Abstract
Radial glial progenitor cells (RGPCs), have been long known to exhibit a striking form of bidirectional nuclear migration. The purpose and underlying mechanism for this unusual cell cycle-dependent “interkinetic” nuclear migration has remained poorly understood. We investigated the basis for this behavior by live imaging of nuclei, centrosomes, and microtubules in embryonic rat brain slices, coupled with blebbistatin and RNAi. We observed nuclei to migrate independent of centrosomes and unidirectionally away from or toward the ventricular surface along microtubules, which we found to be uniformly oriented from the ventricular to the pial surfaces of the brain. Cytoplasmic dynein RNAi specifically inhibited apically-directed nuclear movement. An RNAi screen for kinesin genes identified KIF1A, a member of the kinesin 3 family, as the motor for basally-directed nuclear movement. These observations provide the first direct evidence for a role for kinesins in nuclear migration and neurogenesis, and suggest that a novel cell cycle-dependent switch between distinct microtubule motors drives INM.
Neocortical neurons are born in the germinal zone and migrate over substantial distances to the forming cortical layers. In the latter stages of migration, the cells exhibit a bipolar morphology with a leading process and a trailing axon. The nucleus in these and other migrating neurons invariably trails the centrosome 1-4. However, unlike nonneuronal cells, the centrosome first departs from the nucleus and advances alone into the leading “migratory” process, subsequently followed by the nucleus 1, 4. Cytoplasmic dynein or LIS1 RNAi each inhibited both nuclear and centrosomal movement 4, 5. Inhibition of myosin II expression or activity also interfered with nuclear movement in migrating neurons in brain slices 4 and dissociated neuronal culture 2, 3, 6. Myosin II had no role in centrosome movement in radially migrating neocortical neurons in brain slices 4, though its activity was correlated with centrosome advance in cultured cerebellar neurons 6. The substantial separation between nucleus and centrosome in migrating neocortical neurons implied some ability of nuclei to travel independently to catch up to the centrosomes 4. Based on the organization of microtubules revealed by expression of the microtubule plus-end marker GFP-EB3, the nuclei were deduced to travel toward minus ends of microtubules, similar to vesicular organelles such as lysosomes and late endosomes that use cytoplasmic dynein for their retrograde transport.
The mechanisms involved in the initial stages of neocortical neurogenesis are less well understood. The neuroepithelial cells, referred to as radial glial progenitor cells (RGPCs) as the neocortex thickens, divide rapidly to expand their pool and undergo asymmetric divisions to generate most cortical pyramidal neurons and glia 7. The progenitors span the entire thickness of neural tube and developing neocortex, and exhibit an unusual form of behavior termed interkinetic nuclear migration (INM) 8. Following mitosis, which occurs exclusively at the ventricular surface, the nuclei ascend to the upper region of the ventricular zone (VZ), where they undergo S phase, and then descend back to the ventricular surface. This behavior has been shown to occur in the majority of neuroepithelial cells within the CNS and in some polarized non-neuronal cells.
Although INM was described in the early part of the past century 8, little was known until recently about its biological significance, its role in neurogenesis, and its underlying mechanism. In early studies, cytochalasin B and colchicine were reported to alter the overall distribution pattern of nuclei within the neuroepithelium, suggesting that actin and microtubules might be involved in INM 9-11. We found LIS1 RNAi to block INM in radial glial progenitor cells 5, though, because arrest was complete, it was unclear whether one or both directions of nuclear movement were affected. Intriguingly, the nuclei in the knockdown cells never entered mitosis. Because LIS1 is required for normal mitotic progression 12, 13 these observations suggested that mitotic entry depends on nuclear position. The zebrafish moks309 mutation in the p150Glued subunit of the dynein regulatory complex dynactin has subsequently been reported to alter nuclear migration in retinal neuroepithelial cells 14. More recently, myosin II inhibition was found to affect neuroepithelial cell nuclear migration in zebrafish retina and mammalian cerebral cortex, suggesting a role for this actin-based motor protein as well 15, 16.
We have now carried out a detailed analysis of nuclear migration and microtubule organization in RGPCs and evaluated the contributions of microtubule- and actin-based motors to INM. We find dynein to be required for apical, but not basal, migration, which, instead requires a previously unsuspected unconventional kinesin. Nuclear movement appears completely independent of centrosome behavior and occurs along an array of uniformly oriented microtubules spanning the entire length of the progenitor cell. In contrast to recent reports, we observe no effect of myosin II inhibition in our system. These results lead to a model for INM powered by oppositely directed cell cycle-regulated microtubule motors.
Results
Live nuclear behavior in RGPCs
To understand the role of motor proteins in INM we used in utero electroporation (Fig. 1a) to express CFP-histone H1 (CFP-H1) along with GFP in RGPCs which we monitored in brain slices by fluorescence microscopy every 10-min for 12-18 hr (23 cells in 7 brain slices; Table S1). Individual cells could be monitored throughout most of their migratory and division cycle (Fig. 1b, c; Movie S1), allowing us to identify effects of inhibitory agents at specific stages.
Figure 1. Nuclear and centrosomal dynamics in radial glial progenitor cells throughout cell cycle.
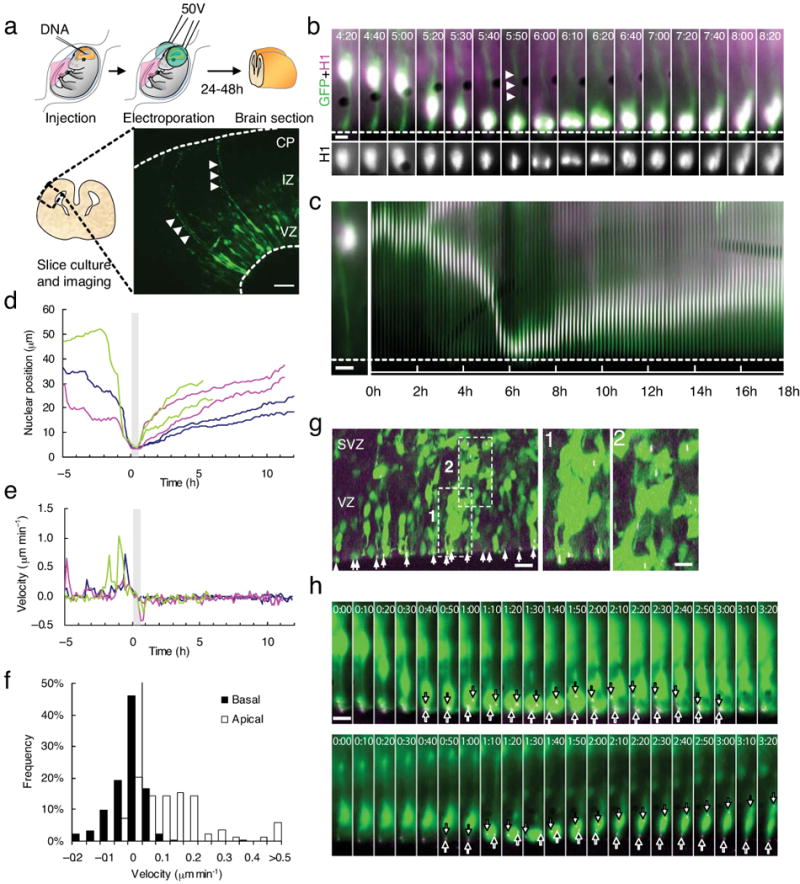
(a) Schematic diagram of transfection using intraventricular injection follow by in utero electroporation. Radial glial progenitors expressing GFP were observed to expand throughout the entire thickness of the neocortex (arrowheads). Bar = 50 μm. (b) Live imaging of radial glial progenitors expressing GFP (green) and CFP-histone H1 (magenta; H1 expression is shown for cell body region in black and white at bottom) in live E18 rat brain slice. During a 4 hr period the radial glial progenitor cell body can be seen to move apically toward the ventricular surface (dashed line), where the cell went through mitosis, during which the basal process (arrowheads) persisted, though thinner (panel 5:40), and the progeny cells moved basally (Movie S1). Time = hh:mm. (c) Kymograph of the same cell imaged using histone H1 for 18 hr showing marked changes in nuclear migration rate in the apical direction, and prolonged, more uniform movement in the basal direction following mitosis. Bars = 5 μm. (d) Tracings of nuclear movements of the radial glial progenitor cell shown in panel A and B (blue tracing) and two other cells (green and magenta tracings). Each cell generated two progeny nuclei which moved basally at comparable rates. (e) Velocities of nuclear movements in (d). Bursts of fast movements up to 1 μm/min could be seen as the nucleus during apically directed (-5 hr to 0 hr) but not during basally directed movement (1hr - 10 hr). The tracings are aligned for a common mitotic start time (grey area). (f) Histograms of the velocity during apically- and basally- directed movements. The velocity distribution of the apical nuclear movements is spread out higher speed movements, whereas the distribution of basal movements is very narrow. (g) Fixed E18 rat brain section two days after in utero electroporation with cDNAs encoding GFP (green) and DsRed-centrin II (magenta). GFP-positive cells expressing DsRed-centrin II-labeled centrosomes can be found within the VZ (box 1) and SVZ (box 2) with centrosomes located throughout both regions, and at the ventricular surface in the endfeet of radial glial progenitor cells. Bar = 10 μm. Bar in insert = 5 μm. (h) Live imaging of a radial glial cell expressing GFP (green) and RFP-centrin II (magenta). Top panels: During downward (apical) nuclear movement, the centrosome remains at the ventricular surface. At the onset of mitosis (0:40) one centrosome (white arrows) departs from the ventricular surface and, following cytokinesis (2;00), returns to this site (3:00). The other centrosome stays close to the ventricular surface throughout mitosis (black arrows). The cell bodies of the daughter cells then migrate upward in a comparably slow and continuous manner (Movie S2). Bottom panels: In some cases, one of the centrosomes departs from the ventricular surface (white arrows) while the other returns to the ventricular surface (Movie S4), as observed in asymmetric cell divisions 20.
