Summary
Ensemble methods (such as Bagging and Random Forests) take advantage of unstable base learners (such as decision trees) to improve predictions, and offer measures of variable importance useful for variable selection. LogicFS (Schwender & Ickstadt, 2008) has been proposed as such an ensemble learner for case-control studies when interactions of single nucleotide polymorphisms (SNPs) are of particular interest. LogicFS uses bootstrap samples of the data and employs the Boolean trees derived in logic regression (Ruczinski et al., 200) is base learners to create ensembles of models that allow for the quantification of the contributions of epistatic interactions to the disease risk. In this article, we propose an extension of logicFS suitable for case-parent trio data, and derive an additional importance measure that is much less influenced by linkage disequilibrium between SNPs than the measure originally used in logicFS. We illustrate the performance of the novel procedure in simulation studies and in a case study of 461 case-parent trios with autistic children.
Keywords: Family-based association study, gene-gene interaction, epistatic interaction, trio logic regression, logicFS, autism
Introduction
In association studies concerned with complex diseases, individual SNPs often only exhibit a small effect size. However, it is hypothesized that interactions of several SNPs and possibly gene-environment interactions might more strongly influence the risk of disease (Garte, 2001). Since the number of possible interactions between genetic markers and between genetic and environmental variables is vast, statistical procedures are required that can cope with this high-dimensional search space. Several methods for tackling this task have been proposed, including exhaustive searches based on multiple testing (Marchini et al., 2005; Goodman et al., 2006) and multifactor dimensionality reduction (Ritchie et al., 2001; Hahn et al., 2003; Ritchie et al., 2003), as well as machine learning methods such as Random Forests (Breiman, 2001; Lunetta et al., 2004; Bureau et al., 2005; Chen et al., 2007) and neural networks (Lucek & Ott, 1997; Ritchie et al., 2003b; North et al., 2003; Tomita et al., 2004). Besides multifactor dimensionality reduction, the restricted partition method proposed by Culverhouse et al. (2004) and logic regression introduced by Ruczinski et al. (2003) have been specifically developed for analyzing SNP data. Overviews and discussions on some of these procedure can be found in Heidema et al. (2006), McKinney et al. (2006), and Musani et al. (2007).
Logic regression has performed well in SNP association studies (Kooperberg et al., 2001; Witte & Fijal, 2001; Etzioni et al., 2004; Ruczinski et al., 2004; Andrew et al., 2008; Harth et al., 2008; Justenhoven et al., 2008; Suehiro et al., 2008), but has also been applied in other biomedical research areas such as the identification of regulatory motifs (Keles et al., 2004), HIV studies (Segal et al., 2004), DNA methylation (Feng et al., 2005), and biomarker detection (Vaidya et al., 2008). In addition, several modifications and extensions of logic regression have been proposed. Logic regression has been embedded into a Bayesian framework (Kooperberg & Ruczinski, 2005; Clark et al., 2007), and the simulated annealing algorithm employed in logic regression to search for interactions has been replaced by other probabilistic search methods such as genetic programming (Nunkesser et al., 2007) and evolutionary algorithms (Clark et al., 2005, 2008). Most recently, Li et al. (2010a) adapted logic regression, which originally has been developed for population-based association studies, to the analysis of case-parent trio data.
Similar to classification and regression trees (Breiman et al., 1984), the Boolean trees used in logic regression models are unstable, i.e. small changes in the data can lead to very different trees. Ensemble methods such as bagging (Breiman, 1996) and Random Forests (Breiman, 2001) take advantage of unstable predictors to improve predictions, and more importantly in the context of SNP association studies, offer measures of variable importance which can improve variable selection. However, none of these methods enable a direct quantification of the importance of combinations of variables. Based on this rationale, Schwender & Ickstadt (2008) proposed a procedure called logicFS (logic Feature Selection) in which a bagging version of the original logic regression is employed to identify disease-associated SNP interactions. The Boolean trees are used as base learners in this ensemble method, which allows for the quantification of the relevance of the detected SNP interactions (i.e. not only of individual SNPs) by providing an importance measure similar to one of the single variable importance measures determined in Random Forests.
