Abstract
In this issue of Molecular Cell, Lee et al. show that Neuropsora crassa uses several Dicer-dependent and -independent pathways to generate miRNA-like RNAs and small-interfering RNAs. Their studies expand the known small RNA biogenesis pathways and suggest the existence of others that are yet to be discovered.
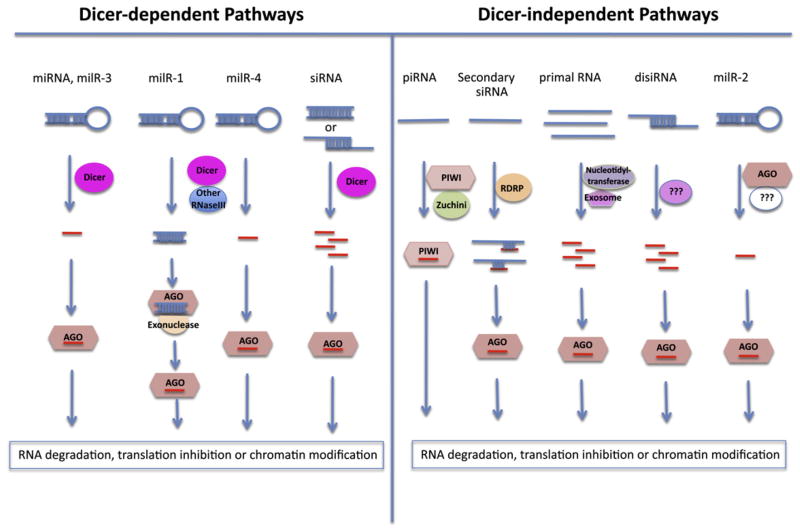
Small noncoding RNAs serve as important regulators of gene expression in almost all eukaryotes. The small RNAs are generated by Dicer-dependent or -independent biogenesis pathways, and are incorporated into the Argonaute (AGO) family of effector proteins to guide sequence-specific gene silencing either transcriptionally or posttranscriptionally by base-pairing with target RNAs (Figure 1). The effector-bound small RNAs can trigger target RNA degradation or translational inhibition, or cause chromatin modifications at target loci (Ghildiyal and Zamore, 2009).
Figure 1. The Many Ways to Generate Small RNAs.
Single-stranded RNAs with stem loop structures can be processed by Dicers and/or other endonuclease III domain-containing enzymes (RNaseIII) to generate miRNAs and miRNA-like small RNAs (milRNAs). Perfectly or partially double-stranded RNAs can also be processed by Dicers and/or other endonucleases to produce siRNAs and disiRNAs (Dicer-independent siRNAs). Other single-stranded RNAs with certain special features may be processed by a variety of enzymes (e.g., PIWI, Zucchini, RDRP, nucleotidyltransferase, exosome, etc.) to generate piRNAs, some secondary siRNAs and primal RNAs. The various small RNAs are loaded onto the AGO family of effector proteins (AGOs and PIWIs) to guide sequence-specific RNA degradation, translational inhibition, or chromatin modification of target transcript RNA or DNA.
Based on their precursor structures, biogenesis pathways, and modes of action, small RNAs are classified as microRNAs (miRNAs), small-interfering RNAs (siRNAs), PIWI-interacting RNAs (piRNAs), and primal small RNAs (priR-NAs) (Ghildiyal and Zamore, 2009; Halic and Moazed, 2010). miRNAs in plants and animals arise from single-stranded RNA precursors with stem-loop structures, which are processed by the RNase III type endoribonuclease Dicer or Dicer-like (DCL) proteins (Bartel, 2004). While most siRNAs are generated from double-stranded RNAs by Dicers, some siRNAs in C. elegans may be generated directly through de novo synthesis by RNA-dependent RNA polymerases (RDRP) (Ghildiyal and Zamore, 2009). The biogenesis of piRNAs and priRNAs is Dicer-independent and requires other RNA processing enzymes (Ghildiyal and Zamore, 2009; Halic and Moazed, 2010).
