Abstract
MLN4924 is a first-in-class experimental cancer drug that inhibits the NEDD8-activating enzyme, thereby inhibiting cullin-RING E3 ubiquitin ligases and stabilizing many cullin substrates. The mechanism by which MLN4924 inhibits cancer cell proliferation has not been defined, although it is accompanied by DNA re-replication and attendant DNA damage. Here we show that stabilization of the DNA replication factor Cdt1, a substrate of Cullins 1 and 4, is critical for MLN4924 to trigger DNA re-replication and inhibit cell proliferation. Even only one hour of exposure to MLN4924, which was sufficient to elevate Cdt1 for 4–5 hours, was found to be sufficient to induce DNA re-replication and to activate apoptosis and senescence pathways. Cells in S phase were most susceptible, suggesting that MLN4924 will be most toxic on highly proliferating cancers. Although MLN4924-induced cell senescence appears to be dependent on induction of p53 and its downstream effector p21Waf1, we found that p53−/−and p21−/− cells were even more susceptible than wild-type cells to MLN4924. Our results suggested that apoptosis, not senescence, may be more important for the anti-proliferative effect of MLN4924. Further, our findings show that transient exposure to this new investigational drug should be useful for controlling p53-negative cancer cells, which often pose significant clinical challenge.
Keywords: MLN4924, re-replication, senescence
Introduction
Duplication of the genetic material is a key event in the cell cycle. In eukaryotes, replication origins are recognized and bound by a six-subunit complex called ORC (Origin Recognizing Complex) (1–3). Cdc6 and Cdt1 are subsequently recruited independently to those sites in late M or early G1 phase (1, 3, 4), followed by the recruitment of MCM2-7 complex to initiate DNA replication (5, 6). It is vitally important that the initiation of replication at replication origins is tightly controlled such that it occurs only once during the cell cycle. Mammalian cells have developed different mechanisms to prevent re-initiation and subsequent re-replication of DNA within the same cell cycle. One such mechanism is the inactivation of Cdt1 during S and G2 phases (7, 8). After replication initiation, Cdt1 is either inhibited by a small protein called Geminin (9, 10) or degraded by two distinct E3 ligases – cdk-dependent SCFskp2 and Cul4-DDB1cdt2 in S or G2/M phase (8, 11). Deregulation of those pathways by depletion of Geminin, Cul4 or Cdt2 activates (or stabilizes) Cdt1 and consequently induces DNA re-replication in different systems (7, 12–14).
Studies have shown that cullin-RING ligases (CRLs), a subclass of E3 ligases that includes both SCFskp2 and CRL4Cdt2, are modified by an ubiquitin-like protein NEDD8, which subsequently facilitates their ligase activities (15–18). Thus, through the modulation of this activity, the NEDD8 pathway regulates the abundance of CRL substrates. MLN4924, a potential cancer drug currently in phase I clinical trials, is a small molecule inhibitor of NEDD8 activating enzyme (NAE) (19, 20). MLN4924 treatment in HCT116 human colon cancer-derived cell line inhibits NAE, and therefore the NEDD8 conjugation pathway, resulting in an increase in protein abundance of CRL substrates such as Cdt1 (21). This is accompanied by an increase in the percentage of cells containing more than 4N DNA, indicating DNA re-replication was occurring. Cells treated with MLN4924 also undergo significant apoptosis contributing to the drug’s anti-proliferative activity. Various CRL substrates play critical functions in cellular growth and survival pathways and the question remained as to which substrates are critical for MLN4924 induced re-replication and apoptosis.
In this paper, we examine whether Cdt1 is the key factor for the induction of DNA re-replication in HCT116 cells treated with MLN4924. Among the different approaches for stimulating Cdt1 activation, MLN4924 shares a similarity with that of Cdt2 depletion in inactivating the CRL4cdt2 E3 ligase, as opposed to Geminin depletion, which activates Cdt1 by a different pathway. We verified this hypothesis and detected a synergistic effect between MLN4924 treatment and Geminin depletion. Transient exposure of cells to MLN4924 led to DNA re-replication, as well as activation of the apoptosis and senescence pathways. This allowed us to test whether a specific part of the cell cycle was particularly susceptible or resistant to MLN4924. Finally, we compared the sensitivity of wild-type (WT) HCT116 cells and isogenic p53−/− or p21−/− HCT116 cells to MLN4924, and discovered that WT HCT116 cells were less susceptible to MLN4924 induced cell death. The results indicate that p53-deficient cancer cells may be more sensitive to MLN4924, emphasizing the therapeutic opportunity with this class of investigational drugs.
Materials and Methods
Cell Lines and Chemicals
Human colorectal cancer cell lines HCT116 (WT, p53−/−, p21−/−) were cultured in McCoy’s 5A modified medium (HyClone) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. Isogenic p21−/− and p53 −/− HCT116 cell lines were described earlier (22). Millennium Pharmaceuticals Inc provided MLN4924, which was then dissolved in DMSO (Sigma). The concentration of Z-Vad-FMK (Calbiochem) used was 50μM. The concentration of Nocodazole (Sigma) used was 40ng/ml.
siRNA
Short interfering (siRNA) oligonucleotides (Invitrogen) were made to the following target sequences (sense): GL2 (control), AACGUACGCGGAAUACUUCGA; Cdt1, GCAAUGUUGGCCAGAUCAA; Cdc6, GAUCGACUUAAUCAGGUAU; Mcm7, GAUGUCCUGGACGUUUACA; Geminin (Gem), UGCCAACUCUGGAAUCAAA (12); Cdt2, GAAUUAUACUGCUUAUCGA. Transfections were performed with 20nM siRNA oligonucleotide duplexes with Lipofectamine RNAiMAX (Invitrogen) according to the instructions of manufacturer.
Antibodies and immunoblotting
Rabbit anti Cdt1, rabbit anti-geminin, and rabbit anti-Cdt2 were raised as described (9, 23). The purchased antibodies were mouse anti-p21 (Lab vision/Neomarkers); mouse anti-β-actin, mouse anti-Chk1, mouse anti-Chk2 (Sigma); rabbit anti-Chk1-P-S317, rabbit anti-Chk2-P-T68; rabbit anti-PARP, rabbit anti-H3-P-S10 (Upstate). Cells were lysed as described (24), and western blot analysis was performed according to standard procedures.
Flow Cytometry Analysis (FACS)
Cells were harvested by trypsinization and fixed with 70% ethanol overnight at −80°C. Cells were then stained and analyzed as described before (12). For FACS analysis with both PI and BrdU double staining, cells were labeled with 10μM BrdU (Sigma) and then harvested as described earlier (12).
Timelapse Movie analysis
HCT116 cells were plated at 15,000 cells per well in 6-well culture plates (Becton Dickinson). For continuous treatment cells were treated with 1μM MLN4924 for 72 hours. For wash-out treatment HCT-116 cells were treated with 1μM MLN4924 for 8 hours then washed with fresh media to remove compound and maintained in fresh compound free media for 8 days. Timelapse movie images were taken at times indicated using an automated TE2000U microscope (Nikon Instruments, Melville, NY) with Hoffman-modulation optics, 20x objective, with environmental control, and an Orca-ER CCD camera (Hamamatsu, Bridgewater, NJ) controlled with MetaMorph imaging software (Molecular Devices, Downingtown, PA).
