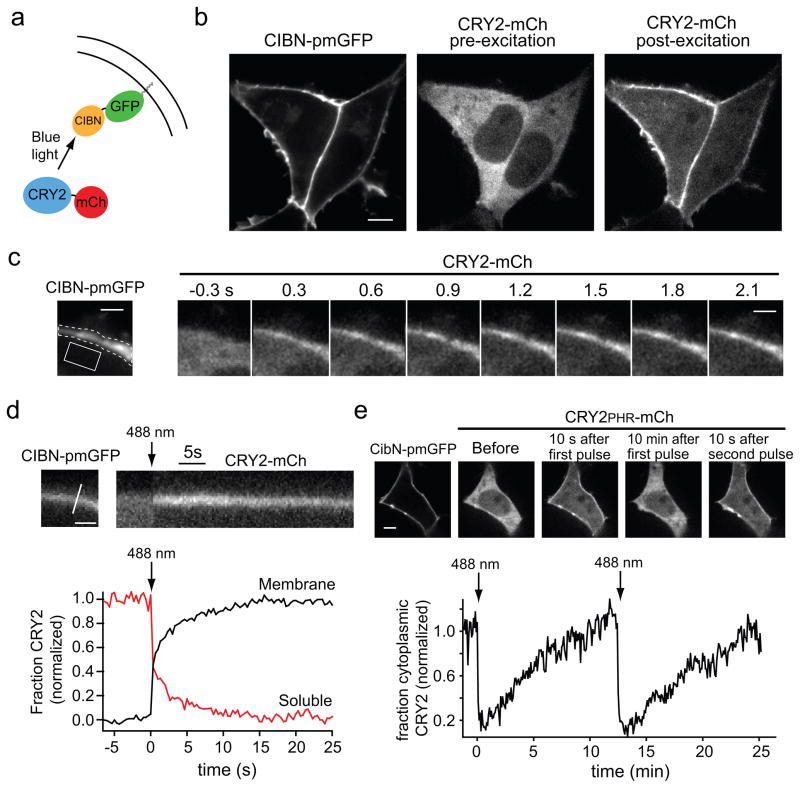
Figure 2.
Light-triggered translocation of CRY2 in mammalian cells. (a) Schematic showing fusion proteins. CIBN-pmGFP contains a CaaX box prenylation motif for targeting to the plasma membrane. (b) Images of CIBN-pmGFP (left panel) and CRY2-mCh (right two panels) coexpressed in HEK293T cells. The localization of CRY2-mCh is shown prior to light excitation (middle panel) and 20 s following a 100 ms pulse of blue light (488 nm, 25μW) (right panel). Scale bar 5 μm. (c) Time course of CRY2-mCh recruitment to the plasma membrane following a single 100 ms pulse of 488 nm light (25 μW). CIBN-pmGFP localization is shown in the left panel. Scale bar 2 μm. (d) CRY2-mCh translocation kinetics following a 100 ms pulse of 488 nm light (arrow). The distribution of CIBN-pmGFP and the line used to generate the CRY2-mCh kymograph is shown in the upper left panel. Scale bar 1 μm. The bottom panel shows quantification of CRY2-mCh in the cytoplasm and at the plasma membrane, using the regions shown in (c) by the dotted and solid lines respectively. Each fraction was normalized between 0 and 1. (e) Activation and reversal time course of the CRY2PHR-CIBN interaction. The top panel shows cells coexpressing the indicated constructs before and after delivery of two 100 ms pulses of blue light (25 μW) spaced 12.5 min apart. The bottom panel shows quantification of cytoplasmic CRY2PHR-mCh, with light pulses delivered at t = 0 and t = 12.5 min (arrows).