Apically directed nuclear movements were unidirectional but very intermittent (Fig. 1c, d), with bursts of fast nuclear translocation (0.5-1 μm/min; Fig. 1e) interrupted by pauses ranging in duration from 0.5 to 2 hr. Nuclear movement away from the ventricular surface was also unidirectional but, surprisingly, much more continuous, with rare or no pauses and so slow as to be almost unnoticeable in our initial analysis (Fig. 1f; avg. basal rate = 0.063 ± 0.009 μm/min; avg. apical rate = 0.14 ± 0.02 μm/min; p < 0.001, student's t test), consistent with the longer duration estimated for G1 in neuroepithelial cells 17, and with differences in the mechanisms responsible for the two directions of nuclear transport.
Centrosome and microtubule behavior in RGPCs
In radially migrating neurons cytoplasmic dynein aided by LIS1 directly transports nuclei toward the centrosome 4. This structure lies at the center of the microtubule cytoskeleton, which we concluded advances as a result of dynein pulling forces exerted from sites within the leading (migratory) process of the cell. Centrosomes are found within the apical endfoot of the radial glial progenitor cell (Fig. 1g) 18-20, but how their behavior is related to that of the oscillating nuclei has not been examined in detail. Direct monitoring of DsRed-centrin II revealed that centrosomes remain at the radial glial endfeet throughout the entirety of interphase, independent of nuclear position (Fig. 1h; Movie S2), indicating that radial glial nuclei travel alone over large distances. When the nuclei reached the ventricular surface, by now duplicated centrosomes separated. One or both could be seen to depart from the ventricular surface in their role as mitotic spindle poles (Fig. 1h, S1, white and black arrows; Movie S2, S3), and clear segregation of centrosomes into the daughter cells could be observed as cytokinesis progressed. In some divisions, each centrosome moved to the endfoot of its respective cell at the ventricular surface and remained at this site. In other divisions, the centrosome of one of the progeny cells moved to the basal (upper) side of the nucleus and then preceded the nucleus as the entire cell migrated away from the ventricular surface (Fig. 1h; Movie S4), consistent with an asymmetric division that produces a radial glial cell and a neuron 20.
To define the organization of microtubules in RGPCs we expressed either GFP-tubulin or GFP-EB3 in RGPCs using in utero electroporation (Fig. 2). Individual GFP-tubulin tagged microtubules could not be resolved in the RGPCs, but microtubule bundles were observed extending throughout the length of both the basal and apical processes (Fig. 2a; arrows in the left panel). During mitosis, the GFP tubulin clearly labeled the mitotic spindle (Fig. 2a, right panels), but, surprisingly, tubulin labeling was not detected within the basal process during this stage. This is despite clear evidence that the basal process extends to the pial surface of the brain throughout mitosis, though thinner than during interphase (Fig. 2a; arrows in right panels).
Figure 2. Microtubule organization in radial glial progenitors throughout the cell cycle.
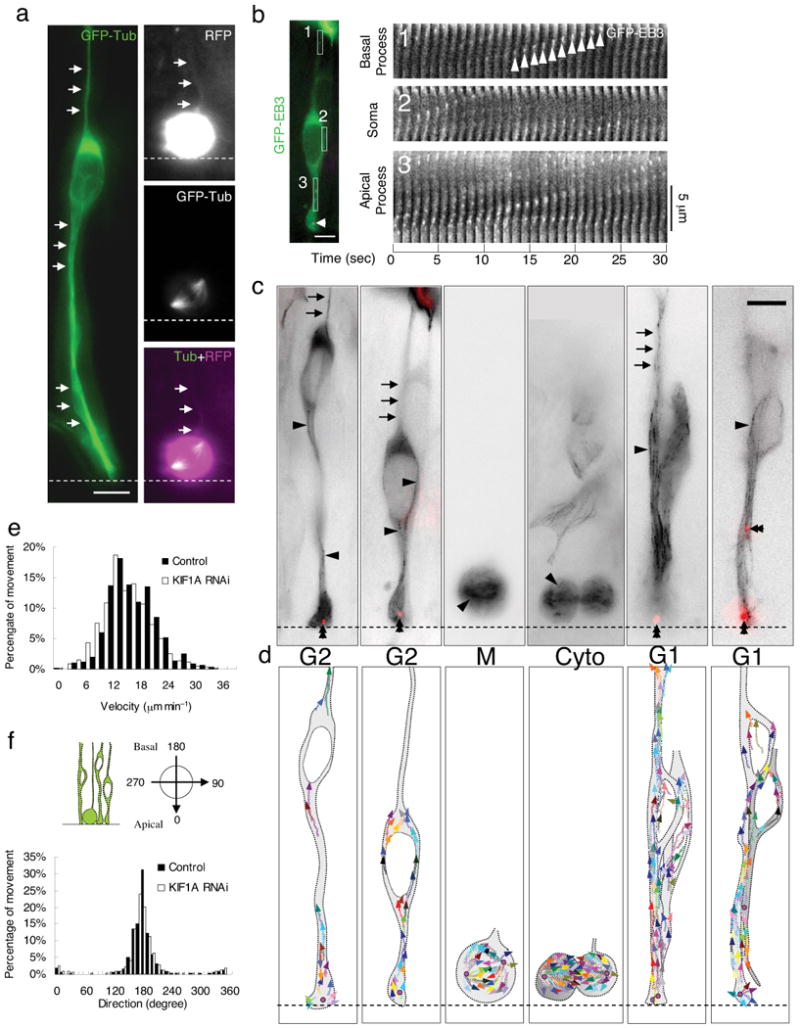
(a) Microtubules of radial glial progenitors expressing GFP-Tubulin (green) in E18 rat brain slices. Left panel: Tubulin is distributed throughout interphase cells, with bundles of microtubules parallel to the long axis of the cell seen in favorable, dilated cell regions (arrows). Right panels: Dividing cell showing mitotic spindle. Basal process remains visible during division (top, arrows), as detected using soluble RFP, whereas tubulin-GFP cannot be found in this region (bottom). (b) Time-lapse images of plus-ends of growing microtubules labeled with GFP-EB3 (green) in an interphase radial glial progenitor cell at E18. (Centrosome is indicated in magenta (arrowhead) by DsRed-centrin co-expression.) Fluorescence images were taken every second. Time series of EB3 images in three regions correspondent to the basal process (box 1), soma (box 2) and apical process (box 3) are aligned to form a kymograph. “Comet tail”-like EB3 streaks are readily observed to move predominantly in the basal direction (arrowheads). (c), (d) GFP-EB3 behavior and microtubules organization in radial glial cells at different cell-cycle stages. (c) Superimposed negative contrast images of EB3 streaks (arrowheads) and centrosomes (double arrowheads, red) from 30 sec - 2 min time-lapse movies. (d) Tracings of the EB3 streaks using multiple colors to distinguish among individual microtubules. When soma is in the upper portion of the VZ (G2), EB3 streaks are mostly found to originate from the centrosomal region within the endfeet, curve around the nucleus, and enter the basal process (arrows; Movie S5, S6). During mitosis (M), EB3 streaks radiate from the two spindle poles to form the mitotic spindle. No detectable EB3 streaks enter the basal process (Movie S7). During cytokinesis (Cyto), the microtubules still radiate from the centrosomes in each daughter cell, with many microtubules aimed toward the midbody. EB3 streaks remain absent from the basal processes at this stage (Movie S8). (Non-radial glial cells are seen in uppar portion of image.) Paired cells following probable symmetric RGPC division (Symm) as evidenced by persistence of centrosomes at the endfeet of both daughter cells (Movie S9). EB3-tipped microtubules are oriented upward in both cells and now re-enter the basal fibers (arrows). Paired cells following probable asymmetric (Asym) RGPC division. The centrosome of daughter cell at right is shifted away with EB3 streaks emerging radially to now form a bidirectional microtubule array (Movie S10). Bars = 5 μm. (e) Velocity distribution shows EB3 movements to be comparable to migrating neurons and other cells. KIF1 A RNAi does not significantly affect the rate of EB3 movements in these cells. (f) Direction distribution of the EB3 movements during interphase indicates that 93% are oriented toward the basal directions. KIF1A RNAi has no obvious effects on the overall orientation of EB3 movements.
GFP-EB3, which labels growing microtubule plus-ends (Fig. 2b-d) 4, revealed the assembly and orientation of individual microtubules. The “comet tail”-like EB3 streaks could be detected throughout the processes and somata of the transfected RGPCs (Fig. 2b; average rate of advance = 15.75 ± 5.25 μm/min; n = 2042 streaks in 18 cells; Fig. 2e) 4, 21, 22. EB3 streaks emerged from the centrosome at the endfeet and extended in a nearly uniform orientation (Fig. 2c; Movie S5, S6; 93% plus-ends basally directed; n = 14 cells in 9 brain slices; Fig. 2f) throughout the length of the cell all the way to the pial surface (data not shown). During mitosis (Fig. 2b), EB3 streaks radiated from the spindle poles and clearly delineated the overall spindle structure (Fig. 2c; Movie S7). EB3 streaks could not be detected within the basal process during mitosis (Fig. 2c, d; Movie S8), but reappeared in the basal fiber of the postmitotic RGPC, extending in advance of the nucleus, which advanced at a considerably slower rate (Fig. 2c, d; Movie S9, S10).