Measure Cell Growth and Clonogenicity
The number of viable cells was estimated with a cell proliferation assay (MTT) kit (Promega) according to the manufacturer’s instructions. Cells were seeded into 96-well plates at 500 cells per well, treated with DMSO or MLN4924 and incubated for 7 days before MTT assay. Cell clonogenecity assay was performed as described (25). Cells were seeded into 6-well plates at 3*103 cells per well. DMSO or 1μM MLN4924 were added for 8 hours. Cells were washed twice with PBS and incubated in fresh medium after the wash-out. Medium was changed every 2–3 days and the Colonies were stained with crystal violet to show cell clonogenicity. OD595 was measured to quantify cell colony numbers and normalized to DMSO treated control sample to obtain cell survival rate.
SA-β-gal Staining Assay for Senescence
Senescence β-galactosidase staining assay was performed in a 6-well plate with staining kit (Cell Signaling Technology, #9860). Cells were washed with PBS, fixed and stained following manufacturer’s instruction. Stained plates were checked under a microscope for development of blue color. For each sample, SA-β-gal positive and total cell numbers were counted from 5 different microscopic fields (roughly >200 cells per field).
Results
Stabilization of Cdt1 protein is critical for MLN4924-induced re-replication in HCT116 cells
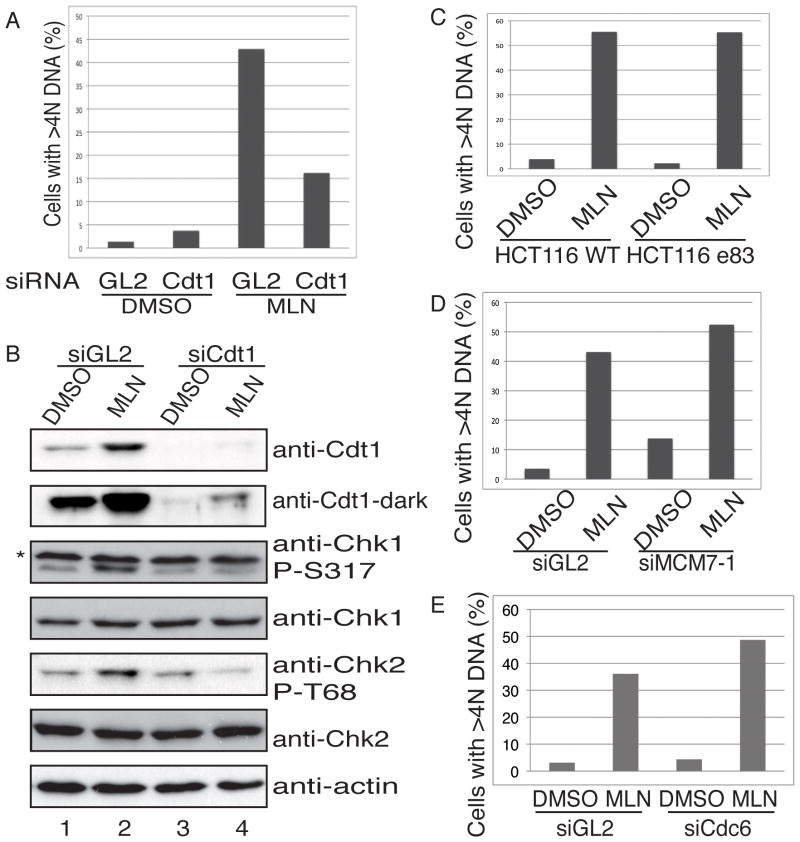
Consistent with previous results (21), we observed re-replication after 20 hours of treatment of HCT116 cells with MLN4924 (Fig. 1A). To investigate whether the regulation of Cdt1 protein level plays a role in MLN4924-induced re-replication, HCT116 cells were treated with siRNA oligonucleotides targeting Cdt1 for 48 hours prior to the addition of MLN4924. After 20 hrs of MLN4924 treatment, Cdt1 protein level increased significantly as expected (Fig. 1B, lane 1,2) and more than 40% of cells were determined to have re-replicated, containing >4N DNA content. However, in the cells depleted of Cdt1 by siRNA, the percentage of re-replicating cells reduced to 15% (Fig. 1A). In these cells, Cdt1 protein expression was effectively repressed (Fig. 1B, lane 1,3 and 4), although at a higher exposure, Cdt1 protein level was observed to be modestly induced in MLN4924 treated cells (Fig. 1B, lane 3,4) indicating that the drug was still inhibiting its degradation, potentially explaining the 15% of cells with >4N DNA.
Figure 1. Cdt1 protein level is important for MLN4924 induced re-replication in HCT116 cells.
(A) HCT116 cells were transfected twice with siGL2 or siCdt1 at 0- and 24-hour time point and incubated for a total of 48 hours before the addition of 0.3μM MLN4924 or DMSO. Cells were harvested for PI FACS after 20 hours of treatment. The percentage of cells containing >4N DNA was shown. (B) Total cell lysates from (A) were blotted with indicated protein antibodies. (*: non-specific band) (C) HCT116 WT or e83 cells were treated with DMSO or 0.3μM MLN4924 for 20 hours before harvested for FACS. The percentage of cells containing >4N DNA was shown. (D) Similar assay as described in (A) was performed with siMcm7. The percentage of cells containing >4N DNA was shown. (E) Similar assay as in (A) was performed with siCdc6.
Re-replication has been shown to induce both single-strand and double-strand DNA breaks, resulting in activation of checkpoint pathways (24, 26). Indeed, we observed both Chk1 and Chk2 phosphorylation in MLN4924 treated cells (Fig. 1B, lane 1, 2). Cdt1 siRNA treatment not only decreased the percentage of cells undergoing re-replication, but also decreased the activation of Chk1 and Chk2 (Fig. 1B, lane 2, 4), indicating that MLN4924 induces DNA damage primarily through Cdt1-dependent re-replication.
To characterize whether other replication initiators contribute to the re-replication induced by MLN4924, we systematically depleted other components of the pre-RC. We treated HCT116 cells hypomorphic for ORC2 (27) with MLN4924 and compared the extent of re-replication with that of WT cells. As shown before, although we could detect ORC2 in 6ug of extract from WT HCT116 cells, it was hard to detect ORC in even 60μg of e83 HCT116 cells (Fig. S1A). Despite this, there was no significant difference in the amount of re-replication between these cell types (Fig. 1C). Furthermore, we performed siRNA knockdown of both MCM7 and Cdc6 (Fig. S1B, S1C) and observed no difference in the amount of MLN4924 induced re-replication in both cases (Fig. 1D, 1E). Interestingly, we noticed that MLN4924 treatment could also induce Cdc6 protein expression (Fig. S1C), which indicated that Cdc6 could be a potential CRL substrate. Overall, these results demonstrate that the ubiquitin-dependent degradation of Cdt1 protein is the rate-limiting step in preventing re-replication, and that stabilization of this component of the pre-RC by MLN4924 induces re-replication in HCT116 cells. The re-replication leads to DNA damage and activates checkpoint pathways.