We tested the ability of blue light to induce interaction between CRY2-mCh and membrane associated CIBN-pmGFP. Within 300 ms after blue light illumination (488 nm), CRY2-mCh began accumulating at the plasma membrane, where it co-localized with CIBN-pmGFP (Fig. 2b,c and Supplementary Video 1). Greater than 95% of cells exhibited robust translocation of CRY2-mCh to the plasma membrane, while the distribution of CIBN-pmGFP did not change following illumination. Translocation was more than 90% complete within 10 s (Fig. 2d). Dim room light (5.2 μW) did not trigger translocation, indicating samples do not require excessive light shielding. CRY2-mCh and CRY2PHR-mCh behaved similarly in translocation experiments, with similar activation and reversal kinetics (Supplementary Fig. 2a). The complete time course of plasma membrane association and dissociation for the CRY2PHR-CIBN interaction is shown in Figure 2e. After an initial pulse of light, CRY2PHR-mCh rapidly translocated to the plasma membrane, then slowly dissociated over ~12 minutes. A subsequent light pulse triggered a second round of translocation nearly identical in magnitude to the first (Fig. 2e), and the interaction could be repeatedly induced at least six times with little or no loss in efficacy (Supplementary Fig. 2b). While blue light has poor tissue penetration ability, multi-photon excitation can allow precise three-dimensional cell targeting in tissues with high cellular densities. We found that the CRY-CIB modules can be activated by two-photon stimulation at 860 nm (range, 820–980 nm) in cell culture and organotypic cultured hippocampal slices (Supplementary Fig. 3), suggesting the potential for precise spatial activation of protein dimerization in whole organisms.
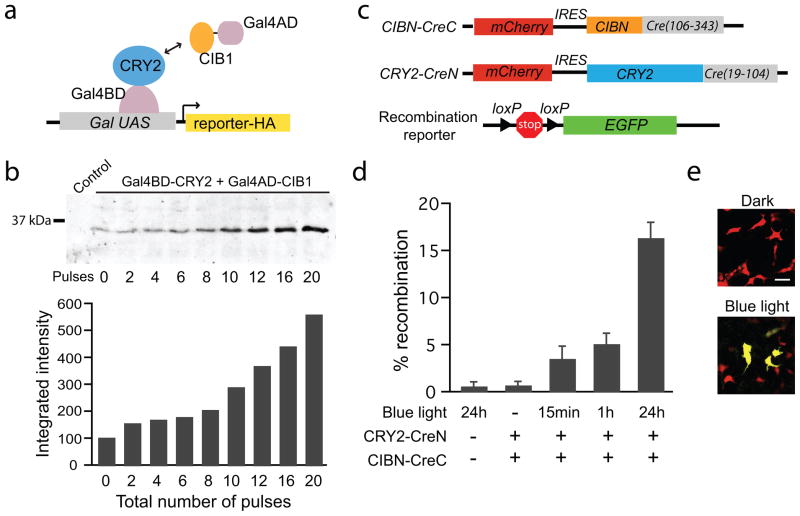
One powerful application of artificial dimerizers is to allow inducible control of a ‘split’ protein, where two inactive fragments are brought together to reconstitute a functional protein activity. In principle, using this approach with optically controlled dimerizers would confer light-dependent activity to a diverse range of target proteins. We used the CRY-CIB modules to reconstitute split versions of two proteins, a Gal4 transcription factor and Cre recombinase. For the former, our approach was similar to a prior light-regulated system using Arabidopsis PHYB and PIF33, but based on the transient nature of the CRY-CIB interaction (Fig. 2e), we speculated that light pulses would induce transcription in a dose-dependent fashion, allowing precise control of protein expression. We coexpressed in yeast the split Gal4 partners Gal4BD-CRY2 and Gal4AD-CIB1, as well as a reporter protein (Snl1) under control of a galactose-inducible promoter (Fig 3a). Yeast were incubated in the dark or subjected to up to 20 blue light pulses (10 s pulses, 8 min apart) over four hours (Fig. 3b). Immunoblot analysis of Snl1 revealed a strong dose-dependence of protein expression in response to light.
Figure 3.
Light induced activation of transcription and DNA recombination. (a) Schematic showing split Gal4 modules (Gal4BD-CRY2 and Gal4AD-CIB1) expressed in yeast cells containing an HA-tagged reporter protein under control of a galactose-inducible promoter. (b) Immunoblot analysis of the HA-tagged reporter (top panel) in response to blue light pulses (10 s pulses, 1.7 mW, 8 min apart). The control lane contains lysates from cells expressing only the reporter. The graph at bottom shows the quantification of western blot bands. (c) Schematic showing the two split Cre constructs (CIBN-CreC and CRY2-CreN) and the Cre reporter. (d) The plot shows % Cre reporter recombination (# GFP expressing cells / # mCherry expressing cells) measured 48 hours after transfection of HEK293T cells with the Cre reporter and indicated constructs. Cells were exposed to blue light pulses (450 nm, 4.5 mW) for indicated times (15 min, 1 hr, or 24 hrs), or kept in the dark for the duration (−). The error bars represent standard deviation for 3 samples from 3 independent experiments. (e) GFP fluorescence images from samples containing both CRY2-CreN and CIBN-CreC that were exposed to 24 hours of blue light or dark. Scale bar 20 μm.