In this manuscript, we adapt logicFS to case-parent trio data to stabilize the search for disease-associated interactions in such family-based data. We note that while much thought and effort has been put into the development of methods for testing candidate SNP interactions in case-parent trios (e.g. Schaid, 1999; Lunetta et al., 2000; Culverhouse et al., 2002; Cordell & Clayton, 2002; Cordell et al., 2004; Baksh et al., 2006, 2007; Kotti et al., 2007), only few methods searching directly for higher order SNP interactions in family-based data have been proposed (Martin et al., 2006; Li et al., 2010a). Moreover, we introduce a new importance measure that takes linkage disequilibrium (LD) between markers into account. The resulting approach called trioFS is then applied to simulated data, and to case-parent trios with autistic children.
Methods
Logic Regression
Logic regression introduced by Ruczinski et al. (2003) is a classification and regression procedure that adaptively searches for Boolean combinations Ck, k = 1,…,K of binary covariates (using the Boolean operators AND, OR, NOT) and incorporates these terms into a generalized linear model
| (1) |
where the choice of the link function g depends on the type of the response Y. If, for example, Y is binary, then g is the logit function, which is typically used for logistic regression models. The logic regression framework also includes many other forms of regression, such as linear models or Cox proportional hazard models.
To find the best logic regression model as described in equation (1), Ruczinski et al. (2003) employ a two-step procedure. First, the best scoring models of different sizes (as determined by the number of covariates used in the Boolean terms) are derived, and then model selection procedures such as permutation tests or cross-validation are used to determine the optimal model size. The search for good scoring models is carried out via simulated annealing, a stochastic search algorithm suitable for global optimization problems, and a tree-representation of the logic expressions Ck. Using a set of moves based on this tree-representation, variables and the AND- and OR-operators can be added to, removed from, or alternated in the logic expressions such that each logic expression can be reached from each other expression in a finite number of moves. In each annealing step, the new model is compared with the current logic regression model by a score function (for example, the deviance if Y is binary). If the newly proposed model is an improvement compared to the current model, it gets accepted. Otherwise, an acceptance probability based on the values of the score function for these two models is computed. This acceptance probability also considers how far the annealing has progressed, that is, it ensures that towards the end of the search newly proposed logic regression models are unlikely to get accepted if they score worse. When logic regression is applied to SNP data, each SNP S is typically split up into two binary variables SD and SR, coding for a dominant and a recessive effect of S, respectively. For a detailed description, see Ruczinski et al. (2003).
Recently, Li et al. (2010a) introduced an extension of logic regression enabling the analysis of case-parent trio data. As in a genotypic transmission disequilibrium test (Schaid, 1996; Cordell et al., 2004), trio logic regression uses the affected proband as a case, and the other Mendelian children (as derived from the parents’ genotypes) as matched pseudo-controls. Since there are 4m – 1 matched pseudo-controls for each case when considering m unlinked SNPs (Cordell et al., 2004), Li et al. (2010a) restrict the analysis to the 1:3 matching typically employed when testing individual SNPs. This is achieved by randomly ordering the genotypes of the three Mendelian children at each marker, and augmenting those genotypes to generate three pseudo-controls. When SNPs are in LD, Li et al. (2010a) take the haplotype structure into account, designate a case phase for each trio, and select three random pseudo-controls under that phase scenario. As in logic regression, the genotypes are then described by two binary variables in dominant and recessive coding, and a conditional logistic likelihood is used in the search for the logic regression model that best discriminates cases and pseudo-controls.