MLN4924 induces re-replication through the inhibition of CRL4Cdt2
Because MLN4924 functions as a NAE inhibitor (21), it is expected to inhibit all cullins, including Cul1 and Cul4 ubiquitin ligases known to degrade Cdt1 (28–32). Cdt2 depletion, and thus inactivation of CRL4Cdt2, in zebra fish, xenopus egg extracts and human cancer cells induces DNA re-replication (13, 33). If CRL4cdt2 inhibition is the primary mechanism by which MLN4924 causes re-replication, one would predict that there should be no synergy between MLN4924 and siCdt2 in induction of re-replication.
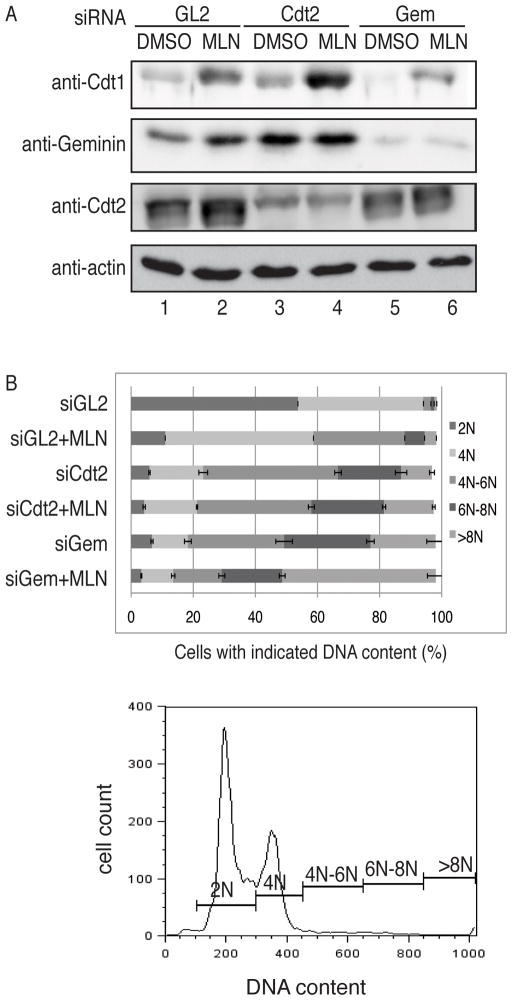
To determine whether MLN4924 acts through the same mechanism to induce re-replication as that of Cdt2 depletion, we compared the Cdt1 protein level in cells depleted of Cdt2 by siRNA or treated with MLN4924. As seen in Figure 2A, Cdt2 depletion caused less Cdt1 accumulation than MLN4924 alone (Fig. 2A, lane 2 and 3) and together the two stabilized Cdt1 more (lane 4). However, the extent of re-replication caused by siCdt2 was more than that observed in cells treated with MLN4924 alone, and adding the two together did not increase re-replication (Figure 2B). This suggests that (a) pure CRL4Cdt2 inhibition with siCdt2 is more effective at inducing re-replication than inhibiting all cullins by MLN4924, but (b) once Cdt1 level has crossed a certain threshold, there is no further increase in re-replication with more Cdt1. In addition, other substrates/pathways affected by MLN4924 may make cells die, thereby decreasing the extent of re-replication observed.
Figure 2. MLN4924 induces re-replication through inhibition of CRL4Cdt2.
(A) HCT116 cells were tranfected with GL2, Cdt2, Geminin siRNA and treated with MLN4924 as described in Fig. 1(A). Cell lysates were harvested and blotted with indicated antibodies. (B) DNA contents of the cells treated in (A) were determined using FACS and plotted in horizontal bar graph. Representative FACS data from siGL2 treated cells indicating different DNA contents measured.
In contrast to the lack of synergy in induction of re-replication by siCdt1 + MLN4924, siGeminin (to activate Cdt1 in S through G2 phase) + MLN4924 caused more re-replication than either agent alone. This is particularly evident when one consider the proportion of cells with >6N DNA content (Fig. 2B).
These results suggest that removal of an inhibitor of Cdt1 (Geminin) will act additively with stabilization of Cdt1 by MLN4924 to cause more re-replication. In contrast, inhibition of CRL4cdt2 by siCdt2 and MLN4924 does not additively cause more re-replication, even though there was more stabilization of Cdt1. This result is consistent with the hypothesis that MLN4924 causes re-replication primarily through the inhibition of CRL4cdt2.
Transient exposure of HCT116 cells to MLN4924 induces re-replication
One advantage of using MLN4924 treatment to induce re-replication lies in its ability to act rapidly. Previous studies have shown that in as little as 5 minutes following MLN4924 treatment, the NEDD8 pathway is inhibited concurrent with the accumulation of Cdt1 protein (20, 21). Therefore we wanted to test whether transient treatment of MLN4924 is sufficient to induce re-replication in HCT116 cells.
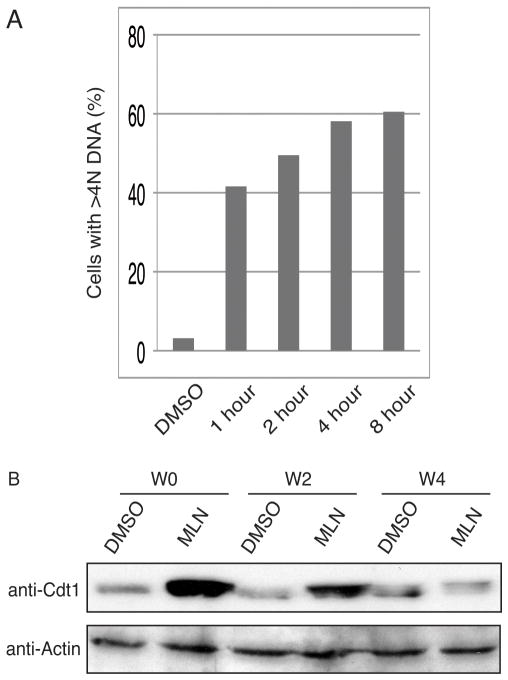
Surprisingly, as shown in Fig. 3A, one-hour exposure to MLN4924 was sufficient to induce re-replication in 40% of the cells. The percentage of cells with re-replication increased with longer pulse of MLN4924, but was close to its maximum after 4 or 8 hours of treatment, with 60 % of cells re-replicating their DNA.
Figure 3. Transient exposure to MLN4924 induces re-replication in HCT116 cells.
(A) HCT116 cells were treated with DMSO or 1μM MLN4924 for indicated hours. Cells were washed with PBS twice, incubated in fresh medium and harvested 24 hours after initial addition of the chemicals. Percentage of cells containing >4N DNA contents was plotted. (B) HCT116 cells were treated with DMSO or 1μM MLN4924 for 4 hours. Cells were then washed and harvested at 0, 2, 4 and 20 hours after the wash-out as indicated. Cell lysates were blotted with Cdt1 or Actin antibodies.