We tested the ability of the CRY-CIB modules to induce dimerization of a split Cre recombinase, allowing light-dependent control of DNA recombination. Based on a previous split Cre recombinase activated by rapamycin10, we fused CRY2 to amino acids 19–104 of Cre (CRY2-CreN), and CIBN to amino acids 106–343 of Cre (CIBN-CreC) (Fig. 3c). The Cre modules showed no toxicity in cells (Supplementary Table 1). As a reporter of Cre recombinase activity, we used a plasmid containing a transcriptional stop sequence flanked by loxP sites preceding EGFP. When transfected into HEK293T cells, the reporter construct alone displayed low levels of recombination (0.6 ± 0.4 % of transfected cells, on average) (Fig. 3d), as did control cells expressing either CIBN-CreC or CRY2-CreN alone (data not shown). Cells containing both CRY2-CreN and CIBN-CreC incubated in the dark showed equivalent levels of recombination as control cells (0.7 ± 0.4 % of transfected cells), indicating minimal or no light-independent CRY2-CIBN interaction. Cells containing CRY2-CreN and CIBN-CreC exposed to continuously pulsed blue light for 24 hours (2 s pulses every 3 min) showed a 158-fold increase in the number of EGFP-positive cells (16.4 ± 1.6 % of transfected cells) compared to dark treated samples, indicating robust light-dependent activation that was comparable to the activity of the rapamycin dimerized split Cre under similar transient transfection conditions10. Importantly, cells exposed to pulsed light for shorter times, mimicking activation of Cre in a tissue or in vivo, showed a substantial increase in activity over dark treated samples. Exposure to light pulses for only 15 mins resulted in a 25-fold increase in EGFP-positive cells over dark treated samples (3.5 ± 1.3 % of transfected cells), while exposure to pulses for one hour gave a ~50-fold increase over dark samples (5.1 ± 1.1 % of transfected cells). We expect that further optimization of these constructs such as linker modification and mutagenesis may allow even more robust light-dependent activation in future iterations.
The above demonstrations of the CRY-CIB modules indicate that they allow temporal, spatial, and dose-dependent optical control of protein dimerization, with time scales in the sub-second range and without the requirement for exogenous ligand seen with chemical dimerizers or the PHY-PIF system. Future improvements to this system will include establishment of minimal domains for efficient packaging in viral vectors, as well as engineering modules with altered dissociation kinetics and spectral sensitivities. Furthermore, as CRY1 has been observed to dimerize11, 12, we are also engineering monomeric versions for homodimerization applications. As this platform is entirely genetically encoded and can be activated using commonly available light sources used for GFP imaging, we expect these modules to be useful for controlling a broad range of biological phenomena.
Methods
Strains and Plasmids
For yeast two-hybrid studies, strains AH109 (MATa, trp1-901, leu2-3, 112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2::GAL1UAS-GAL1TATA-HIS3, GAL2UAS-GAL2TATA- ADE2, URA3::MEL1UAS-MEL1TATA-lacZ, MEL1) and Y187 (MATα, ura3-52, his3-200, ade2-101, trp1-901, leu2-3, 112, gal4Δ, gal80Δ, met-, URA3::GAL1UAS-GAL1TATA-lacZ, MEL1) were used (Clontech). The light inducible transcription study used yeast strain PJ69-4a (MATa trp1-901 leu2-3,112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2::GAL1-HIS3, GAL2-ADE2, met2::GAL7-lacZ).