Logic Feature Selection (logicFS)
In logicFS (Schwender & Ickstadt, 2008), logic regression is applied to several bootstrap samples drawn from the subjects in a case-control study to detect high-order SNP interactions associated with the case-control status. Thus, bagging (Breiman, 1996) with base learner logic regression is used in logicFS to stabilize the search for such interactions. To identify the interactions composing the logic expressions, and hence the logic regression models, each logic expression Ckb, b = 1,…,B, in each of the B models (or the complement of Ckb, if the respective parameter estimate is negative) is transformed into a disjunctive normal form, i.e. an OR-combination of AND-combinations. Each of these AND-combinations in this disjunctive normal form represents one of the interactions.
Since some of the detected interactions will have a larger effect on the disease risk than others, Schwender & Ickstadt (2008) also propose an importance measure for quantifying the relevance of each identified interaction, which is related to one of the variable importance measures (VIM) used in Random Forests (Breiman, 2001). For each of the interactions comprised by a logic regression model, the value of the importance measure is computed by predicting the case-control status of the out-of-bag observations, i.e. the subjects that are not part of the bootstrap sample used to fit this model. This is done for both the original model as it has been found by logic regression, and a reduced model, which is derived by removing the interaction of interest from the original model and refitting the parameters in the model with the reduced logic expressions. For each iteration b = 1,…,B and each interaction Pj, j = 1,…,J, appearing in at least one of the B logic regression models, this leads to two numbers of correctly classified out-of-bag observations, denoted by Nb and The importance of Pj is then quantified as
where is the set of all interactions comprised by the b-th logic regression model.
Logic Feature Selection for Trios (trioFS)
Similar to the analysis of data from case-control studies, applying trio logic regression to several subsets of the case-parent trio data can strengthen the identification of disease-associated SNP interactions. When randomly drawing these subsets, it is necessary to take the matching into account. Thus, we do not sample from the cases and pseudo-controls per se, but sample the case-pseudo-control status within each of the case-parent trios. Furthermore, we decided to use subsampling, i.e. to randomly draw a certain percentage of trios (typically, 63.2% of the trios, as this is the percentage of subjects expected to be in a bootstrap sample) instead of using bootstrap sampling (i.e. to randomly draw with replacement). This is foremost for computational efficiency, as sub-sampling works as good as bagging, but is computationally cheaper (Buehlmann & Yu, 2002).
In the computation of an importance measure, it would again be possible to employ the number of correct predictions of the case-(pseudo-)control status. However, since the controls are artificial, and their number is three times as large as the number of cases, we use another statistic to measure the goodness of the fitted model, namely the predictive log-likelihood ℓpred (Schmid & Hothorn, 2008). Thus, the parameter estimate for the b-th trio logic regression model and the cases and matched pseudo-controls in the set containing the out-of-bag observations of the b-th iteration are employed to compute the predictive log-likelihood
where ∈ {0,1} is the value of the logic expression C1b in the b-th model for the case in the i-th trio, and (p = 1, 2, 3) are the values of C1b for the matched pseudo-controls. This predictive log-likelihood is calculated for all B logic regression models, and the importance of an interaction Pj is calculated by removing this interaction from the models that contain Pj and computing the log-likelihood of the respective reduced model. The importance of Pj is then given by
where the factor −2 is used to be in accordance with a likelihood ratio test.
A problem with importance measures such as the ones of Random Forests and logicFS, which is similar to the multicollinearity problem in linear regression, is that the importance can be lowered substantially when the corresponding SNPs are in strong LD (Lunetta et al., 2004; Nicodemus & Malley, 2009). If, for example, an interaction between the SNPs S1, S2, and S3 is disease-associated, and S1 is in strong LD with S4, then the actual interaction will appear in some of the logic regression models, and the interaction of S4, S2, and S3 is contained in other models. Hence, the actual interaction will show a reduced importance. To adjust for LD, we identify for each interaction Pj the logic regression models that contain interactions of the same number of terms as Pj, that only differ from Pj by SNPs that are in tight LD with the SNPs in Pj, where each of the replaced SNPs in Pj must have exactly one counterpart in the other interaction. If the b-th model contains such a neighbor interaction, we replace it by Pj, and refit the changed model to estimate . We then remove Pj from the model to compute , and the improvement
| (2) |
of the b-th model due to Pj. The adjusted importance measure is then given by
| (3) |
where is the set containing all neighbor interactions of the interactions composing the b-th logic regression model.