To determine the rate of Cdt1 turnover following MLN4924 wash-out, we treated HCT116 cells with MLN4924 for 4 hours, and harvested the cells after different time periods. Cdt1 protein level increased following MLN4924 treatment (Fig. 3B, W0). However, by 4 hours after wash-out, Cdt1 level decreased to basal levels (Fig. 3B, W4). These results indicate that although Cdt1 stability returns to normal in roughly 4 hours after the removal of MLN4924, this short period of Cdt1 stabilization is sufficient to induce irreversible DNA re-replication.
S phase cells are more susceptible to MLN4924 induced re-replication
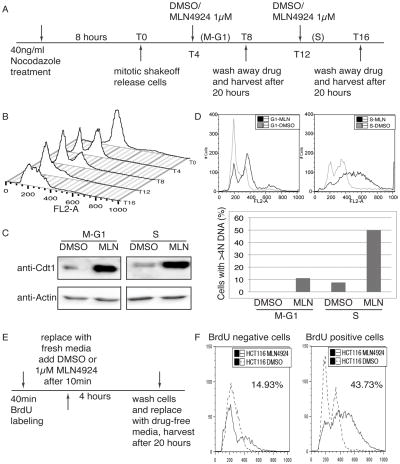
Since a mere 4-hour pulse treatment of MLN4924 leads to re-replication, we decided to test which portion of the cell cycle was more susceptible to the drug exposure. HCT116 cells synchronized by nocodazole block/mitotic shake-off were exposed to MLN4924 at the indicated times post-mitosis (Fig. 4A). Cells were then collected for flow cytometry analysis (FACS). As shown in Fig. 4B, cells started to enter G1 around 4 hours after nocodazole release and began S phase after approximately 12 hours. This suggests that the majority of the cells were in M-G1 at T4 and in either S phase or at the G1/S transition at T12. We saw a similar increase in Cdt1 protein level after MLN4924 treatment in both populations (Fig. 4C). However, whereas only 10% of the cells re-replicated when Cdt1 was stabilized in M-G1 phase cells, 50% of the cells showed re-replication upon stabilization of Cdt1 in S phase cells (Fig. 4D). These results demonstrate that S phase cells are more susceptible to MLN4924-induced DNA re-replication.
Figure 4. S phase cells are more susceptible to MLN4924 induced re-replication.
(A) Schematic of experimental procedures of (B) to (D). (B) FACS profiles of control samples harvested at indicated time points. (C) Total lysates from cells harvested 20 hours after the drug wash-out were blotted with indicated antibodies. (D) FACS profiles from above cells were shown. The percentage of cells with >4N DNA is plotted in the bar graphs below. (E) Schematic of experiment of (F). (F) FACS profiles were shown as indicated. (Dashed line: DMSO; solid line: MLN4924) Percentages of re-replicating cells after MLN4924 treatment are indicated.
To ask whether cells must be in S-phase for MLN4924 to induce re-replication, we labeled the cells with bromodeoxyuridine (BrdU) for 40 minutes to mark cells in active S phase, washed out BrdU and added MLN4924 for 4 hours. Cells were collected for FACS after 20 hours (Fig. 4E). BrdU positive cells, which were actively replicating when exposed to MLN4924, showed 44% of cells re-replicating, whereas only 14% of the BrdU negative cells re-replicated (Fig. 4F). These results further demonstrate that actively replicating cells are more susceptible to MLN4924–induced re-replication.
Both checkpoint and apoptosis pathways are activated upon short exposure of cells to MLN4924
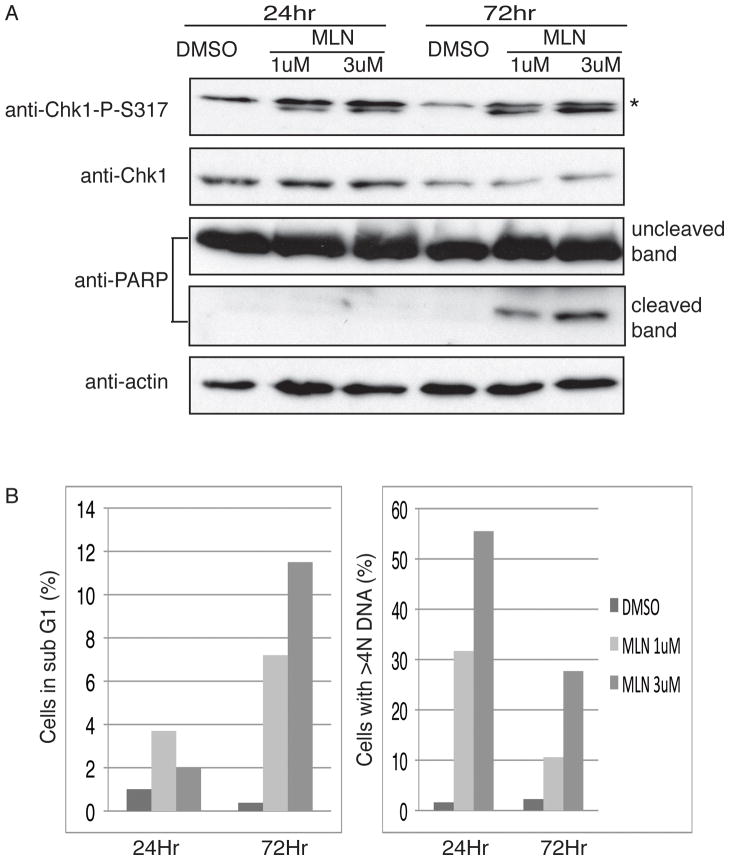
Re-replication induces DNA damage and checkpoint activation (24, 26, 34). The initiation of DNA re-replication by a short exposure to MLN4924 led us to test whether checkpoint pathways were similarly activated in those cells. We treated HCT116 cells with MLN4924 for 8 hours and harvested cells 24 or 72 hours after wash-out. Chk1 was activated 24 hours after drug wash-out while DNA re-replication was seen in 30 to 55% of cells (at 1 and 3μM MLN4924, respectively). The DNA damage checkpoint pathway still persisted even at 72 hours after wash-out, when re-replication was observed in 10 to 25% of cells (Fig. 5A, 5B). In addition, we noticed that PARP cleavage happened only at the later time point, suggesting that apoptosis was not activated until 72 hours after wash-out. This was further confirmed by the increase of sub-G1 population cells (Fig. 5B). Overall, these results were consistent with the idea that even transient exposure of MLN4924 leads to re-replication, activates checkpoint pathways and eventually induces apoptosis following irreparable DNA damage.
Figure 5. Both checkpoint and apoptosis pathways are activated upon short exposure of cells to MLN4924.
(A) HCT116 cells were treated with DMSO, 1μM or 3μM MLN4924 for 8 hours. Cells were harvested 24 or 72 hours after the drug wash-out. Cell lysates were blotted with indicated protein antibodies. (*: nonspecific band) (B) Cells from above were harvested for PI FACS. Percentages of cells containing <2N (left) or >4N DNA (right) are shown.