Plasmids used in this study are described in Supplementary Table 2 and will be available from Addgene (http://www.addgene.com). Oligos are listed in Supplementary Table 2. Gal4 binding domain fusions (Gal4BD-X) were in pDBTrp, a version of pDBLeu (Invitrogen) with a Trp+ selection marker. Gal4 activation domain fusion proteins (Gal4AD-Y) were in pGADT7rec (Clontech). To generate Gal4BD and Gal4AD constructs, Cry2 and Cib1 were PCR amplified from Arabidopsis thaliana cDNA using gene specific primers. PCR products from this amplification were then used as template for a second round of PCR using oligos designed to allow expression (via homologous recombination in yeast) of full length CRY2 or CRY2PHR (amino acids 1–498) at the C–terminus of Gal4BD, or full length CIB1 or CIBN (amino acids 1–170) at the C-terminus of Gal4AD.
For expression in mammalian cell lines, full length Cry2 and amino acids 1-498 of Cry2 were PCR amplified and ligated into vector pmCherry-N1 (Clontech) at Xho I and Xma I sites. CibN was cloned in a similar manner using Nhe I and Age I sites into a version of eGFP-C1 that contained a CaaX polybasic sequence from KRas4B (KKKKKKSKTKCVIM) at the C-terminus. To mutate the NLS sequences, we used oligos CRY2dNLSf and CRY2dNLSr for CRY2 and CIBdNLSf and CIBdNLSr for CIB1. PCR amplification was carried out using mutagenic oligos and forward and reverse oligos from two-hybrid cloning to generate two overlapping fragments of DNA, which were joined via homologous recombination in yeast. Constructs were tested for interaction in yeast, then moved to mammalian vector systems as previously described.
The Cre recombination constructs were first assembled in yeast (in vector p414ADH) via homologous recombination of two overlapping fragments that had been generated by PCR. For the CRY2-CreN construct, the first fragment contained (in order) 33 bp of homology to p414ADH, a Sac I site, a Kozak sequence, full length (+NLS) CRY2, a flexible linker (GGGGSGGGGSGG). The second PCR fragment contained the flexible linker, a Not I site, amino acids 19–104 of Cre recombinase, a stop codon, a Xma I site, followed by 33 base pairs of homology to the yeast vector. The CIBN-CreC construct was assembled identically, except CIBN was used in place of CRY2, and amino acids 106–343 of Cre were used in place of 19–104. After recombination in yeast, inserts containing fusion proteins were cut out of p414ADH using Sac I and Xma I, and cloned into the MCS (Sac I / Xma I sites) of pmCherryC1, downstream from an IRES2 element that was placed between mCherry and the MCS.
Yeast two-hybrid experiments
Gal4BD plasmids containing vector only, CRY2, or CRY2PHR were expressed in strain AH109 and patched on YPD plates. On top of patches, Y187 yeast expressing Gal4AD fusions with CIB1, CIBN, or empty vector control were patched. Yeast were mated overnight at 30° C, then streaked on SD -Trp/-Leu plates to select for diploid cells that contained both Gal4AD and Gal4BD plasmids. Colonies that grew on SD -Trp/-Leu were then streaked on SD -Trp/-Leu/-His/ + 3 mM 3-aminotriazole (3-AT) plates. β-galactosidase assays were performed by growing yeast cells overnight in SD -Trp/-Leu medium, followed by dilution to 0.2 OD600 in SD -Trp/-Leu medium the next morning. Following an initial 3 hour growth period in the dark, cultures were either kept in the dark or continuously exposed to an LED blue light source (461 nm, 1.9 mW) for 4 hours. At the conclusion of the 4 hour period, cultures were harvested and lysed with Y-PER reagent (Thermo Scientific). The assay for β-galactosidase activity was then carried out following a standard protocol for liquid cultures (Clontech Laboratories, protocol #PT3024-1) using ONPG (Sigma-Aldrich) as a substrate. Experiments were carried out at least three times with similar results to those shown. We observed that samples incubated in dim room light (0.25 μW) gave results indistinguishable from samples incubated in total darkness. In constrast, bright room light (34 μW) activated reporters ~30% as well as blue LED treated samples.