Dynein RNAi blocks apically directed but not basally directed nuclear movement
To determine the extent to which dynein contributes to INM in RGPCs it was necessary to perform dynein RNAi under conditions of partial as well as complete inhibition and to monitor cells live through successive cell cycle stages (Fig. 3). At 5 days following in utero electroporation with dynein heavy chain shRNAs, we observed complete arrest of radial glial nuclei at a range of distances from the ventricular surface (Fig. 3a, e; Movie S11; Table S1), and a complete loss of mitotic divisions as we previously reported for LIS1 RNAi 5. We observed a comparable arrest of nuclei following 24-48 hr expression of dynamitin, which inhibits dynactin function 23(Fig. 3d; Movie 12). At 3 days of dynein RNAi, apically-directed nuclear movement could be detected, but motile events were more brief (52.9 ± 28.7 min vs. 288.3 ± 68.4 min, n = 22) and slower (0.27 ± 0.32 μm/min vs.0.46 ± 0.21 μm/min, n = 19) than for controls (Fig. 3b; Movie S13. Basally-directed nuclear migration was at rates similar to controls and with comparable continuity (Fig. 3c; Movie S14; 0.064 ± 0.016 μm/min; Table S1). These data support a role for cytoplasmic dynein in apically-, but not in basally-directed nuclear movement, strongly consistent with the orientation of microtubules indicated by EB3 labeling.
Figure 3. Effects of dynein functional inhibition on interkinetic nuclear migration in radial glial progenitors.
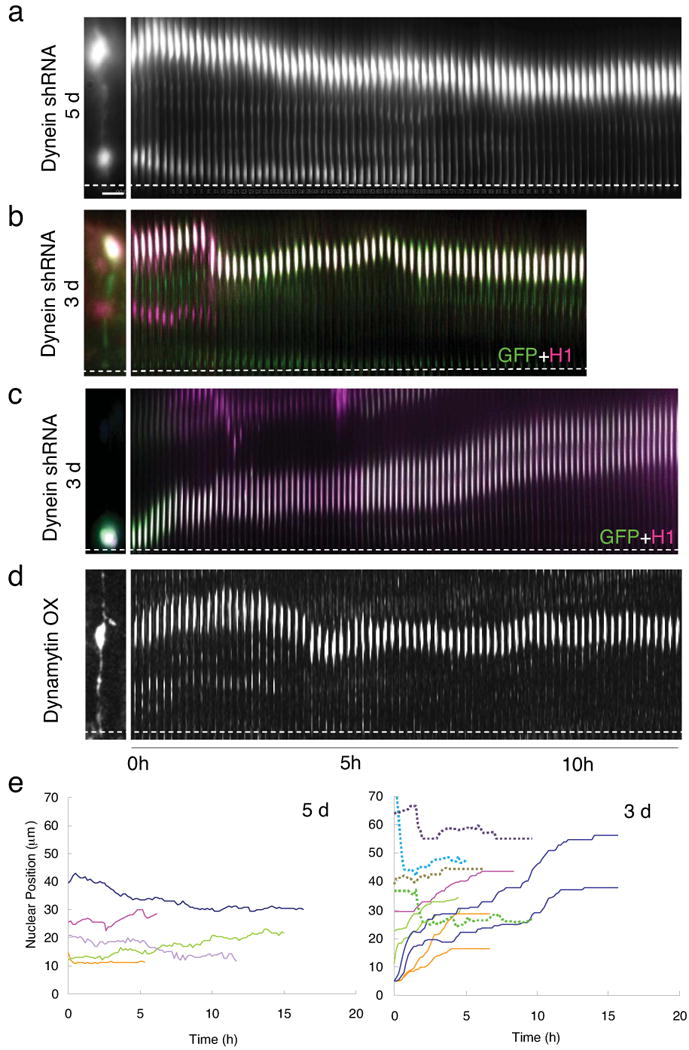
(a) A radial glial progenitor cell in E21 rat brain slice imaged 5 days after in utero electroporation with cDNA construct expressing cytoplasmic dynein HC shRNA and GFP. The cell exhibited normal bipolar morphology (shown at left), but with nuclear motility limited to a gradual drifting toward the ventricular surface (Movie S11). (b) Radial glial cell expressing dynein HC shRNA construct plus CFP-histone-H1 (magenta) at E19, 3 days following in utero electroporation. The nucleus migrated toward the ventricular surface. but with shorter and more intermittent movements, at a slower than normal average rate (see text for details; Movie S13). (c) Normal basally directed nuclear movement of paired radial glial cell nuclei at E19, following 3 days of HC RNAi (conditions as in panel B; Movie S14). (d) Severe inhibition of INM in cells overexpressing dynamytin for 1.5 days (Movie 12). (e) Tracings of the nucleus in cells expressing dynein shRNA for 5 days (left panel) and 3 days (right panel). Most of the nuclei were immobile after 5 days of dynein RNAi, so neither apically- or basally directed movement is observed. However, in cells expressing dynein HC shRNA for 3 days, the apically directed movements were largely inhibited (dashed lines), whereas the basally directed movements were comparable to controls (Table S1). Bar = 5 μm
Effects of myosin II inhibition on interkinetic nuclear migration
Myosin II inhibition has been reported to interfere with either basally directed 16 or both directions of nuclear migration 15 during INM in neuroepithelial cells. Expression of myosin IIB shRNA produces a substantial 65-70% decrease in myosin IIB protein levels in cultured cells (Fig. 4a) 24 and blocks radial neuronal migration in brain slices 4. However, it had no apparent effect on INM in our embryonic rat brain slices (Fig. 4b; Movie S15; n = 16 in 5 slices), with average rates of movement in both directions comparable to those in control cells (Fig. 4c, h; Movie S16; and Table S1). We also applied blebbistatin acutely to cells while they were undergoing either apically- or basally-directed nuclear movement (n = 18 cells in 5 slices; Fig. 4d, e; Movie S17, S18; Table S1), and observed no detectable interruption of the normal nuclear trajectory. We did, nonetheless, observe a clear increase in the duration of mitosis (Fig. 4g; 90 ± 11 min, n = 6 vs. control 48.9 ± 4.2 min, n = 9, p < 0.01) and inhibition of cytokinesis (Fig. 4f; Movie S19) using myosin IIB RNAi or blebbistatin, respectively, well-established effects of myosin inhibition in dividing cells.
Figure 4. Myosin 2B is not essential for interkinetic nuclear migration.
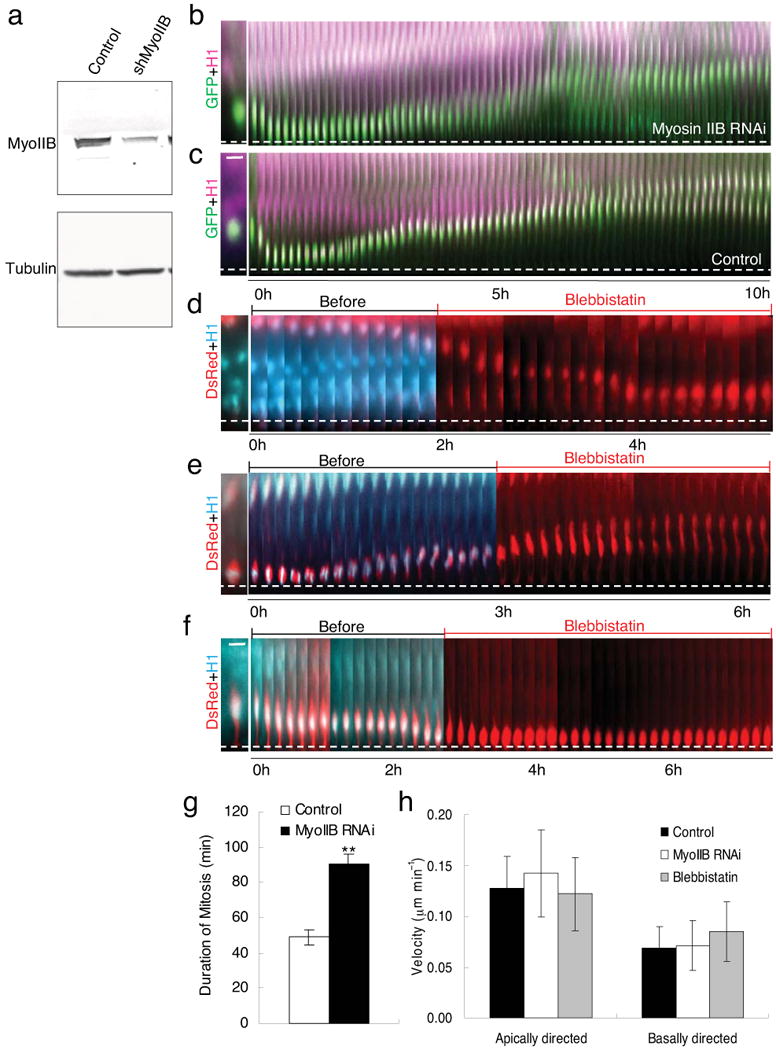
(a) Western blot showing a substantial 65-70% decrease in myosin IIB expression in Rat2 cells transfected with myosin IIB shRNA for 3 days. (b)-(e) E18 rat brain slices monitored by live time-lapse imaging following myosin II RNAi or exposure to the myosin II inhibitor blebbistatin. Nuclei were imaged using CFP-histone-H1 and either GFP for RNAi or DsRed for use with blebbistatin. GFP (green) and CFP-histone-H1 (magenta) were co-expressed with and without myosin IIB shRNA (Movies S15, S16). No apparent effect on basally directed movement of radial glial nucleus is observed relative to control (b, c). Cells co-transfected with DsRed (red) and CFP-histone-H1 (blue) were treated with 50 μm blebbistatin for 3-5 hrs during live cell imaging. No effect was observed on apically directed (d) or basally directed (e) nuclear movement in radial glial cells. However, cell in (f) was blocked in mitosis as expected. (White dashed lines indicate ventricular surface; see Movies S17-S19). Bar = 5 μm. (g) Duration of mitosis increases about 2-fold in cells subjected to myosin IIB RNAi. **: p<0.01. (H) Velocities of apically and basally directed movements in cells treated with myosin IIB shRNA and blebbistatin showing no significant difference between these groups and the control.