Senescence is induced after transient exposure to MLN4924 through the induction of p53 and p21
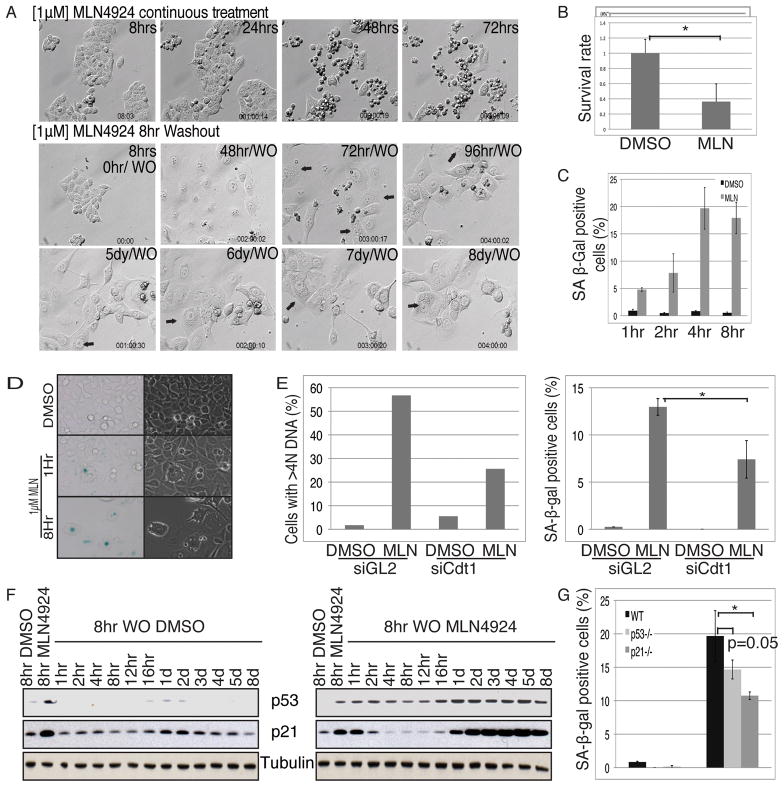
When culturing cells after transient exposure to MLN4924, we noticed changes in cell morphology starting approximately 72 hours post-wash-out, including an increase in cell size, intracellular vesicle accumulation and flatness. As shown in the upper panel of Fig. 6A, after 48 hours or more of continuous exposure to MLN4924, cells shrank and became round, suggesting those cells were undergoing apoptosis. However, after a transient 8-hour exposure to MLN4924, cells exhibited the flattened, vesiculated morphology, often noticed when cellular senescence pathway is activated.
Figure 6. The senescence pathway is induced in HCT116 cells after transient exposure to MLN4924.
(A) HCT116 cells were treated with 1μM MLN4924 continuously (top panel) or only for 8 hours (middle and lower panels). Movie images of the cells at indicated time points are shown. (B) HCT116 cells were treated as described in the text. Cell survival rate in the colony formation assay is shown. Error bar represents three independent experiments. (C) HCT116 cells were treated with 1μM MLN4924 for indicated hours before wash-out. SA β-Gal staining assay was then performed after 72 hours. Positive stained cells were counted and plotted as percentage of total cell numbers. Mean±standard deviation of three different experiments. (D) Representative SA-β-gal staining for indicated samples. (E) HCT116 cells were transfected twice with siGL2 or siCdt1 as described in Figure 1(A). 48 hours after initial transfection, cells were treated with 1μM MLN4924 for 8 hours. Cells were either harvested for FACS analysis after 24 hours, or subjected to SA-β-gal staining assay after 72 hours. (F) HCT116 cells were treated with 1μM MLN4924 8 hours. Cells were harvested for western blots of p53 and p21 at different time points after wash-out. (G) HCT116 WT, p53−/− or p21−/− cells were treated with 1μM MLN4924 for 8 hours. SA-β-gal staining assay was performed 72 hours after the wash-out. Percentage of positive stained cells is shown. Mean±standard deviation of 3 experiments. * indicates statistical significance (p<0.01).
Senescence is marked by permanent withdrawal from the cell cycle. To test whether MLN4924 induces senescence, we first performed colony formation assays to determine the clonogenicity of the cells (35). We added MLN4924 to HCT116 cells for 8 hours and cultured cells for 7 days after wash-out for colony formation as measured by crystal violet staining (Fig. S2A). Quantitation of the optical density of staining (Fig. 6B) showed that MLN4924 treatment suppressed the clonogenicity, a characteristic of senescent cells.
Senescence Associated β-gal (SA-β-gal) staining is a well-accepted biomarker of senescence (36). Transient treatment of cells with MLN4924 increased the percentage of re-replicating cells (Fig. 3A) and the percentage of SA-β-gal staining (Fig. 6C, 6D). This suggested that the senescence pathway was activated upon transient exposure to MLN4924. These results were consistent with the earlier findings that re-replication can activate the DNA damage response leading to cellular senescence (37).
We next examined whether reduction of re-replication by Cdt1 depletion could decrease senescence following MLN4924 treatment. We performed a similar assay as that displayed in Fig. 1A, except that the HCT116 cells were exposed to MLN4924 for only 8 hours and cells collected for FACS after 24 hours or SA-β-gal staining after 72 hours (Fig. 6E). Consistent with our hypothesis, Cdt1 depletion reduced both re-replication and senescence to a similar degree. Together, these data suggest that re-replication induced by transient exposure to MLN4924 leads to senescence. 8 hr exposure to MLN4924 also induced apoptosis, as measured by the cleavage of PARP (Fig. 5A). Thus transient treatment with MLN4924 induces senescence or apoptosis (also evident in Fig. 6A, 72hr/WO), while continuous treatment with the drug leads mostly to apoptosis (Fig. 6A, 72 hrs).
Both p53 and p21 can have a function in the cellular senescence pathway (38, 39). We therefore examined protein expression levels of p53 and p21 over 8 days following an 8-hour treatment with MLN4924. Both p53 and p21 were induced 24 hour post-wash-out and their expression persisted thereafter for the entire time course (Fig. 6F). We then performed the SA-β-gal staining assay in p53−/− or p21−/− HCT116 cells to determine the level of senescence in the absence of these proteins. As shown in Fig. 6G, the number of SA-β-gal stained cells was only half in the p21−/− HCT116 compared to those of WT HCT116, indicating that p21 plays an important role in the senescence pathway. p53 appeared to be less essential than p21, which was consistent with results from other studies (40). The p16 gene is silenced in these cells (41), so the residual senescence in the p21−/− cells was most likely by a p21- and p16- independent pathway.