Live cell imaging
Live cell imaging was performed on a custom built spinning disc confocal microscope with a Yokogawa CSU10 scan head mounted on a Nikon TE300 inverted stand as previously described13. Images were acquired using a 60× Plan Apochromat 1.4 NA objective. A 1.5× tube lens between the filter wheel and camera focused light on the chip of a Hamamatsu C9100 EM-CCD camera giving a pixel size of 86 × 86 nm. The focal plane was controlled by a piezo-driven Z-stage (Applied Scientific Instruments). The EM-CCD Camera, filter wheel, stage, and AOTF laser line switching were controlled by Metamorph software (Molecular Devices). An environmental chamber (In Vivo Scientific) enclosing the microscope stand maintained the temperature at 37° C. HEK293T cells were grown on glass coverslips (Deckgläser #1, 18 mm) and maintained in DMEM containing 10% FBS. Cells were transfected with Lipofectamine 2000 (Invitrogen) when 50–80% confluent according to the manufacturers protocols and imaged 24 hours following transfection.
Excitation was provided by solid state 488 nm (Coherent) or 561 nm (Spectraphysics) lasers shuttered via an acousto-optical tunable filter (AOTF) (Neos Technologies), with emission directed through a filter wheel (Applied Scientific Instrumentation) holding either band pass or long pass filters (Chroma). Power used for stimulation of translocation was equivalent to that used for imaging GFP (25 μW measured 1 cm from the objective). Wavelengths tested for triggering translocation were consistent with the absorbance profile of cryptochrome, which responds to UVA/blue light with a peak at 450 nm, and weakly above 500 nm14, 15 — i.e. 405 nm illumination triggered interaction, but illumination at 561 nm did not. Illumination at 514 nm (which would be used with YFP for two-color imaging) triggered translocation at high intensity illumination, but not at lower intensity (under 2 μW) (Supplementary Fig. 4).
Reports in the plant literature suggest that light activation of cryptochromes may be reversed with green light14, 16, however we did not observe any effect of illumination with green light (514 or 540 nm) on the reversal time of the CRY2-CIBN interaction. We also tested the interaction of CIBN-mCh with CRY2-pmGFP. While these proteins expressed well, they did not show light-induced interaction. It is possible that membrane association of CRY2 sterically hinders the association with CIB in this conformation, and adjustment of the linker length may remedy this.
Dose-dependent activation of transcription
pDBTrp-CRY2 and pGBKT7rec-CIB1 constructs, along with a plasmid from a galactose-inducible yeast overexpression library17 expressing the protein Snl1 from a galactose-inducible promoter, were co-transformed into strain PJ694-a and plated on SD -Trp/-Leu/-Ura plates. The triple transformed yeast were grown overnight at 30 C in media containing SD -Trp/-Leu/-Ura, then diluted to 0.1 OD600 in SD -Trp/-Leu/-Ura and placed in the dark. Following an initial 3 hour growth period in the dark, yeast cells were exposed to pulses of blue light from a fluorescent microscope beam (Leica MZFLIII) equipped with a GFP filter (10 s in duration, spaced 8 min apart, 1.7 mW). Cultures remained in the dark a total of four hours following the initial light exposure, at which point they were harvested for immunoblotting. Yeast were lysed in 2% SDS by glass bead disruption (425–600 μm beads, Sigma), after which samples were boiled for 3 minutes, placed in 2× Laemmli Sample Buffer, boiled for 1 minute, and centrifuged at 14,000 rpm for 5 minutes. Equal amounts of total protein were run on a 12% SDS-PAGE gel and immunoblotted using standard procedures using a mouse anti-HA primary antibody (Covance) and an IRDye 700CW goat anti-mouse IgG secondary antibody (Li-COR). Proteins were visualized using an Odyssey infrared imaging system (Li-COR).
Light activation of split Cre recombinase
HEK293T cells were transfected with the Cre reporter and indicated constructs, and % Cre reporter recombination (# of GFP expressing cells / # of mCherry expressing cells) was measured 48 hours after transfection. For light treated samples, blue light pulses (2 s pulse delivered every 3 min, 450 nm, 4.5 mW) were administered by a custom LED array light source. For 24 hour experiments, light was administered from 24 to 48 hours following transfection. For pulse experiments, samples were exposed to pulsed light (15 min or 1 hr) at 24 hours post transfection, then incubated in the dark until 48 hours post transfection to allow reporter expression. Nontreated (−) samples were kept in the dark for the duration. Samples were fixed in 4% paraformaldehyde and the ratio of cells expressing GFP to cells expressing mCherry was calculated, based on the average counts from three wells. The results reported are the average and standard deviation from three independent experiments. To calculate the fold activation, the ratio of the percent of cells activated in the light to the percent activated in the dark was calculated after the background was subtracted. For the trypan blue cell counts described in Supplementary Table 1, harvested cell cultures were incubated with 0.4% trypan blue solution (Sigma) for 3 minutes, then the number of blue vs. white cells was determined using a hemocytometer.