Another problem is that an interaction might be identified by logic regression that consists of the actual disease-associated interaction and one or rarely more additional SNPs that only slightly increase the disease risk in the sample. While over-fitting is in general not a problem for the computation of the improvements as they are determined based on the out-of-bag observations, the importance of the actual interaction itself will be decreased nonetheless. This problem can be solved in a similar way as the LD problem, i.e. by replacing the extended interactions in the logic regression models, and analogous to equation (2), by calculating the improvements that would have been due to the actual interaction, had it been in the model instead of the extended interaction. The resulting importance measure adjusted for both LD and too large interactions is given by
| (4) |
where εb is the set containing all interactions comprised in any of the B logic regression models that in interaction with another SNP make up one of the interactions in the b-th model. We typically restrict εb to interactions containing one additional variable, since we only rarely observe that an interaction intended to be disease-associated is extended by more than one interaction term in our simulation studies. However, the publicly available software also allows for the extension of more than one additional variable in the interaction terms.
To demonstrate the proposed importance measures (3) and (4) in an example, assume that trioFS with B = 5 iterations is applied to case-parent trio data, and that the disease-associated interaction P1 consisting of the SNPs S1, S2, and S3 is found in iterations 1 and 3, whereas interaction P2 composed of S4, S2 and S3 is identified in iterations 2 and 5, as S1 and S4 are in strong LD. In this case, will be zero for b = 2, 4, 5, and will be zero for b = 1, 3, 4, but will be larger than zero for b = 2, 5, and will be larger than zero for b = 1, 3. If we, moreover, assume that in the fourth iteration an interaction P3 consisting of P1 and S5 is detected, then , but = 0, as P3 is not an extension of P2.
We note that while the main goal of the trioFS procedure is to generate hypothesis, not to carry out hypothesis tests per se, it is possible to define and generate permutation-based p-values for the SNP interactions based on the importance measures. In such a permutation test, the 1:3 matching has to be taken into account by randomly assigning the case status in a trio to one of the four Mendelian children derived from the parents’ genotypes. A simple approach for the p-value estimation is to apply trioFS to a sufficiently large number of permutations of the case-pseudo-control status across trios, and compare the values of the importance measures in these permutations with the observed values from the original application. However, such an approach would be computationally challenging even in small data sets. An alternative and much less time-consuming procedure, which typically leads to almost identical p-values (see the supplementary material to Schwender et al., 2010), is to employ the logic expressions found in the original analysis in all applications to the permuted case-pseudo-control status. In each iteration of this procedure, we permute the case-pseudo-control status, apply a conditional logistic regression to each of the B bootstrap samples using the respective logic expression from the original analysis as predictor, and compute the values of the importance measures based on these refitted models and the corresponding out-of-bag observations. The permutation-based p-values are then given by the fraction of the importances for an interaction determined in these iterations that are larger than or equal to the original importance.
The computation of the p-values can be further accelerated by making use of the alternative representation of the conditional likelihood proposed by Li et al. (2010b). Instead of considering all n trios in the maximization of the log-likelihood separately, one aggregates all trios showing the same value of the logic expression for the case and the same number of pseudo-controls for which the logic expression is true. Since there are only eight such case-pseudo-control combinations, and two of those do not contribute to the log-likelihood, and thus to the maximization procedure, the conditional log-likelihood can be maximized by considering six instead of n components, leading to a substantial reduction in required computing time.
Results
To illustrate the performance of trioFS, we applied the method to data from a simulation study considering different effect and sample sizes, and to a case study of parents with autistic children.