p21 and p53 deficient HCT116 cells are more sensitive to transient treatment with MLN4924
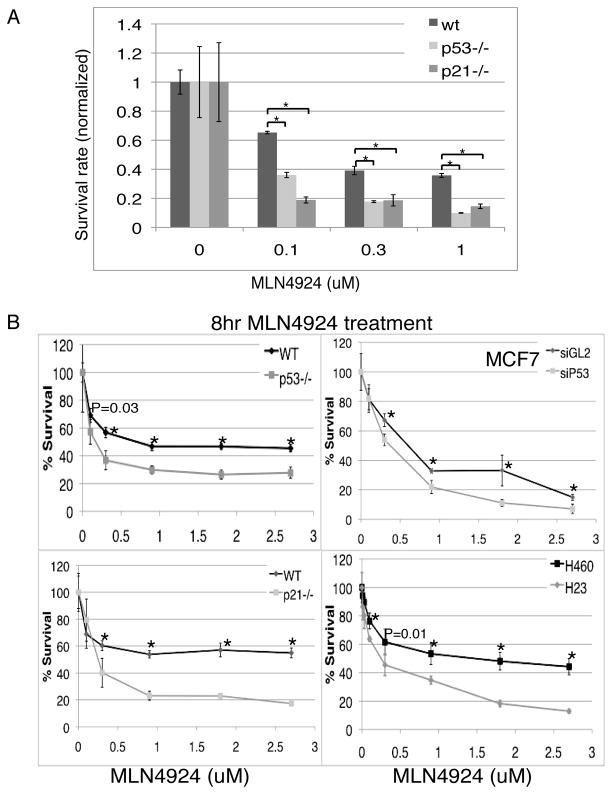
Since transient exposure to MLN4924 causes both senescence and apoptosis, but senescence is attenuated in the p21−/− cells, we could ask how important is the senescence for the toxicity of MLN4924 on cancer cells. We noticed that although p21- and p53-deficient cells exhibited less senescence after transient treatment with MLN4924, the total cell numbers observed were much less than in WT cells, which suggested that p53−/−or p21−/− HCT116 cells might be more susceptible to overall cell death or growth inhibition by MLN4924. Colony formation assays following an 8-hour treatment of MLN4924 in all three cell-lines confirmed this (Fig. 7A). Consistent with our previous results (Fig. 6B), colony formation was decreased by MLN4924 treatment in a dose-dependent manner in WT HCT116 cells, but the p53- and p21- deficient cells formed fewer colonies than WT cells, indicating that the absence of p53 or p21 sensitized the cells to the drug treatment. Since senescence is attenuated in the mutant cells (Fig. 6G), the result suggests that apoptosis pathways are important for cell killing after transient exposure to MLN4924.
Figure 7. p53 mutant cells are susceptible to transient treatment with MLN4924.
(A) HCT116 WT, p53−/− or p21−/− cells were treated with 0, 0.1, 0.3, 0.9, 1.8 or 2.7μM of MLN4924. Cell survival rates were measured as described in Figure 6(B). Mean and standard deviation from triplicates. * indicates statistical significance (p<0.01). (B) Viable HCT116, MCF7 (MLN4924 treatment 24hrs after control or p53 siRNA), H460 and H23 cells after 8hr MLN4924 treatment at different doses were measured using MTT assay as described. The points indicate mean and standard deviation of triplicates. * indicates statistically significant difference at various MLN4924 concentrations between the two cell lines (p<0.01).
We also performed an MTT cell growth assay to compare cell survival rate upon either transient or 72-hour continuous exposure to MLN4924. After 8-hour treatment, the IC50 for WT, p53−/− and p21−/− cells were 0.9μM, 0.18μM and 0.25μM, respectively (Fig. 7B), which was consistent with the results of the colony formation assays. However, the difference was much smaller upon 72-hour continuous exposure: IC50 of 0.08μM, 0.07μM and 0.07μM in WT, p53−/− and p21−/− cells, respectively (Fig. S4A).
To confirm that cells with mutant p53 were more susceptible to cell death by MLN4924, we performed MTT assay in MCF7 cells after MLN4924 transient exposure. After p53 knockdown, IC50 decreased from 0.68μM to 0.4μM (Fig 7B, S4B). Similar results were obtained when we compared IC50 in two lung cancer cells NCI-H23 (p53 mutant, IC50 0.28μM) and NCI-H460 (wild type p53, IC50 1.5μM) (Fig 7B). Intriguingly, when we compared IC50 in more than 20 cancer cell lines upon 72hr MLN4924 treatment, after dividing them into p53 WT (wild type) group and p53 MT (mutant) group, we found the median value of WT group was significantly higher than MT group (465nM vs. 280nM, Fig. S4C) despite all the different genetic backgrounds. It must be noted, however, that there areother genetic factors that affect MLN4924 sensitivity besides p53 status. For example we obtained opposite results when comparing MCF7 (WT p53) with MDA-MB-231 (MT p53) cells where the MCF7 cells were more susceptible to MLN4924 (Fig. S4D). Despite this exception, our data suggests that p53 mutant cells are generally more susceptible to MLN4924.
Thus there is a clear therapeutic advantage of transient MLN4924 treatment, particularly considering that the p53 mutant cells are more susceptible to cell death than the p53 WT cells. Since up to 50% of human tumors have a mutant p53 gene, our results suggest that the kinetics of MLN4924 administration might alter the therapeutic index.
Discussion
As a potential anti-cancer drug, MLN4924 was discovered to inhibit NAE activity, inhibit cullins, increase the expression of CRL substrates, induce re-replication and cause cell death (21). In this paper, we demonstrated that the regulation of Cdt1 protein level is the rate-limiting step for the induction of re-replication upon MLN4924 treatment. It is noteworthy that even transient exposure of HCT116 colon cancer cells to MLN4924 leads to DNA re-replication. Once re-replication is induced, DNA damage checkpoint pathways are activated, which then lead to apoptosis and cellular senescence. We found that p53−/− and p21−/− HCT116 cells are both more sensitive to MLN4924 exposure than wild type cells, indicating that cancer cells with p53 mutations, are likely more susceptible to transient exposure to the drug.
Various CRL substrates accumulate upon MLN4924 treatment, including Cdt1, p27, NRF2(21) and possibly Cdc6 (Fig. S1C). However, our data suggests that the deregulation of Cdt1 protein level plays an essential role in DNA re-replication induction, demonstrated by the decline in re-replication when Cdt1 is knocked down. That depletion of Orc2, MCM7 and Cdc6 did not prevent re-replication should not be interpreted to say that pre-RC components are not required for re-replication. The more likely hypothesis is that these proteins are in vast excess and so do not become rate limiting for re-replication after siRNA depletion.
We noticed a high G2 peak and a residual 15% of cells re-replicating after MLN4924 treatment in Cdt1 depleted cells (Supplementary Fig. S1C). The 15% of cells labeled as re-replicating could arise from the tail of the large G2/M peak observed and may not be real re-replication that leads to DNA damage, as there was no activation of either Chk1 or Chk2 in these cells (Fig. 1B). Taken together, these data show that MLN4924 cause a G2/M arrest, consistent with the report that siCdt2 can induce G2/M arrest (13). This hypothesis was further confirmed by the increased phosphorylation of Cdc2 on Y15 and the loss of phosphorylation of H3 on S10 in cells treated with MLN4924 (Supplementary Fig. S1E), indicating that cells cannot enter mitosis. None of these changes, increase in G2 population, increase in Cdc2-P-Y15, and decrease of H3 phosphorylation, were relieved by decreasing Cdt1. Thus unlike re-replication, the G2/M block seen with MLN4924 may be due to stabilization of substrates other than Cdt1.