Although various classes of miRNAs and siRNAs have been described in animals and plants, our knowledge of fungal small RNAs has been rather limited. No miRNAs were found in fungi. The only siRNAs reported in the filamentous fungus Neurospora crassa are DNA damage induced, the AGO protein QDE- 2-interacting small RNAs (qiRNAs) (Lee et al., 2009). Lee et al. identified several new classes of Neurospora small RNAs that also interact with QDE-2, including miRNA-like RNAs (milRNA) and Dicer-independent siRNAs (disiRNAs) (Lee et al., 2010). Their analysis revealed several novel small RNA biogenesis pathways (Figure 1). The study indicates that small RNA biogenesis is much more diverse in fungi than previously thought.
By analyzing the small RNAs coimmunoprecipitated with QDE-2 by deep sequencing, Lee et al. identified 25 miRNA-like loci that can generate RNA precursors with stem-loop structures, giving rise to miRNA-like RNAs with a strong strand bias. These milRNAs have a clear preference for U at their 5′ termini and show a size distribution that peaks at 19 nt and 25 nt. Genetic analysis of four of the most abundant milRNAs (milR-1 to milR-4) suggests that milRNAs have surprisingly diverse biogenesis pathways (Figure 1). The production of milR-1 is Dicer dependent and also requires the activity of a QDE-2-interacting exonuclease, QIP, and QDE-2 itself. However, QDE-2 slicer activity was not required. QDE-2 binds 33 nt pre-milRNAs in a qip mutant but associated with mature milRNAs in a wild-type strain, suggesting that QDE-2 functions to recruit QIP for milR-1 processing rather than to directly slice the precursors. The biogenesis of milR-3 is Dicer dependent but does not require QDE-2, so milR-3 biogenesis resembles miRNA formation in plants. Although milR-4 maturation requires Dicer but not QDE-2 or QIP, milR-4 is reduced but not completely eliminated in the dcl mutant, suggesting a partial dependence on Dicer and the possible involvement of another endoribonuclease. Most strikingly, the biogenesis of milR-2 is Dicer independent and requires the slicer activity of QDE-2 but not the QDE-2-interacting protein QIP for its maturation. Thus, the fungus N. crassa uses a distinct biogenesis pathway to generate each of these four milRNAs (Figure 1).
The finding of Dicer-independent pathways for milRNA biogenesis prompted Lee et al. to search for new ribonucleases involved in milRNA formation. An RNase III domain-containing protein homologous to the yeast mitochondrial ribosomal protein MRPL3 was identified as a candidate for further analysis. A heterokaryotic strain of mrpl3ko and wild-type nuclei with reduced mrpl3 mRNA was used because the mrpl3 deletion strain is lethal. The level of both milR-1 and milR-4 but not of milR-2 was reduced in the mrpl3ko(het) strain, indicating a role of MRPL3 in the biogenesis of some milRNAs. Although MRPL3s from other eukaryotic organisms are known to be present in the large subunit of the mitochondrial ribosomes, it would be important to determine the subcellular localization of Neurospora MRPL3. If the effect of Neurospora MRPL3 on milRNAs biogenesis is direct, MRPL3 is likely to be functional also in the cytoplasm or nucleus.
To test the biological functions of the milRNAs, Lee et al. constructed artificial milRNA targets using a Myc-tagged reporter containing milR-1 complementary sequences, and transformed these constructs into wild-type and qde-2 mutant strains. Protein levels of the reporter were much higher in the qde-2 mutant than in the wild-type, whereas the RNA levels of the reporter were only increased modestly in the qde-2 mutant. These results indicate that like animal miRNAs and some plant miRNAs, milRNAs in Neurospora may induce silencing of their targets mainly by translational inhibition. Using a mammalian miRNA target-prediction program, Lee et al. identified candidate endogenous targets for these milRNAs. Some of the putative target mRNAs of milR-1 were elevated in milR-1ko and qde-2 mutants, and some candidate target mRNAs were associated with QDE-2, supporting that these milRNAs do regulate gene expression in vivo. Although the milRNAs are generated by different Dicer-dependent or Dicer-independent pathways, the hairpin structures of their precursors, their distribution on the stem loop structure and strong strand bias, the presence of miRNA*-like sequences, and their role in suppressing target gene expression indicate that for all purposes these milRNAs could be considered as genuine miRNAs.