Simulation
As a first set-up, we simulated 100 data sets each consisting of genotypes for 100 unlinked SNPs, typed in 1000 case-parent trios. In each of these data sets, the SNPs S3 and S7 were simulated such that the chance of being a case was 3 times larger for subjects exhibiting at least one copy of the variant allele at both S3 and S7. Thus, a subject showing the interaction S3D ⋀S7D had a 3-fold increase in the chance of being a case, where the symbol ⋀ denotes the logic AND-operator. The genotypes of the other 98 SNPs were drawn under the assumption of no association with the outcome.
We applied trioFS with B = 20 iterations to these 100 data sets, and computed VIMTrio, VIMLD, and VIMAdj. S3D ⋀ S7D was identified as the (usually by far) most important interaction in all applications of trioFS when considering VIMAdj, and in all but one application when using VIMTrio to rank the interactions. In fact, S3D ⋀ S7D is the only interaction that was detected in all applications. In general, S3D⋀S7D was considered very important by all three metrics VIMTrio, VIMLD, and VIMAdj. However, with the exception of one application in which VIMTrio = VIMAdj, the value of VIMAdj was typically substantially larger than the value of VIMTrio (Figure 1). The reason for this is that although S3D ⋀ S7D exhibits the by far largest importance, interactions composed of S3D⋀S7D and one other SNP were also frequently identified in some of the iterations of trioFS, reducing the improvement due to S3D ⋀ S7D as quantified in equation (2) in such an iteration to zero, and thus, decreasing the overall importance of S3D ⋀ S7D (see Table 1 for an example output of trioFS). Adjusting for LD has no effect on VIMTrio in this application, since all SNPs were simulated independently from each other. Thus, no pair of SNPs found in interaction with S3D ⋀ S7D exhibited an r2-value larger than 0.7, which was used as the defining threshold for LD in equation (3) to quantify VIMLD (see Table 1).
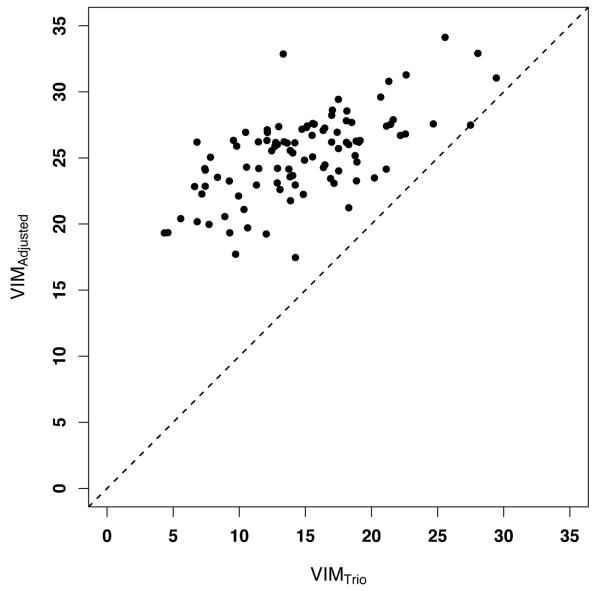
Figure 1.
Scatter plot for the values of VIMTrio and VIMAdj for the interaction term S3D ⋀ S7D, derived from the applications of trioFS to 10 simulated data sets where disease risk is determined by said interaction. The values for importance measure VIMTrio are always smaller than the corresponding values for VIMAdj, illustrating the benefit of accounting for potentially over-fitted interactions. Since SNPs were not in linkage disequilibrium, the values for the importance measure VIMLD are the same as the ones for VIMTrio, and are omitted from the plot.
Table 1.