Two-photon microscopy
Two-photon microscopy was performed using a Zeiss LSM 710 confocal scanhead mounted on an Axio-observer microscope a with Chameleon II ultra laser source using a 20 × 1.0 NA Apochromat objective (Zeiss). IR laser power was normalized by measuring the power at the sample using a FiedMaxII power meter (Coherent) tuned to the corresponding wavelength. All two-photon experiments were performed using equivalent power, unless otherwise stated. HEK293T cells expressing CRY2PHR-mCh and CIBN-pmGFP were imaged using 561 nm excitation with emission collected through a band-pass filter set for 570–610 nanometers. We collected 10 images of CIBN-mCh prior to exposure with 800–1000 nm excitation (~15% AOM at 930 nm for 30 iterations). Two-photon excitation was repeated every 25 images (acquired at 1 Hz) for the duration of the experiment. Translocation occurred within the range of 830–980 nm. After the time series completion, the localization of CIBN-pmGFP was determined by acquiring a single image with 488 nm exposure (~1.5 μs pixel dwell time for 1 s) with emission collected through a band pass filter set for 500–550 nm.
Organotypic slice culture was carried out as previously described except that rat pups were used instead of mouse pups18. Briefly, hippocampi from postnatal day 5–6 rat pups were dissected and slices (350 μm) were prepared using a McIlwain tissue chopper and cultured on 0.4 μm millicell membrane inserts (Millipore). After 6 days in culture, slices were biolistically transfected with CIBN-pmGFP/CRY2PHR-mCh constructs (Helios gene gun, Biorad). Following biolistic transfection, slices were either maintained in darkness or photostimulated 3–4 days following transfection. Following light stimulation, slices were immediately fixed in PBS containing 4% paraformaldehyde in PBS for 10–15 min on ice. The CRY2PHR-mChsignal was enhanced with a rabbit polyclonal anti-RFP antibody (MBL) prior to mounting the slices for imaging.
Supplementary Material
Supplementary Figure 1 Mutation of putative NLS sequences in CRY2 and CIB1.
Supplementary Figure 2 Reversibility and repeated induction of CRY2-CIBN interaction.
Supplementary Figure 3 CRY2PHR-CIBN interaction can be activated by two-photon excitation.
Supplementary Figure 4 YFP can be imaged without activating CRY2-CIBN interaction.
Supplementary Video 1 Light-triggered translocation of CRY2-mCh to the plasma membrane. HEK293T cells expressing CRY2-mCh were exposed to a 100 ms pulse of 488 nm light at t = 0. The mCh channel was recorded at 3 Hz. The dimensions of the movie are 35 μm × 35 μm.
Supplementary Table 1 CRY2-CIB modules and light treatments show no toxicity in cultured cells.
Supplementary Table 2 Plasmids and oligos used in CRY2-CIB experiments
Acknowledgments
We thank B. Arenkiel and E. Spana for comments on the manuscript, and I. Davison for design of a custom LED light pulsing device. This work was supported in part by grants from the National Institutes of Health R01 NS039402 and R01 MH064748 and the Howard Hughes Medical Institute (to M.D.E.) and National Institutes of Health grant R01 DK81584 (to C.L.T.).
Footnotes
Author Contributions
C.L.T. conceived the idea and directed the work. M.J.K., R.M.H., and C.L.T. designed experiments. M.J.K, R.M.H., L.A.P., J.W.S., and C.L.T. performed experiments. M.D.E. supervised microscopy experiments. M.J.K., R.M.H., and C.L.T. wrote the manuscript. M.D.E. and C.L.T. edited the manuscript and reviewed the data.