RNAi screen for kinesins involved in neuronal migration pathway
These results, the uniform orientation of microtubules (Fig. 2), and the unidirectional movement of nuclei in the basal direction in RGPCs (Fig. 1) suggested a potential role for a plus-end directed microtubule motor protein. To explore this possibility, we performed a functional shRNA screen of kinesin heavy chains. The rat genome encodes 45 different kinesin heavy chains 25, precluding a complete screen. We limited the number of candidates by focusing on microtubule plus-end directed kinesins. This approach also eliminated mitotic kinesins, all of which are either minus-end directed or serve in microtubule assembly regulation 25, 26. Of the 13 plus-end directed kinesin heavy chain genes, 11 are already known to be expressed in the embryonic mouse neocortex 27. These genes represent isoforms from 5 of the ∼14 known kinesin families: kinesin 1 (KIF5A, B, C), kinesin 2 (KIF3A, B, C), kinesin 3 (KIF1A, B, C), kinesin 8 (KIF18A), and kinesin 11 (KIF26A). For each kinesin isoform, 3 sequences were selected, which, with controls, totaled 36 RNAi sequences for which shRNAi-encoding constructs were generated (Table S2).
In utero eletroporation was performed at E16, and the effects of each of the 36 shRNAs were tested in fixed E20 brain slices. 11 of the 33 kinesin-directed shRNAs had no effect on the distribution of neuronal precursors (e.g., KIF1B and 5A in Fig. S2; Table S2), whereas clear effects were observed with the remaining 22 shRNAs, typically manifested as an accumulation of GFP-positive cells within the VZ and subventricular zone (SVZ) and a moderate to severe reduction in bipolar GFP-positive cells in the intermediate zone and cortical plate (e.g., KIF5B, KIF3B, KIF18A and KIF3A in Fig. S2; Table S2). The severity of these effects differed between shRNAs for the same kinesin gene, as well as between kinesin isoforms within the same family and between families (Table S2). Nonetheless, strong effects were observed for kinesins within each of the 5 families, in each case using shRNAs directed against multiple independent sequences.
Role of KIF1A in interkinetic nuclear migration
Of the positive kinesins, one exhibited particular characteristics potentially associated with defects in radial glial nuclear migration, KIF1A (Fig. 5). The misdistribution of cell somata within the VZ was relatively severe, and, in addition, accumulation of radial glial cell bodies near or at the ventricular surface could be observed (Fig. 5b-e), potentially consistent with a defect in the earliest stages of the neuronal migration pathway. Phosphovimentin immunostaining revealed no accumulation of radial glial cells in mitosis (Fig. 6a; mitotic index: KIF1 RNAi, 5.1 ± 1.5 % vs. control, 5.3 ± 1.0 %, n = 3, p = 0.89), supporting a function for KIF1A at some other cell cycle stage. KIF1A protein levels were reduced by 70% by 48hr (Fig. 5a), consistent with the severity of the physiological effects.
Figure 5. KIF1A is required for basally directed nuclear movement.
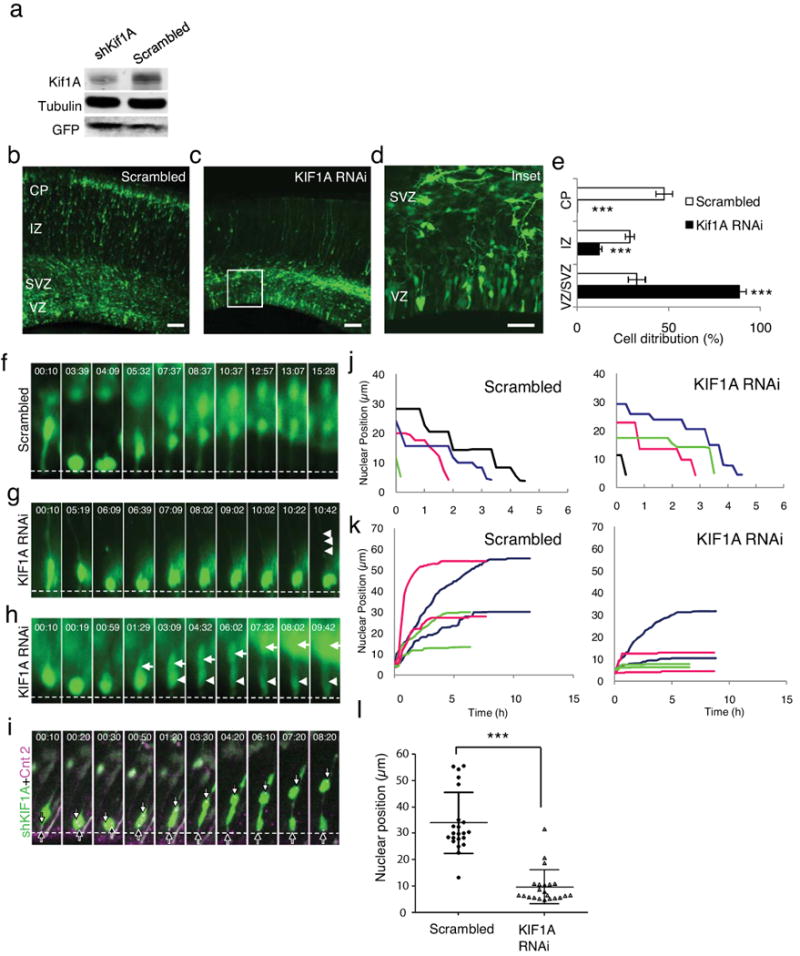
(a) KIF1A RNAi reduced KIFA levels by 70% as judged by immunoblotting of PC12 cells exposed to shRNA construct for 2 days. (b-d) E16 rat embryonic brains were electroporated with scrambled (b) or KIF1A shRNAs (c, d), fixed, and then imaged at E20. Inset from panel c is shown at higher power in d. KIF1A RNAi caused clear and potent inhibition of cell redistribution relative to control, with almost complete loss of cells from intermediate zone (IZ) and cortical plate (CP), and accumulation of radial glial cell somata near ventricular surface. Persistence of radial glial basal process staining in IZ indicates retention of overall cell morphology. Scale bar=50μm. (e) Quantification of cell distribution in scrambled (blue bar) vs. KIF1A (red bar) shRNA-expressing brain slices showing significant decreases in neural cell redistribution (p<0.001; student's t-test, compare with scrambled). (f-h) Apically directed nuclear movement was recorded by time-lapse imaging of radial glial cells expressing scrambled shRNA (F; Movie S20), and KIF1A shRNA (g, h). Basally directed nuclear movement was inhibited in both (g) or one (h) of the daughter cells. (i) The cnetrosome of the mobile daughter moved away from ventricular surface, a behavior consistent with newborn neurons (Fig. 1h) 20. (Dashed lines indicate ventricular surface; time = hh:mm; Movie S21, S22). Bar = 5 μm. (j) Tracings of nuclear migration show no apparent effect of KIF1A RNAi in apical direction. (k) In contrast, marked effects are observed in basal direction for all by one nucleus (blue line), which exhibits rapid movement characteristic of newly-formed neuron. Note potent inhibition of nuclear migration in sibling of this cell (also in blue). (l) Final nuclear position following recording of basally directed nuclear movements. Average nuclear positions were is 34.096 ± 11.6 vs. 9.6 ± 6.5 μm for control and KIF1A RNAi conditions, respectively. (0 of Y-axis indicates ventricular surface; p<0.001; student's t-test, n = 22).
Figure 6. Effects of KIF1 RNAi on cell cycle progression and cell fate determination.