Values of VIMTrio and the adjusted importance measures VIMLD and VIMAdj for the top five interactions found in the application of trioFS to one of the simulated trio data sets that resulted in the value of VIMTrio closest to the median of the 100 VIMTrio values. The interaction term S3D ⋀ S7D that specifies disease risk in this simulation is the most important finding. The other interactions also contain this term, and thus, the variable importance measure VIMAdj that allows for this type of over-fitting is boosted. Since SNPs were not in linkage disequilibrium in this simulation, the values for the importance measures VIMLD and VIMTrio are identical
| Interaction | VIMTrio | VIMLD | VIMAdj |
|---|---|---|---|
| S3D ⋀ S7D | 14.23 | 14.23 | 22.95 |
| 1.37 | 1.37 | 1.37 | |
| 1.18 | 1.18 | 1.18 | |
| 0.95 | 0.95 | 0.95 | |
| S1D ⋀ S3D ⋀ S7D | 0.89 | 0.89 | 0.89 |
To investigate whether trioFS is also able to detect S3D⋀ S7D when this interaction has a smaller effect size, we simulated S3 and S7 such that the odds of being a case were 2.5, 2, or 1.5 times larger for subjects showing S3D ⋀ S7D. For each of these three odds ratio, 100 data sets were generated, each consisting of 1000 case-parent trios typed at S3, S7 and 98 additional independent SNPs intended to have no effect on the disease risk. Additionally, we simulated 100 data sets consisting of 500 trios for each of the four odds ratios 3, 2.5, 2, and 1.5. We then applied trioFS with B = 20 iterations to each of these data sets.
The simulation reveals that sample sizes of 1000 trios or fewer had to be considered insufficient for a study to detect interactions with odds ratios of 1.5 or smaller (Table 2).
Table 2.
Results of the applications of trioFS to the 100 data sets from each of 8 simulation scenarios in which the interaction S3D ⋀ S7D is intended to be disease-associated. This tables contains the numbers of applications in which this interaction is found, is identified as the most important interaction or amongst the five most important interactions, respectively, the numbers of applications in which an extension of S3D ⋀ S7D is identified as most important interaction or amongst the five most important interactions, respectively, and the numbers of applications in which S3D ⋀ S7D shows a Bonferroni corrected p-value p smaller than 0.05 or equal to zero, and an extensions exhibits a p-value pExt smaller than 0.05, using VIMAdj or, in brackets, VIMTrio as importance measure
| OR = 1.5 | OR = 2.0 | OR = 2.5 | OR = 3.0 | |||||
|---|---|---|---|---|---|---|---|---|
| Trios | 500 | 1000 | 500 | 1000 | 500 | 1000 | 500 | 1000 |
| Found | 8 | 33 | 87 | 100 | 99 | 100 | 100 | 100 |
| Top 1 | 0 (0) | 0 (0) | 44 (20) | 97 (72) | 99 (75) | 100 (96) | 97 (91) | 100 (99) |
| Top 5 | 1 (1) | 0 (0) | 54 (37) | 100 (89) | 99 (87) | 100 (98) | 100 (100) | 100 (100) |
| Ext. Top 1 | 0 (0) | 0 (0) | 13 (20) | 0 (22) | 1 (24) | 0 (4) | 1 (7) | 0 (1) |
| Ext. Top 5 | 1 (1) | 1 (1) | 67 (67) | 98 (98) | 98 (98) | 99 (99) | 100 (100) | 99 (99) |
| p ≤ 0.05 | 0 (0) | 0 (0) | 0 (0) | 67 (60) | 59 (64) | 100 (99) | 99 (96) | 100 (100) |
| p = 0 | 0 (0) | 0 (0) | 0 (0) | 46 (36) | 32 (37) | 100 (99) | 94 (92) | 100 (99) |
| pExt ≤ 0.05 | 0 (0) | 0 (0) | 0 (0) | 7 (7) | 1 (1) | 61 (72) | 18 (47) | 90 (97) |
trioFS detected S3D ⋀ S7D only in 8 of the 100 applications to the data sets with 500 trios and 33 times in the data sets consisting of 1000 trios, where in only the former applications S3D ⋀ S7D ranked once under the five interactions with the largest values of either VIMTrio or VIMAdj. Interactions composed of S3D ⋀ S7D and another SNP were detected in 40 or 83 of the applications, respectively, but they were just once amongst the five top-ranking interactions in both simulation scenarios.