In addition to milRNAs, a new class of siRNAs, namely the disiRNAs (for Dicer-independent siRNAs), was also identified by Lee et al. These disiRNAs are generated mainly from loci that produce both sense and antisense transcripts. They do not show a strand bias, are predominantly 22 nt in length, and have a 5′ U preference. Although most of the small RNAs from Neurospora are Dicer dependent, which is evidenced by a significant reduction of small RNAs in dcl mutants (Lee et al., 2010), the levels of the examined disiRNAs were not altered in the dcl mutants, indicating that these disiRNAs are not dependent on Dicer. Nor are they dependent on QDE-2, MRPL3, QDE-1 (a RDRP), or QDE-3 (a RecQ DNA helicase). The authors speculated that the disiRNAs are generated by as-yet-unidentified Dicer-independent pathway(s). siRNAs derived from overlapping sense and anti-sense transcripts were first reported in plants (Borsani et al., 2005; Katiyar-Agarwal et al., 2006), and recently were found also in animals (Okamura and Lai, 2008). The plant and animal nat-siRNAs (for natural-antisense-transcripts-derived siRNAs) are Dicer dependent, and function in the cleavage of complementary target RNA transcripts (Borsani et al., 2005; Katiyar-Agarwal et al., 2006; Okamura and Lai, 2008). The fact that the disiRNAs are associated with the AGO protein QDE-2 suggests that they can function in gene regulation, although their mode of action is not known.
Analysis of AGO-associated small RNAs by deep sequencing has identified various new classes of small RNAs, including a number of Dicer-independent small RNAs, such as piRNAs from mammals and Drosophila, 22G-RNAs from C. elegans, and primal RNAs from fission yeast (Ghildiyal and Zamore, 2009). Dicer-independent small RNAs have not been reported in plants, although they are likely also present in plants. All of the described small RNAs are ultimately loaded onto an effector protein for carrying out their regulatory functions (Figure 1). Most of the small RNAs reported to date appear to associate with AGOs. It should be noted that some AGO proteins have the ability to bind longer RNAs, such as 30- to 40-nt small RNAs in the fission yeast AGO1 complex (Halic and Moazed, 2010) and the milRNA precursors in the QDE-2 complex (Lee et al., 2010), suggesting that more complex functions of these AGO proteins remain to be discovered. In addition, some small RNAs may associate with other types of RNA-binding proteins, which may confer different functions (Zheng et al., 2008).
The work of Lee et al. in this issue of Molecular Cell provides an excellent example of how new small RNAs can be discovered by examining AGO-interacting small RNAs. Importantly, this study revealed previously unsuspected diversity and complexity in small RNA biogenesis in fungal systems. It begs the question, how many more small RNA bio-genesis pathways are there?
References
- Bartel DP. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Borsani O, Zhu JH, Verslues PE, Sunkar R, Zhu JK. Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic M, Moazed D. Cell. 2010;140:504–516. doi: 10.1016/j.cell.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Villegas A, Zhu JK, Staskawicz BJ, Jin HL. Proc Natl Acad Sci USA. 2006;103:18002–18007. doi: 10.1073/pnas.0608258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Chang SS, Choudhary S, Aalto AP, Maiti M, Bamford DH, Liu Y. Nature. 2009;459:274–277. doi: 10.1038/nature08041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Li L, Gu W, Xue Z, Crosthwaite SK, Pertsemlidis A, Lewis ZA, Freitag M, Selker EU, Mello CC, Liu Y. Mol Cell. 2010;38:803–814. doi: 10.1016/j.molcel.2010.04.005. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Lai EC. Nat Rev Mol Cell Biol. 2008;9:673–678. doi: 10.1038/nrm2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XW, Pontes O, Zhu JH, Miki D, Zhang F, Li WX, Iida K, Kapoor A, Pikaard CS, Zhu JK. Nature. 2008;455:1259–1270. doi: 10.1038/nature07305. [DOI] [PMC free article] [PubMed] [Google Scholar]