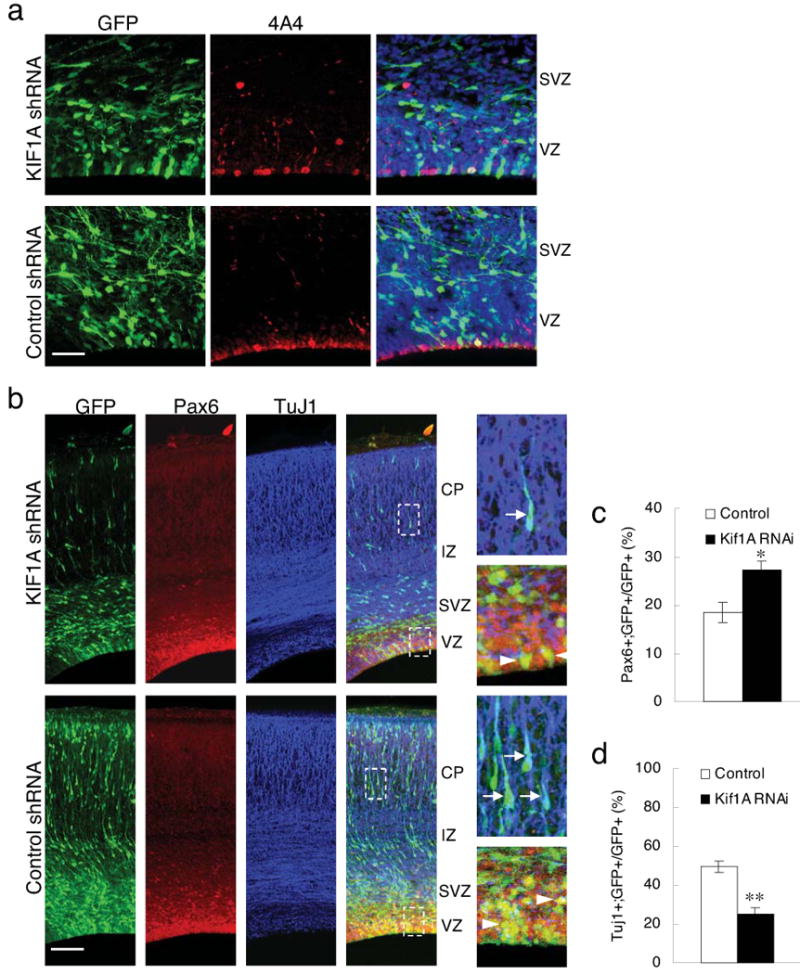
(a) Brain sections stained with phosphovimentin (4A4) antibody (Red) 4 days after electroporation showing mitotic cells. Bar = 50 μm. (b) Brain sections stained with progenitor marker Pax6 (Red) and neuronal marker TuJ1 (Blue). The right panels shows examples of TuJ1 (arrows) and Pax6 (arrowheads) positive cells in the boxed areas. Bar = 100 μm. (c) Quantification of mitotic index of the transfected GFP+ cells within the VZ. There is no significant difference between cells electroporated with KIF1A and control shRNA. (d) Duration of mitosis in radial glial progenitors in live brain slices showing no obvious effects of KIF1A shRNA on mitosis. (e), (f) Quantifications of the percentage of Pax6+ or TuJ1+ cells, respectively, among the transfected GFP+ cells in brain sections. *: p<0.05; **: p<0.01, student's t-test.
To test the role of KIF1A in nuclear migration directly, we performed live imaging of radial glial progenitor cells subjected to KIF1A RNAi. We also tested cells subjected to KIF1B RNAi, which had shown a phenotype in fixed brain sections. The Kif1B effect on the overall distribution of transfected cells was weaker than for KIF1A, possibly reflecting less efficient protein knockdown (Fig. S2B). Nonetheless, KIF1B is closely related throughout its length to KIF1A, and the two genes could well have overlapping functions 28. We also tested KIF3A and KIF3B because of their strong phenotypes in fixed brain sections, and in view of the role of KIF3A in primary cilia-mediated neural cell signaling 29. Finally, we examined KIF5B, which exhibited only a mild brain developmental phenotype, but is a prominent neuronal and nonneuronal kinesin involved in vesicular transport, and could serve as a good control for indirect effects on nuclear migration.
Cells expressing kinesin shRNAs were monitored live for 9-15 hours. Defects in INM were observed only for Kif1A (Fig. 5f-h). Nuclei in these cells exhibited normal apically-directed movement toward the ventricular surface, with similar velocity and punctuated by pauses of similar duration to those we observed in wild-type radial glial cells (Fig. 5j). Importantly, these cells exhibited no obvious delay in mitosis (mitotic duration: KIF1 RNAi, 53.8 ± 6.0 min, n = 8; control, 48.9 ± 4.2 min, n = 9, p = 0.52). In contrast, two patterns of behavior were detected following mitosis at the ventricular surface that were indicative of a defect in basally directed nuclear migration. In one pattern, the rates of basally directed nuclear movement for both progeny of division were dramatically reduced relative to controls (0.025 ± 0.022 μm/min vs. 0.069 ± 0.021 μm/min; Fig. 5g, k, and Movie S20, S21). As a result, the nuclei remained close to the ventricular surface for prolonged periods of at least 6-9 hours representing the remainder of the time-lapse period, vs. 0.5-1 hr for controls (Fig. 5k, KIF1A RNAi and Table S1), with a markedly decreased average nuclear travel distance relative to controls (Fig. 5l; 9.57 ± 6.47 vs. 33.96 ± 11.56 μm, n = 22).
A second pattern emerged in cases of what may be asymmetric division; the nucleus from one of the two progeny cells migrated away from the ventricular surface (Fig. 5h arrows, and Movie S22), while the other remained immobile (Fig. 5h, arrowheads). The latter cells were judged to be RGPCs and the former newly generated migrating neurons based in part on their morphology. We also examined centrosome behavior in brains coelectroporated with cDNAs encoding Kif1A shRNA and DsRed-Centrin II (Fig. 5i). In the post-mitotic RGPCs the centrosome returned to the ventricular surface, whereas in the presumptive neurons, the centrosome shifted to the basal side of the nucleus and preceded it as the cell migrated away from the ventricular surface20 (Fig. 1h). In contrast to these results, live analysis of Kif1B, Kif3A (Fig. S3; Movie S25, S26), Kif3B and Kif5B (data not shown) revealed no apparent effect on basal or apical nuclear migration, despite the effect of these treatments on overall neuronal redistribution. Our results, therefore, suggest that KIF1A in particular is required for radial glial nuclear migration. It is not apparently involved in mitosis or migration of newly generated neurons.
As a further test for a role for KIF1A in nuclear migration, we co-expressed RNAi-insensitive human myc- or DsRed-tagged KIF1A with a KIF1A shRNA (Fig. 7a-d). KIF1A expression alone produced no detectable phenotype as judged in fixed brain sections (data not shown). However, clear rescue of neuronal distribution by DsRed-HuKIF1A was observed (Fig. 7b, c; Table S3). Rescue was also evaluated by live imaging, which revealed virtually complete restoration of basally directed nuclear migration (Fig. 7e, f, lower panels; Fig. 7g; Movie S23, S24).
Figure 7. Rescue of KIF1A RNAi, and model for bidirectional nuclear migration.
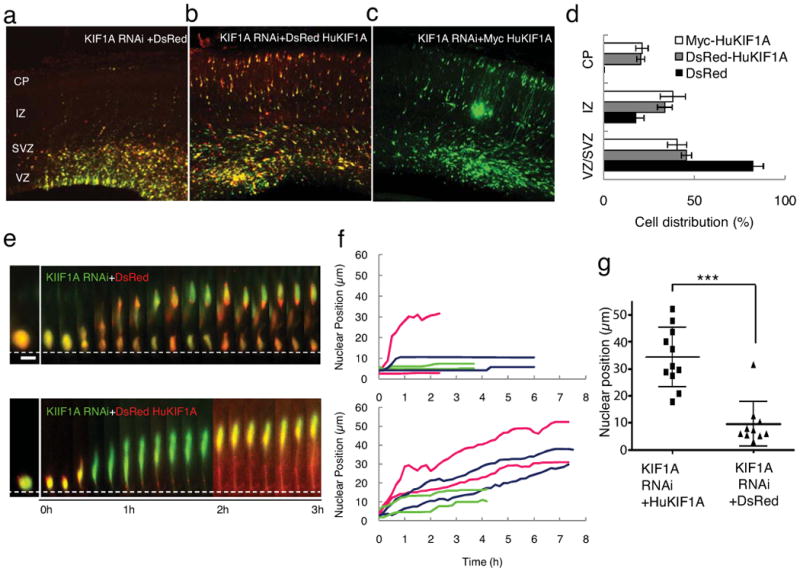
(a-c) E16 rat brain transfected by in utero electroporation with KIF1A shRNA (green) tested for rescue by co-expression of DsRed (red); DsRed-human KIF1A (red), and myc-human KIF1A, and fixed at E20. Clear rescue of neuronal misdistribution phenotype is produced by co-expression of DsRed-human KIF1A (red) or (c) myc-human KIF1A, but not by DsRed alone. Bar=50μm. (d) Statistical analysis of rescue experiments reveals recovery of cell number within the three brain regions shown (see Table S3 for details). For rescue by myc-huKIF1A all GFP-positive cells were counted, whereas on the DsRed-huKIF1A-expressing GFP-positive cells (87% of total) were counted. Significant rescue was observed with DsRed-human KIF1A (grey bar, p<0.001 compare with DsRed; student's t-test) and Myc-humanKIF1A (white bar, P<0.001 compare with DsRed, student's t-test). (e) Live recording of basally directed nuclear movement in KIF1A RNAi cells (green) co-expressing DsRed (red, also see Movie S23), or recued by co-expression of DsRed-human KIF1A (red, also see Movie S24). Bar = 5 μm (f) Tracings of basally directed nuclar movement in control and DsRed-human KIF1A rescued cells. Each color represents progeny of one radial glial progenitor cell division. (g) Final nuclear position following recording of basally directed nuclear movements Nuclear localization from apical directed nuclear movement were plotted. Average nuclear position is 34.22±3.31 vs. 9.49±2.61 μm for DsRed-huKIF1A and DsRed rescues of KIF1A RNAi, respectively. (0 μm in Y-axis indicates ventricular surface p<0.001; student's t-test, n = 11).
To rule out the possibility that KIF1A might contribute to radial glial nuclear migration indirectly through effects on the microtubule cytoskeleton or overall cell organization we co-expressed a KIF1A shRNA with GFP-centrin or GFP-EB3. We observed no detectable effect of KIF1A RNAi on centrosome distribution or behavior, radial glial cell morphology (Fig. 5g), or microtubule organization or orientation (Fig. 2e, f).
Finally, to examine how KIF1A RNAi might affect cell fate, we stained brain sections using the neural progenitor marker Pax6 and the neuronal marker TuJ1 4 days after KIF1A shRNA electroporation (Fig. 6b). We observed an increase in the percentage of Pax6-positive cells (Fig. 6c; 27.1 ± 2.1 % vs. 18.5 ± 2.0 % for control cells, n = 3 brains, p < 0.05) and a striking decrease in that of TuJ1+ cells (Fig. 6d; 25.3 ± 3.0 % vs. 49.5 ± 2.8 % for control cells, n = 3 brains, p < 0.01) among all electroporated GFP-positive cells. These results suggested a dramatic shift in progenitor cell differentiation.