In all but one applications to the data sets from the simulation scenarios with odds ratios of 2.5 and 3, S3D ⋀ S7D was detected and ranked first when considering VIMAdj. In a few of the analyses, S3D ⋀ S7D ranked not first, but typically second or third when basing this ranking on VIMTrio, where the scenarios with the 500 trios performed worse than the scenarios with the 1000 trios. Usually, all top five interactions contained S3D ⋀ S7D (Table 2).
Employing VIMAdj in the applications to the data sets from the simulation scenario with 1000 case-parent trios and an odds ratio of 2 led to the detection of S3D ⋀ S7D as the top-ranking interaction in 97% of the cases, whereas S3D ⋀S7D itself or an extension of it was found as most important interaction in 72 or 22 of the applications, respectively, if VIMTrio was used. In the six remaining applications, three-way interactions of the SNPs with no main effect showed up as most important, but in these cases at least three of the other four top ranking SNPs were either S3D ⋀ S7D or an extension of it, i.e. a three-way interaction containing S3D ⋀ S7D. When considering the data sets consisting of 500 case-parent trios, S3D ⋀ S7D itself was found in 87 of the applications, and in another 12 analyses it was identified in interaction with another SNP. When the ranking is based on VIMTrio, both S3D ⋀ S7D itself and extensions of it ranked first in 20 of the applications, and represented the most important interaction in 44 and 13 of the analyses, when considering VIMAdj, respectively.
We also computed p-values based on 10,000 permutations of the case-pseudo-control status for all identified interactions, and adjusted for multiple comparisons using the Bonferroni correction. The term S3D⋀S7D was identified as significant in virtually all analyses based on 1000 trios, when the effect size was assumed to be 2.5 or larger (Table 2). Frequently, none of the 10,000 permuted importances were actually larger than the observed (un-permuted) importance of S3D ⋀S7D. The p-value was smaller than 0.05 in about 60% (VIMTrio) or 67% (VIMAdj) of the applications, when an odds ratio of 2 was assumed. When analyzing 500 trios, an odds ratio of 3 was necessary to systematically achieve significance. Virtually all interactions with a p-value smaller than 0.05 were either S3D ⋀ S7D or an extension of it. An exception is S7D, which showed up significant in some of the applications.
To evaluate whether trioFS is also able to detect three-way interactions, S3D ⋀ S5D ⋀ S7D was simulated such that it exhibits an odds ratio of 3, 2.5, or 2, and 97 SNPs were randomly drawn under the assumption of no association with the outcome. In this way, six sets consisting of 100 data sets were generated, where the data sets in three of these sets contained 500 case-parent trios, and in the other sets 1000 trios. We then applied trioFS with B = 20 iterations to all of these data sets and computed VIMTrio and VIMAdj.
Not surprisingly, even larger effect sizes are required to detect the higher order interaction (Table 3). Even for odds ratios of 2, neither S3D⋀S5D⋀S7D nor extensions of it were identified. However, in all but one simulation scenario with odds ratio of 2.5 and 3, S3D ⋀ S5D ⋀ S7D was detected by trioFS. In almost any application to the data sets with 1000 trios, this interaction ranked first when employing VIMAdj, and it ranked first in most analyses when considering VIMTrio. Only in a few of the applications, the value of VIMAdj for S3D ⋀S5D ⋀S7D was substantially larger than the value of VIMTrio, as just a few extensions of S3D⋀S5D⋀S7D appeared in the applications of trioFS. Instead the two-way interactions contained in S3D ⋀ S5D ⋀ S7D, i.e. S3D ⋀ S5D, S3D ⋀ S7D, and S5D⋀S7D, showed up in almost any application of trioFS. In the settings with 500 trios, frequently at least one of these two-way interactions had a higher importance than S3D⋀S5D⋀S7D, whereas the studies with 1000 trios reliably identified the three-way interaction as the most important one. This can also be summarized using the permutation-based p-values for S3D ⋀ S5D ⋀ S7D, which were smaller than 0.05 in virtually any application to the 1000 trios, and zero in most of the applications. On a positive note for the smaller studies, in many instances more than just one of the two-way interactions and S3D ⋀ S5D ⋀ S7D appeared amongst the top five SNPs and with a p-value smaller than 0.05, suggesting that this three-way interaction might be important for the disease risk prediction.