In vivo data suggested that Cdt1 protein level peaked at 2–4 hours after injection of MLN4924 into tumor-bearing mice and started to decrease by 4–8 hours post-injection (21). Therefore we wished to evaluate the effect of transient exposure of cancer cells to MLN4924. Amazingly even one-hour exposure was sufficient to induce re-replication in 40% of a colon cancer cell population in culture. With short treatment, we discovered that S phase cells were more susceptible to MLN4924 induced re-replication, which is consistent with the idea that S phase cells have already licensed origins (and fired many of them), so that relicensing by transient stabilization of Cdt1 would cause re-replication. The observations that transient exposure can lead to re-replication and S phase cells are more susceptible to this exposure are positive indicators for the clinical usefulness of this compound.
In addition to activation of apoptosis, we observed activation of senescence pathway after transient exposure of MLN4924. As previously stated, this was not due to a reduction in re-replication, as HCT116 cells displayed an equivalent increase in cells with a >4N DNA content even after short treatment with MLN4924 compared to continuous treatment (Fig. 3). This re-replication subsequently led to DNA damage and activated checkpoint and apoptosis pathways (Fig. 5). Unexpectedly, we observed that the senescence phenotype did not appear in continuously treated cells (Fig. 6A). Although there was barely any difference in the extent of re-replication between the two treatments, DNA damage signals (DDS) were possibly different, resulting in a different choice of cell fate between apoptosis and senescence (42). Upon short exposure, no new DDS occurred from persistent origin re-firings, which likely occurred in the continuously treated cells. This lower level of DNA damage signaling perhaps is not great enough in duration or extent to trigger cells apoptosis, though it is sufficient to induce p21 and p53.
One remaining question is what activates senescence. Is it related to re-replication induced DNA damage? Previous papers suggested DNA damage caused by re-replication could activate the senescence pathway (37, 43, 44). In our hands, depletion of Geminin or Emi1 in HCT116 cells similarly induced senescence after 3–4 days (data not shown). Consistent with this idea, decrease in re-replication by depletion of Cdt1 reduced cellular senescence (Fig. 6E). Thus the senescence is triggered by the re-replication induced DNA damage. Another intriguing question is, once cell fate has been determined, is it reversible? We treated cells with Z-VAD-FMK together with MLN4924 to inhibit cells from entering apoptosis (Supplementary Fig. S3). However, there was no significant increase in senescence, which suggested an irreversible commitment to apoptotic, non-senescent pathways.
It has already been suggested that p53 and p21 level are increased during cellular senescence (45, 46). Multiple studies have shown that the p53-p21 pathway is critical for senescence to occur in human fibroblasts and cancer cells (38, 42, 47, 48). However, some researchers have observed that although p53 and p21 are positive factors in senescence they are not necessary (49). Our results suggest that p53 and p21 have important functions in initiating cellular senescence upon MLN4924 treatment in tumor cells, but they are dispensable given that p53−/− or p21−/− cells showed decreased but not absent SA-β-gal staining (Fig. 6G).
Although both p53−/− and p21−/− HCT116 cells underwent less senescence than WT cells, both were more susceptible to cell death after transient treatment with MLN4924 (Fig. 7), suggesting a shifting of the balance towards a more apoptotic phenotype upon intermittent treatment in those cells. This p53-independent susceptibility to MLN4924 is potentially critical for clinical applications, where nearly half of human tumors have mutated their p53 gene. Conventional chemotherapy is less effective in p53 mutant cells. Thus MLN4924 is exceptional in its ability to target p53 mutant tumors.
Supplementary Material
Acknowledgments
This work was supported by the Research Project Grant from National Institutes of Health (R01-CA60499 and CA89406). Jie Lin is supported by the Predoctoral Traineeship Award from Department of Defense Breast Cancer Research Program (W81XWH-08-1-0286).
We are grateful to Dr. Fred Bunz for providing us isogenic p21−/− and p53−/− HCT116 cells. We wish to thank members of the Dutta lab for reading the manuscript and helpful discussions.
References
- 1.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–74. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 2.Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–34. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 3.Dutta A, Bell SP. Initiation of DNA replication in eukaryotic cells. Annu Rev Cell Dev Biol. 1997;13:293–332. doi: 10.1146/annurev.cellbio.13.1.293. [DOI] [PubMed] [Google Scholar]
- 4.Lei M, Tye BK. Initiating DNA synthesis: from recruiting to activating the MCM complex. J Cell Sci. 2001;114:1447–54. doi: 10.1242/jcs.114.8.1447. [DOI] [PubMed] [Google Scholar]
- 5.Maiorano D, Moreau J, Mechali M. XCDT1 is required for the assembly of pre-replicative complexes in Xenopus laevis. Nature. 2000;404:622–5. doi: 10.1038/35007104. [DOI] [PubMed] [Google Scholar]
- 6.Nishitani H, Lygerou Z, Nishimoto T, Nurse P. The Cdt1 protein is required to license DNA for replication in fission yeast. Nature. 2000;404:625–8. doi: 10.1038/35007110. [DOI] [PubMed] [Google Scholar]
- 7.Arias EE, Walter JC. Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 2007;21:497–518. doi: 10.1101/gad.1508907. [DOI] [PubMed] [Google Scholar]
- 8.Kim Y, Kipreos ET. Cdt1 degradation to prevent DNA re-replication: conserved and non-conserved pathways. Cell Div. 2007;2:18. doi: 10.1186/1747-1028-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wohlschlegel JA, Dwyer BT, Dhar SK, Cvetic C, Walter JC, Dutta A. Inhibition of eukar yotic DNA replication by geminin binding to Cdt1. Science. 2000;290:2309–12. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- 10.Tada S, Li A, Maiorano D, Mechali M, Blow JJ. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat Cell Biol. 2001;3:107–13. doi: 10.1038/35055000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Machida YJ, Hamlin JL, Dutta A. Right place, right time, and only once: replication initiation in metazoans. Cell. 2005;123:13–24. doi: 10.1016/j.cell.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 12.Zhu W, Chen Y, Dutta A. Rereplication by depletion of geminin is seen regardless of p53 status and activates a G2/M checkpoint. Mol Cell Biol. 2004;24:7140–50. doi: 10.1128/MCB.24.16.7140-7150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell. 2006;23:709–21. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 14.Zhong W, Feng H, Santiago FE, Kipreos ET. CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature. 2003;423:885–9. doi: 10.1038/nature01747. [DOI] [PubMed] [Google Scholar]
- 15.Gong L, Yeh ET. Identification of the activating and conjugating enzymes of the NEDD8 conjugation pathway. J Biol Chem. 1999;274:12036–42. doi: 10.1074/jbc.274.17.12036. [DOI] [PubMed] [Google Scholar]