Discussion
The vertebrate neocortex has long been known to originate from a pseudostratified epithelium, as is the case in much of the central nervous system, the vertebrate heart, Drosophila imaginal disks, and other tissues and organs. The mechanism and purpose of the oscillatory nuclear behavior in neuroepithelial cells has remained a critical but poorly understood aspect of brain development with important consequences for the organization and function of the cortical neuronal layers. Our results identify a highly unusual mechanism for this behavior involving bidirectional nuclear transport by microtubule minus-end and plus-end motors along a uniformly directed microtubule cytoskeleton.
Organization of the RGPC microtubule cytoskeleton
Centrosomes are found throughout the VZ ad SVZ of the developing neocortex, and have been demonstrated in particular within the endfeet of RGPCs 18, 19. Based on most other cases of neuronal and nonneuronal cell and nuclear migration, centrosomes might reasonably be expected to participate in INM in RGPCs. Nonetheless, extensive fixed tissue and live brain slice analysis carried out as part of the current study revealed no instance of a nucleus following an associated centrosome in these cells. Despite these findings, centrosome integrity has been found to be important for INM and neural progenitor self-renewal 19, and recent results have implicated the mother centriole, in particular, in the latter activity 20. Our current results suggest that these effects might be indirect, conceivably reflecting roles for RGPC centrosomes in microtubule organization or in primary ciliogenesis. Curiously, we find that both GFP-tubulin and GFP-EB3 disappear from the basal process of RGPCs during late G2, though the process persists throughout mitosis (Fig. 1)7, behavior quite unusual for dividing cells. The process is reinvaded by microtubules following mitosis at rates similar to the advance of EB3 streaks, and, as a result, we suspect it may be driven primarily by microtubule reassembly.
Evidence for a dual motor system for INM
We observed RGPC nuclei to move unidirectionally toward or away from the ventricular surface, though with distinct characteristics in each case. These observations and the uniformity of microtubule orientation we determined in RGPCs suggested a role for oppositely-directed microtubule motors in the two directions of nuclear movement. Components of the cytoplasmic dynein pathway, have been implicated in INM by recent studies 5, 14, but the direction of movement in which dynein participates has remained uncertain. This situation was partly a consequence of the complete immobilization of RGPC nuclei by LIS1 RNAi in our own previous study5. A study of retinal neuroepithelial cells in mok mutant zebrafish, which are defective in the p150Glued subunit of the dynactin complex, 14 reported that nuclei traveled faster and also further in the basal direction, though apically directed movement was slower, as in the current study. These differences could reflect the ability of dynactin to regulate some kinesins as well as cytoplasmic dynein 30, 31, and dynactin inhibition to disrupt the microtubule cytoskeleton 32, but further studies will be needed to resolve this issue.
In view of a role of myosin II in nuclear movement late in the neuronal migration pathway during glial-guided migration of bipolar neurons 4 and in cerebellar granule cells in vitro 6, and other aspects of cell migration 33, we tested for a role for myosin in RGPC behavior. We observed no effect of myosin II RNAi or of blebbistatin on INM either in the apical or basal directions. These results directly contradict recent reports in zebrafish retinal epithelium 15 and in cultured rat telencephalon 16. The former used a diffusion-based, rather than a directed migration model to analyze nuclear movement, which we argue would underestimate the contribution of microtubule motors. The latter study deduced a blebbistatin-induced defect in basal RGPC nuclear migration from analysis of brain sections fixed at a series of time points following BrdU-labeling 16. Live imaging may be of value in resolving differences between that study and the current results.
Our data strongly support the identification of a kinesin as the basal nuclear transport motor. A screen of plus-end directed kinesins implicated several in brain development, a finding that should serve as a basis for much future work. We found that KIF1A, a member of the kinesin III family protein, was essential, in particular, for basally directed nuclear migration in radial glial progenitor cells. Our results, therefore, favor a simple model to account for bidirectional nuclear movement in these cells, involving distinct plus-end and minus-end directed microtubule motors (Fig. S4). Unlike cytoplasmic dynein, which is a very fast minus-end directed microtubule motor 34, KIF1A is plus-end directed and moves slowly and processively as a monomer 35, though faster and more processively as a dimer 36, 37. It is highly expressed in the vertebrate CNS 38, and its C. elegans homologue, UNC-104, has been implicated in anterograde synaptic vesicle transport 39. Knockout mice were reduced in size, showed severe neurological abnormalities, and died within 24 h after birth 40. Compensatory up-regulation of kinesin I expression was reported, and therefore, the suitability of this model for analysis of the specific role of Kif1A in INM is uncertain. Kinesin 1 and kinesin 2 RNAi each had pronounced effects on overall neuronal redistribution in our hands (Fig. S2), though neither affected INM. Kinesin-1 mutations have been reported to rescue dynactin-mediated nuclear mislocation in Drosophila photoreceptors 41, but how these results might relate to the current results is unclear. We note that our most complete analysis, including RNAi rescue, was conducted for Kif1A, and that roles for other kinesins in INM remain to be more definitively ruled out. This is a particular issue for those kinesins which had no effect on INM, but for which biochemical confirmation of knockdown could not readily be performed.
Our model implies that the oppositely directed motor proteins likely act from the nuclear surface to account for the observed centrosome-independent nuclear migration along a parallel array of microtubules. Dynein, dynactin, LIS1, and Ndel1 are known to be recruited to the nuclear envelope during late G2 and early M-phase to pull on microtubules emanating from the centrosome and contribute to nuclear envelope breakdown 42-45. Conceivably, this mechanism for dynein recruitment could be activated early in radial glial progenitor cells to drive nuclear movement toward the ventricular surface. Based on similar arguments, KIF1A most likely acts from the nuclear surface. How this may occur is uncertain, but KIF1A contains a C-terminal pleckstrin homology (PH) domain, with a potential role in membrane binding. Like other such domains, the KIF1A PH domain has been directly demonstrated to interact with membrane lipids 46. Conceivably, this region of KIF1A may play a role in its recruitment to the radial glial nuclear surface to participate in nuclear migration. A recent study reported evidence for dynein and kinesin immunoreactivity associated with neural progenitor or migrating neuronal nuclei 47. How these results may relate to those in the current study remains to be explored.
A particularly fascinating aspect of our model (Fig. S4) is the reciprocal activation of dynein and kinesin pathways during the cell cycle. Migration away from the ventricular surface occurs during G1, whereas migration back to the ventricular surface occurs during G2. We suspect that KIF1A and cytoplasmic dynein are reciprocally recruited to the nuclear surface as a function of cell cycle progression, though alteration in dynein and KIF1A expression or activation could also be involved.
KIF1A RNAi had no effect on mitotic progression or the early stages of neuronal migration (Fig. 5, 6), but resulted in a striking increase the population of Pax6-positive progenitor cells, with a reciprocal decrease in neurons. We suspect this effect results directly from a failure in basally-directed INM. The extent to which the Pax6-positive cells remain pluripotent, and their ultimate fate remain important questions for further investigation.
Materials and Methods
Molecular and Immunonological Reagents
For RNAi, shRNA constructs based on the pRNAT-U6.1/Neo (GenScript, NJ) or pRetro-U6G vectors (Cellogenetics, MD) were used, each of which coexpress GFP with an shRNA. RNAi for the cytoplasmic dynein HC was as reported previously 4. The construct for myosin IIb RNAi (pSuper-MHC2b) was a gift from Dr. Morgan Sheng (MIT)24. For the kinesins screen 3 RNAi sequences were chosen for each kinesin heavy chain gene. Kinesin RNAi screening was done by electroporation in E16 embryos and brains in E20 were fixed and imaged on LSM510 confocal microscope (Zeiss, Germany). The most potent KIF1A shRNA identified in our in utero electroporation assays in addition to KIF1B, and myosin IIB shRNAs were tested for knockdown by immunoblotting in PC12 rat phaeochromocytoma cells plated on collagen I-coated dishes to 60% confluency, and transfected using Lipofectamine 2000 (Invitrogen). Cells were lysed in 50mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 1 mM EGTA with protease inhibitor cocktail for mammalian tissues (Sigma). Monoclonal Anti-KIF1A, (BD Bioscience, CA), polyclonal anti-KIF1B (Bethyl Laboratories, Inc.), polyclonal anti-myosin IIB (Cell Signaling Technology), anti-α tubulin and polyclonal anti-GFP antibodies (Chemicon) were used for immunoblotting. To label the centrosome, the nucleus, the microtubules, DsRed-Centrin II 4, CFP-H1 48 (a gift from Dr. Rafael E. Herrera, Baylor College of Medicine, Houston), GFP-EB3 22 (a gift from Dr. Niels Galjart, Erasmus University, The Netherlands) and YFP-Tubulin constructs were used. Myc-KIF1A was a gift from Dr. Geri Kreitzer at Weill Medical College of Cornell University. DsRed KIF1A was generated from human KIF1A full-length clone (Open Biosystem), ligation to DsRed expression vector with EcoRI and XhoI sites.