References
- 1.Spencer DM, Wandless TJ, Schreiber SL, Crabtree GR. Controlling signal transduction with synthetic ligands. Science. 1993;262:1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- 2.Bishop A, et al. Unnatural ligands for engineered proteins: new tools for chemical genetics. Annu Rev Biophys Biomol Struct. 2000;29:577–606. doi: 10.1146/annurev.biophys.29.1.577. [DOI] [PubMed] [Google Scholar]
- 3.Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. A light-switchable gene promoter system. Nat Biotechnol. 2002;20:1041–1044. doi: 10.1038/nbt734. [DOI] [PubMed] [Google Scholar]
- 4.Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yazawa M, Sadaghiani AM, Hsueh B, Dolmetsch RE. Induction of protein-protein interactions in live cells using light. Nat Biotechnol. 2009;27:941–945. doi: 10.1038/nbt.1569. [DOI] [PubMed] [Google Scholar]
- 6.Tyszkiewicz AB, Muir TW. Activation of protein splicing with light in yeast. Nat Methods. 2008;5:303–305. doi: 10.1038/nmeth.1189. [DOI] [PubMed] [Google Scholar]
- 7.Leung DW, Otomo C, Chory J, Rosen MK. Genetically encoded photoswitching of actin assembly through the Cdc42-WASP-Arp2/3 complex pathway. Proc Natl Acad Sci U S A. 2008;105:12797–12802. doi: 10.1073/pnas.0801232105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khanna R, et al. A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell. 2004;16:3033–3044. doi: 10.1105/tpc.104.025643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu H, et al. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science. 2008;322:1535–1539. doi: 10.1126/science.1163927. [DOI] [PubMed] [Google Scholar]
- 10.Jullien N, Sampieri F, Enjalbert A, Herman JP. Regulation of Cre recombinase by ligand-induced complementation of inactive fragments. Nucleic Acids Res. 2003;31:e131. doi: 10.1093/nar/gng131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sang Y, et al. N-terminal domain-mediated homodimerization is required for photoreceptor activity of Arabidopsis CRYPTOCHROME 1. Plant Cell. 2005;17:1569–1584. doi: 10.1105/tpc.104.029645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenfeldt G, Viana RM, Mootz HD, von Arnim AG, Batschauer A. Chemically induced and light-independent cryptochrome photoreceptor activation. Mol Plant. 2008;1:4–14. doi: 10.1093/mp/ssm002. [DOI] [PubMed] [Google Scholar]
- 13.Kennedy MJ, Davison IG, Robinson CG, Ehlers MD. Syntaxin-4 defines a domain for activity-dependent exocytosis in dendritic spines. Cell. 2010;141:524–535. doi: 10.1016/j.cell.2010.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banerjee R, et al. The signaling state of Arabidopsis cryptochrome 2 contains flavin semiquinone. J Biol Chem. 2007;282:14916–14922. doi: 10.1074/jbc.M700616200. [DOI] [PubMed] [Google Scholar]
- 15.Ahmad M, et al. Action spectrum for cryptochrome-dependent hypocotyl growth inhibition in Arabidopsis. Plant Physiol. 2002;129:774–785. doi: 10.1104/pp.010969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouly JP, et al. Cryptochrome blue light photoreceptors are activated through interconversion of flavin redox states. J Biol Chem. 2007;282:9383–9391. doi: 10.1074/jbc.M609842200. [DOI] [PubMed] [Google Scholar]
- 17.Gelperin DM, et al. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 2005;19:2816–2826. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gogolla N, Galimberti I, DePaola V, Caroni P. Preparation of organotypic hippocampal slice cultures for long-term live imaging. Nat Protoc. 2006;1:1165–1171. doi: 10.1038/nprot.2006.168. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 Mutation of putative NLS sequences in CRY2 and CIB1.
Supplementary Figure 2 Reversibility and repeated induction of CRY2-CIBN interaction.
Supplementary Figure 3 CRY2PHR-CIBN interaction can be activated by two-photon excitation.
Supplementary Figure 4 YFP can be imaged without activating CRY2-CIBN interaction.
Supplementary Video 1 Light-triggered translocation of CRY2-mCh to the plasma membrane. HEK293T cells expressing CRY2-mCh were exposed to a 100 ms pulse of 488 nm light at t = 0. The mCh channel was recorded at 3 Hz. The dimensions of the movie are 35 μm × 35 μm.
Supplementary Table 1 CRY2-CIB modules and light treatments show no toxicity in cultured cells.
Supplementary Table 2 Plasmids and oligos used in CRY2-CIB experiments