- 16.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 17.Read MA, Brownell JE, Gladysheva TB, et al. Nedd8 modification of cul-1 activates SCF(beta(TrCP))-dependent ubiquitination of IkappaBalpha. Mol Cell Biol. 2000;20:2326–33. doi: 10.1128/mcb.20.7.2326-2333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiba T, Tanaka K. Cullin-based ubiquitin ligase and its control by NEDD8-conjugating system. Curr Protein Pept Sci. 2004;5:177–84. doi: 10.2174/1389203043379783. [DOI] [PubMed] [Google Scholar]
- 19.Petroski MD. Mechanism-based neddylation inhibitor. Chem Biol. 17:6–8. doi: 10.1016/j.chembiol.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 20.Brownell JE, Sintchak MD, Gavin JM, et al. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol Cell. 37:102–11. doi: 10.1016/j.molcel.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 21.Soucy TA, Smith PG, Milhollen MA, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–6. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 22.Bunz F, Dutriaux A, Lengauer C, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 23.Abbas T, Sivaprasad U, Terai K, Amador V, Pagano M, Dutta A. PCNA-dependent regulation of p21ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 2008;22:2496–506. doi: 10.1101/gad.1676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin JJ, Dutta A. ATR pathway is the primary pathway for activating G2/M checkpoint induction after re-replication. J Biol Chem. 2007;282:30357–62. doi: 10.1074/jbc.M705178200. [DOI] [PubMed] [Google Scholar]
- 25.Montes de Oca R, Andreassen PR, Margossian SP, et al. Regulated interaction of the Fanconi anemia protein, FANCD2, with chromatin. Blood. 2005;105:1003–9. doi: 10.1182/blood-2003-11-3997. [DOI] [PubMed] [Google Scholar]
- 26.Zhu WDA. An ATR-and BRCA1-Mediated Fanconi Anemia Pathway Is Required for Activating the G2/M Checkpoint and DNA Damage Repair upon Rereplication. MCB. 2006:26. doi: 10.1128/MCB.02141-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dhar SK, Yoshida K, Machida Y, et al. Replication from oriP of Epstein-Barr virus requires human ORC and is inhibited by geminin. Cell. 2001;106:287–96. doi: 10.1016/s0092-8674(01)00458-5. [DOI] [PubMed] [Google Scholar]
- 28.Sugimoto N, Tatsumi Y, Tsurumi T, et al. Cdt1 phosphorylation by cyclin A-dependent kinases negatively regulates its function without affecting geminin binding. J Biol Chem. 2004;279:19691–7. doi: 10.1074/jbc.M313175200. [DOI] [PubMed] [Google Scholar]
- 29.Takeda DY, Parvin JD, Dutta A. Degradation of Cdt1 during S phase is Skp2-independent and isrequired for efficient progression of mammalian cells through S phase. J Biol Chem. 2005;280:23416–23. doi: 10.1074/jbc.M501208200. [DOI] [PubMed] [Google Scholar]
- 30.Li X, Zhao Q, Liao R, Sun P, Wu X. The SCF(Skp2) ubiquitin ligase complex interacts with the human replication licensing factor Cdt1 and regulates Cdt1 degradation. J Biol Chem. 2003;278:30854–8. doi: 10.1074/jbc.C300251200. [DOI] [PubMed] [Google Scholar]
- 31.Hu J, McCall CM, Ohta T, Xiong Y. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat Cell Biol. 2004;6:1003–9. doi: 10.1038/ncb1172. [DOI] [PubMed] [Google Scholar]
- 32.Senga T, Sivaprasad U, Zhu W, et al. PCNA is a cofactor for Cdt1 degradation by CUL4/DDB1-mediated N-terminal ubiquitination. J Biol Chem. 2006;281:6246–52. doi: 10.1074/jbc.M512705200. [DOI] [PubMed] [Google Scholar]
- 33.Sansam CL, Shepard JL, Lai K, et al. DTL/CDT2 is essential for both CDT1 regulation and the early G2/M checkpoint. Genes Dev. 2006;20:3117–29. doi: 10.1101/gad.1482106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGarry TJ. Geminin deficiency causes a Chk1-dependent G2 arrest in Xenopus. Mol Biol Cell. 2002;13:3662–71. doi: 10.1091/mbc.E02-04-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stein GH, Beeson M, Gordon L. Failure to phosphorylate the retinoblastoma gene product in senescent human fibroblasts. Science. 1990;249:666–9. doi: 10.1126/science.2166342. [DOI] [PubMed] [Google Scholar]
- 36.Dimri GP, Lee X, Basile G, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92:9363–7. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Micco R, Fumagalli M, Cicalese A, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–42. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 38.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 39.Stein GH, Drullinger LF, Soulard A, Dulic V. Differential roles for cyclin-dependent kinase inhibitors p21 and p16 in the mechanisms of senescence and differentiation in human fibroblasts. Mol Cell Biol. 1999;19:2109–17. doi: 10.1128/mcb.19.3.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zou X, Ray D, Aziyu A, et al. Cdk4 disruption renders primary mouse cells resistant to oncogenic transformation, leading to Arf/p53-independent senescence. Genes Dev. 2002;16:2923–34. doi: 10.1101/gad.1033002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Myohanen SK, Baylin SB, Herman JG. Hypermethylation can selectively silence individual p16ink4A alleles inneoplasia. Cancer Res. 1998;58:591–3. [PubMed] [Google Scholar]
- 42.Han Z, Wei W, Dunaway S, et al. Role of p21 in apoptosis and senescence of human colon cancer cells treated with camptothecin. J Biol Chem. 2002;277:17154–60. doi: 10.1074/jbc.M112401200. [DOI] [PubMed] [Google Scholar]
- 43.Liontos M, Koutsami M, Sideridou M, et al. Deregulated overexpression of hCdt1 and hCdc6 promotes malignant behavior. Cancer Res. 2007;67:10899–909. doi: 10.1158/0008-5472.CAN-07-2837. [DOI] [PubMed] [Google Scholar]
- 44.Verschuren EW, Ban KH, Masek MA, Lehman NL, Jackson PK. Loss of Emi1-dependent anaphase-promoting complex/cyclosome inhibition deregulates E2F target expression and elicits DNA damage-induced senescence. Mol Cell Biol. 2007;27:7955–65. doi: 10.1128/MCB.00908-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alcorta DA, Xiong Y, Phelps D, Hannon G, Beach D, Barrett JC. Involvement of the cyclin-dependent kinase inhibitor p16 (INK4a) in replicative senescence of normal human fibroblasts. Proc Natl Acad Sci U S A. 1996;93:13742–7. doi: 10.1073/pnas.93.24.13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Atadja P, Wong H, Garkavtsev I, Veillette C, Riabowol K. Increased activity of p53 in senescing fibroblasts. Proc Natl Acad Sci U S A. 1995;92:8348–52. doi: 10.1073/pnas.92.18.8348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown JP, Wei W, Sedivy JM. Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts. Science. 1997;277:831–4. doi: 10.1126/science.277.5327.831. [DOI] [PubMed] [Google Scholar]
- 48.Kang JY, Kim JJ, Jang SY, Bae YS. The p53-p21(Cip1/WAF1) pathway is necessary for cellular senescence induced by the inhibition of protein kinase CKII in human colon cancer cells. Mol Cells. 2009;28:489–94. doi: 10.1007/s10059-009-0141-9. [DOI] [PubMed] [Google Scholar]
- 49.Roninson IB, Broude EV, Chang BD. If not apoptosis, then what? Treatment-induced senescence and mitotic catastrophe in tumor cells. Drug Resist Updat. 2001;4:303–13. doi: 10.1054/drup.2001.0213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.