In utero electroporation and imaging of radial glial cells in situ
Plasmids or oligonucleotides were transfected by intraventricular injection of embryonic rats in utero followed by electroporation as previously described 5. Animals were maintained according to protocols approved by the Institutional Animal Care and Use Committee at Columbia University. For centrosome and nucleus imaging, coronal slices were prepared 24-48 h after electroporation as described previously 4, 5. Briefly, slices were placed on Millicell-CM inserts (Millipore, MA) in the brain slice culture medium containing 25% Hanks balanced salt solution, 47% Basal modified Eagle's medium, 25% normal horse serum, 1×Pen/Strep/Glutamine (Gibco), and 0.66% glucose, and incubated at 37 °C in 5% CO2. GFP-positive cells were imaged on an inverted microscope DMIRB (Leica) or IX2 (Olympus) with a 40× objective (N.A. = 0.6). Time-lapse images were captured by Coolsnap HQ camera (Roper Scientific) or Electron Multiplier CCD Camera (Hamamatsu) using MetaMorph software (Universal Imaging) at intervals of 10-15 min for 12-18 h. The duration of mitosis was measured from the onset of mitosis when the cell body reaches the ventricular surface and starts to round up to the completion of cytokinesis. For centrosome imaging, cells that contain supernumerary RFP-centrin spots (∼10%) were omitted from our quantitative analysis. The extra spots were judged not to represent centrosomes based on their very rapid and randomly oriented movements 4. For EB3 and microtubule imaging, coronal slices were placed directly on top of a coverslip bottom culture dish (MatTek, MA) and imaged with a high N.A. 40× oil objective (N.A. = 1.3) at intervals of 1 s form 1-2 min as previously described 4. Some of the brains were fixed, sliced imaged with confocal microscopy as previously described for static analysis 4, 5.
Brian slices were imaging for 2-3 h before incubation with 50μM blebbistatin (Calbiochem, San Diego, CA) were used to incubate with brain slices for extra 3 h during time-lapsed recording. Blebbistatin is known to be inactivated when exposure <490nm from previous studies 49, 50. Therefore, after we applied blebbistatin, brain slices were only exposure on 543nm for DsRed imaging.
Immunocytochemistry
Immunocytochemistry was performed as previously described. Rat embryos were perfused transcardially with ice-chilled saline followed by 4% PFA (EMS) and sectioned on a Vibrotome (Leica Microsystems). Primary antibodies were applied overnight at the following concentrations: anti-TuJ1, 1:400 (Covance); 4A4, 1:500 (MBL International Corporation); and anti-Pax6, 1:200 (Covance). Sections were then washed with PBS and incubated in Alexa 546 or 647-conjugated secondary antibodies (1:400; Molecular Probes).
Supplementary Material
Supplemental Movie 1. Dynamics of normal interkinetic nuclear migration of radial glial progenitors.
Supplemental Movie 2. Centrosomal dynamics in radial glial prenitors throughout cell cycle.
Supplemental Movie 3. Centrosomal dynamics in radial glial progenitors throughout cell cycle.
Supplemental Movie 4. Centrosomal dynamics in radial glial progenitors throughout cell cycle.
Supplemental Movie 5. EB3 movements in premitotic radial glial progenitors.
Supplemental Movie 6. EB3 movements in premitotic radial glial progenitors.
Supplemental Movie 7. EB3 movements in radial glial progenitors during mitosis.
Supplemental Movie 8. EB3 movements in radial glial progenitors during cytokinesis.
Supplemental Movie 9. EB3 movements in radial glial progenitors after symmetric cell division.
Supplemental Movie 10. EB3 movements in radial glial progenitors after asymmetric cell division.
Supplemental Movie 11. Behavior of a radial glial progenitor after 5 days of dynein HC RNAi.
Supplemental Movie 12. Behavior of a radial glial progenitor after 1.5 days of dynamytin overepxression.
Supplemental Movie 13. Behavior of a radial glial progenitor after 3 days of dynein HC RNAi.
Supplemental Movie 14. Basally directed nuclear movment of a pair of radial glial progenitor progeny after 3 days of dynein HC RNAi.
Supplemental Movie 15. Basally directed nuclear movement of a radial glial progenitor after 2 days of myosin IIB RNAi.
Supplemental Movie 16. Basally directed nuclear movement of a radial glial progenitor after 2 days of myosin IIB control RNAi.
Supplemental Movie 17. Apically directed nuclear movement of a blebbistatin treated radial glial progenitor cell.
Supplemental Movie 18. Basally directed nuclear movement of a blebbistatin treated radial glial progenitor cell.
Supplemental Movie 19. Nuclear movement of a blebbistatin treated mitotic radial glial progenitor cell.
Supplemental Movie 20. Basally directed nuclear movement of a radial glial progenitor cells after 2 days of RNAi using scrambled KIF1A sequence.
Supplemental Movie 21. Failure in basally directed nuclear movement of a postmitotic radial glial progenitor cell after 2 days of KIF1A RNAi.
Supplemental Movie 22. Differential effects following 2 days of KIF1A RNAi on postmitotic progeny of asymmetrically divided radial glial progenitor cell.
Supplemental Movie 23. Failure of basally directed nuclear movement in postmitotic KIF1A shRNA-/DsRed-co-expressing radial glial progenitor cell.
Supplemental Movie 24. Rescue of basally directed nuclear movement in KIF1A shRNA-/DsRed-human KIF1A-co-expressing radial glial progenitor cell.
Supplemental Movie 25. Almost normal bidirectional nuclear migration behavior in KIF3A shRNA-expressing radial glial progenitor cell.
Supplemental Movie 26. Normal basally directed nuclear migration in KIF1B shRNA-expressing radial glial progenitor cell.
Acknowledgments
We would like to thank Dr. K. Helen Bremner for assistance in surgical procedures, Dr. Xiaoqun Wang for his help in molecular cloning, Geri Kreitzer for reagents, and Drs. Wang, Kreitzer, Wes Gruber, and Julie Canman, Silvia Cappello for very helpful comments and suggestions throughout the project. This work was supported by National Institutes of Health (NIH) grant HD40182 to R. B. Vallee and New York State SCIRB fellowship to J.-W. Tsai.
Footnotes
Author contributions: J.-W. T. and R. B. V. conceived and designed the project. J.-W. T. and W.-N. L. performed most of the experiments. S. K. assayed biochemical effects of RNAi. The study was performed in lab of R. B. V. and revisions in lab of A. R. K.
References
- 1.Solecki DJ, Model L, Gaetz J, Kapoor TM, Hatten ME. Par6alpha signaling controls glial-guided neuronal migration. Nat Neurosci. 2004;7:1195–1203. doi: 10.1038/nn1332. [DOI] [PubMed] [Google Scholar]
- 2.Bellion A, Baudoin JP, Alvarez C, Bornens M, Metin C. Nucleokinesis in tangentially migrating neurons comprises two alternating phases: forward migration of the Golgi/centrosome associated with centrosome splitting and myosin contraction at the rear. J Neurosci. 2005;25:5691–5699. doi: 10.1523/JNEUROSCI.1030-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schaar BT, McConnell SK. Cytoskeletal coordination during neuronal migration. Proc Natl Acad Sci U S A. 2005;102:13652–13657. doi: 10.1073/pnas.0506008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsai JW, Bremner KH, Vallee RB. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat Neurosci. 2007;10:970–979. doi: 10.1038/nn1934. [DOI] [PubMed] [Google Scholar]
- 5.Tsai JW, Chen Y, Kriegstein AR, Vallee RB. LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. J Cell Biol. 2005;170:935–945. doi: 10.1083/jcb.200505166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Solecki DJ, et al. Myosin II motors and F-actin dynamics drive the coordinated movement of the centrosome and soma during CNS glial-guided neuronal migration. Neuron. 2009;63:63–80. doi: 10.1016/j.neuron.2009.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- 8.Sauer FC. Mitosis in the neural tube. J Comp Neurol. 1935;62:377–405. [Google Scholar]
- 9.Webster W, Langman J. The effect of cytochalasin B on the neuroepithelial cells of the mouse embryo. Am J Anat. 1978;152:209–221. doi: 10.1002/aja.1001520204. [DOI] [PubMed] [Google Scholar]
- 10.Messier PE, Auclair C. Inhibition of nuclear migration in the absence of microtubules in the chick embryo. J Embryol Exp Morphol. 1973;30:661–671. [PubMed] [Google Scholar]
- 11.Watterson RL, Veneziano P, Bartha A. Absence of a true germinal zone in neural tubes of young chick embryos as demonstrated by the colchicine technique. Anat Rec. 1956;124:379–380. [Google Scholar]
- 12.Faulkner NE, et al. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat Cell Biol. 2000;2:784–791. doi: 10.1038/35041020. [DOI] [PubMed] [Google Scholar]
- 13.Siller KH, Serr M, Steward R, Hays TS, Doe CQ. Live imaging of Drosophila brain neuroblasts reveals a role for Lis1/dynactin in spindle assembly and mitotic checkpoint control. Mol Biol Cell. 2005;16:5127–5140. doi: 10.1091/mbc.E05-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Del Bene F, Wehman AM, Link BA, Baier H. Regulation of neurogenesis by interkinetic nuclear migration through an apical-basal notch gradient. Cell. 2008;134:1055–1065. doi: 10.1016/j.cell.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Norden C, Young S, Link BA, Harris WA. Actomyosin is the main driver of interkinetic nuclear migration in the retina. Cell. 2009;138:1195–1208. doi: 10.1016/j.cell.2009.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schenk J, Wilsch-Brauninger M, Calegari F, Huttner WB. Myosin II is required for interkinetic nuclear migration of neural progenitors. Proc Natl Acad Sci U S A. 2009;106:16487–16492. doi: 10.1073/pnas.0908928106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi T, Nowakowski RS, Caviness VS., Jr Cell cycle parameters and patterns of nuclear movement in the neocortical proliferative zone of the fetal mouse. J Neurosci. 1993;13:820–833. doi: 10.1523/JNEUROSCI.13-02-00820.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chenn A, Zhang YA, Chang BT, McConnell SK. Intrinsic polarity of mammalian neuroepithelial cells. Mol Cell Neurosci. 1998;11:183–193. doi: 10.1006/mcne.1998.0680. [DOI] [PubMed] [Google Scholar]
- 19.Xie Z, et al. Cep120 and TACCs control interkinetic nuclear migration and the neural progenitor pool. Neuron. 2007;56:79–93. doi: 10.1016/j.neuron.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, et al. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature. 2009;461:947–955. doi: 10.1038/nature08435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mimori-Kiyosue Y, Shiina N, Tsukita S. The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr Biol. 2000;10:865–868. doi: 10.1016/s0960-9822(00)00600-x. [DOI] [PubMed] [Google Scholar]
- 22.Stepanova T, et al. Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein) J Neurosci. 2003;23:2655–2664. doi: 10.1523/JNEUROSCI.23-07-02655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Echeverri CJ, Paschal BM, Vaughan KT, Vallee RB. Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J Cell Biol. 1996;132:617–633. doi: 10.1083/jcb.132.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryu J, et al. A critical role for myosin IIb in dendritic spine morphology and synaptic function. Neuron. 2006;49:175–182. doi: 10.1016/j.neuron.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 25.Miki H, Okada Y, Hirokawa N. Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol. 2005;15:467–476. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 26.Wozniak MJ, Milner R, Allan V. N-terminal kinesins: many and various. Traffic. 2004;5:400–410. doi: 10.1111/j.1600-0854.2004.00191.x. [DOI] [PubMed] [Google Scholar]
- 27.Visel A, Thaller C, Eichele G. GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 2004;32:D552–556. doi: 10.1093/nar/gkh029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niwa S, Tanaka Y, Hirokawa N. KIF1Bbeta- and KIF1A-mediated axonal transport of presynaptic regulator Rab3 occurs in a GTP-dependent manner through DENN/MADD. Nat Cell Biol. 2008;10:1269–1279. doi: 10.1038/ncb1785. [DOI] [PubMed] [Google Scholar]
- 29.Spassky N, et al. Primary cilia are required for cerebellar development and Shh-dependent expansion of progenitor pool. Dev Biol. 2008;317:246–259. doi: 10.1016/j.ydbio.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deacon SW, et al. Dynactin is required for bidirectional organelle transport. J Cell Biol. 2003;160:297–301. doi: 10.1083/jcb.200210066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ligon LA, Tokito M, Finklestein JM, Grossman FE, Holzbaur EL. A direct interaction between cytoplasmic dynein and kinesin I may coordinate motor activity. J Biol Chem. 2004;279:19201–19208. doi: 10.1074/jbc.M313472200. [DOI] [PubMed] [Google Scholar]
- 32.Quintyne NJ, et al. Dynactin is required for microtubule anchoring at centrosomes. J Cell Biol. 1999;147:321–334. doi: 10.1083/jcb.147.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vallee RB, Seale GE, Tsai JW. Emerging roles for myosin II and cytoplasmic dynein in migrating neurons and growth cones. Trends Cell Biol. 2009 doi: 10.1016/j.tcb.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paschal BM, Shpetner HS, Vallee RB. MAP 1C is a microtubule-activated ATPase which translocates microtubules in vitro and has dynein-like properties. J Cell Biol. 1987;105:1273–1282. doi: 10.1083/jcb.105.3.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okada Y, Hirokawa N. A processive single-headed motor: kinesin superfamily protein KIF1A. Science. 1999;283:1152–1157. doi: 10.1126/science.283.5405.1152. [DOI] [PubMed] [Google Scholar]
- 36.Tomishige M, Klopfenstein DR, Vale RD. Conversion of Unc104/KIF1A kinesin into a processive motor after dimerization. Science. 2002;297:2263–2267. doi: 10.1126/science.1073386. [DOI] [PubMed] [Google Scholar]
- 37.Hammond JW, et al. Mammalian Kinesin-3 motors are dimeric in vivo and move by processive motility upon release of autoinhibition. PLoS Biol. 2009;7:e72. doi: 10.1371/journal.pbio.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okada Y, Yamazaki H, Sekine-Aizawa Y, Hirokawa N. The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell. 1995;81:769–780. doi: 10.1016/0092-8674(95)90538-3. [DOI] [PubMed] [Google Scholar]
- 39.Hall DH, Hedgecock EM. Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans. Cell. 1991;65:837–847. doi: 10.1016/0092-8674(91)90391-b. [DOI] [PubMed] [Google Scholar]
- 40.Yonekawa Y, et al. Defect in synaptic vesicle precursor transport and neuronal cell death in KIF1A motor protein-deficient mice. J Cell Biol. 1998;141:431–441. doi: 10.1083/jcb.141.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whited JL, Cassell A, Brouillette M, Garrity PA. Dynactin is required to maintain nuclear position within postmitotic Drosophila photoreceptor neurons. Development. 2004;131:4677–4686. doi: 10.1242/dev.01366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beaudouin J, Gerlich D, Daigle N, Eils R, Ellenberg J. Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell. 2002;108:83–96. doi: 10.1016/s0092-8674(01)00627-4. [DOI] [PubMed] [Google Scholar]
- 43.Salina D, et al. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell. 2002;108:97–107. doi: 10.1016/s0092-8674(01)00628-6. [DOI] [PubMed] [Google Scholar]
- 44.Hebbar S, et al. Lis1 and Ndel1 influence the timing of nuclear envelope breakdown in neural stem cells. J Cell Biol. 2008;182:1063–1071. doi: 10.1083/jcb.200803071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Splinter D, et al. Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLoS Biol. 8:e1000350. doi: 10.1371/journal.pbio.1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klopfenstein DR, Vale RD. The lipid binding pleckstrin homology domain in UNC-104 kinesin is necessary for synaptic vesicle transport in Caenorhabditis elegans. Mol Biol Cell. 2004;15:3729–3739. doi: 10.1091/mbc.E04-04-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang X, et al. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 2009;64:173–187. doi: 10.1016/j.neuron.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Contreras A, et al. The dynamic mobility of histone H1 is regulated by cyclin/CDK phosphorylation. Mol Cell Biol. 2003;23:8626–8636. doi: 10.1128/MCB.23.23.8626-8636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kolega J. Phototoxicity and photoinactivation of blebbistatin in UV and visible light. Biochem Biophys Res Commun. 2004;320:1020–1025. doi: 10.1016/j.bbrc.2004.06.045. [DOI] [PubMed] [Google Scholar]
- 50.Medeiros NA, Burnette DT, Forscher P. Myosin II functions in actin-bundle turnover in neuronal growth cones. Nat Cell Biol. 2006;8:215–226. doi: 10.1038/ncb1367. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Movie 1. Dynamics of normal interkinetic nuclear migration of radial glial progenitors.
Supplemental Movie 2. Centrosomal dynamics in radial glial prenitors throughout cell cycle.
Supplemental Movie 3. Centrosomal dynamics in radial glial progenitors throughout cell cycle.
Supplemental Movie 4. Centrosomal dynamics in radial glial progenitors throughout cell cycle.
Supplemental Movie 5. EB3 movements in premitotic radial glial progenitors.
Supplemental Movie 6. EB3 movements in premitotic radial glial progenitors.
Supplemental Movie 7. EB3 movements in radial glial progenitors during mitosis.
Supplemental Movie 8. EB3 movements in radial glial progenitors during cytokinesis.
Supplemental Movie 9. EB3 movements in radial glial progenitors after symmetric cell division.
Supplemental Movie 10. EB3 movements in radial glial progenitors after asymmetric cell division.
Supplemental Movie 11. Behavior of a radial glial progenitor after 5 days of dynein HC RNAi.
Supplemental Movie 12. Behavior of a radial glial progenitor after 1.5 days of dynamytin overepxression.
Supplemental Movie 13. Behavior of a radial glial progenitor after 3 days of dynein HC RNAi.
Supplemental Movie 14. Basally directed nuclear movment of a pair of radial glial progenitor progeny after 3 days of dynein HC RNAi.
Supplemental Movie 15. Basally directed nuclear movement of a radial glial progenitor after 2 days of myosin IIB RNAi.
Supplemental Movie 16. Basally directed nuclear movement of a radial glial progenitor after 2 days of myosin IIB control RNAi.
Supplemental Movie 17. Apically directed nuclear movement of a blebbistatin treated radial glial progenitor cell.
Supplemental Movie 18. Basally directed nuclear movement of a blebbistatin treated radial glial progenitor cell.
Supplemental Movie 19. Nuclear movement of a blebbistatin treated mitotic radial glial progenitor cell.
Supplemental Movie 20. Basally directed nuclear movement of a radial glial progenitor cells after 2 days of RNAi using scrambled KIF1A sequence.
Supplemental Movie 21. Failure in basally directed nuclear movement of a postmitotic radial glial progenitor cell after 2 days of KIF1A RNAi.
Supplemental Movie 22. Differential effects following 2 days of KIF1A RNAi on postmitotic progeny of asymmetrically divided radial glial progenitor cell.
Supplemental Movie 23. Failure of basally directed nuclear movement in postmitotic KIF1A shRNA-/DsRed-co-expressing radial glial progenitor cell.
Supplemental Movie 24. Rescue of basally directed nuclear movement in KIF1A shRNA-/DsRed-human KIF1A-co-expressing radial glial progenitor cell.
Supplemental Movie 25. Almost normal bidirectional nuclear migration behavior in KIF3A shRNA-expressing radial glial progenitor cell.
Supplemental Movie 26. Normal basally directed nuclear migration in KIF1B shRNA-expressing radial glial progenitor cell.