Table 3.
Results of the applications of trioFS to the 100 data sets from each of 8 simulation scenarios in which the interaction S3D⋀S5D⋀S7D is intended to be disease-associated. This tables contains the number of applications in which this interaction is found, is identified as the most important interaction or amongst the five most important interactions, respectively, the number of applications in which one of the three two-way interactions contained in S3D ⋀ S5D ⋀ S7D is identified as most important interaction or amongst the five most important interactions, respectively, and the numbers of applications in which S3D ⋀ S5D ⋀ S7D shows a Bonferroni corrected p-value p smaller than 0.05 or equal to zero, and an extensions exhibits a p-value pExt smaller than 0.05, using VIMAdj or, in brackets, VIMTrio as importance measure
| OR = 2.0 | OR = 2.5 | OR = 3.0 | ||||
|---|---|---|---|---|---|---|
| Trios | 500 | 1000 | 500 | 1000 | 500 | 1000 |
| Found | 0 | 0 | 100 | 99 | 100 | 100 |
| Top 1 | 0 (0) | 0 (0) | 35 (47) | 98 (89) | 2 (36) | 99 (100) |
| Top 5 | 0 (0) | 0 (0) | 79 (70) | 99 (96) | 89 (90) | 100 (100) |
| Pruned Top 1 | 0 (0) | 0 (0) | 62 (44) | 1 (10) | 97 (63) | 1 (0) |
| Pruned Top 5 | 0 (0) | 0 (0) | 100 (100) | 99 (99) | 100 (100) | 98 (98) |
| p ≤ 0.05 | 0 (0) | 0 (0) | 37 (55) | 99 (98) | 46 (75) | 100 (100) |
| p = 0 | 0 (0) | 0 (0) | 6 (10) | 96 (90) | 0 (3) | 99 (100) |
| pPruned ≤ 0.05 | 0 (0) | 0 (0) | 59 (42) | 45 (41) | 93 (66) | 7 (7) |
In the final simulation set-up, we investigated the performance of trioFS and the differences between the importance measures when SNPs are in strong LD. We examined two specific settings for this simulation study, but also refer the reader to the autism case study discussed in the following section, which we believe is a particularly nice illustration of the differences between VIMTrio and VIMLD when SNPs are in strong LD. For each of these settings, one considering an odds ratio of 2.5 for S3D ⋀ S7D, the other an odds ratio of 3, we simulated 100 data sets consisting of 100 SNPs typed at 1000 trios. This time, we generated two LD-blocks of SNPs, one consisting of S2, S3, and S4, and the other of S6, S7 and S8. The pairwise r2-values within these blocks were larger than 0.99. The remaining 94 SNPs were randomly drawn.
Usually a minimum of three, and in most applications four of the top five interactions were composed of two SNPs, one from each of the two LD-blocks containing S3 and S7, with permutation-based p-values typically equal zero, but always less than 0.05 (see Table 4 for an example). The other top five interactions were always three-way interactions consisting of two SNPs from these LD-blocks and another SNP which only slightly contributed to the effect of the interaction. The term S3D ⋀ S7D is sometimes found as the most important interactions, however, frequently another interaction consisting of either S2D, S3D or S4D, and S6D, S7D or S8D ranks first.
Table 4.
The five interactions with the largest values for VIMTrio determined in an application of trioFS to one of the simulated data sets in which S3D ⋀ S7D shows an odds ratio of 3. Here, S3 is in strong LD with S2 and S4, and S7 is in strong LD with S6 and S8. The numbers in the brackets are the Bonferroni corrected p-values corresponding to the respective importance measure
| Interaction | VIMTrio | VIMLD | VIMAdj |
|---|---|---|---|
| S3D ⋀ S7D | 3.69 (0.000) | 9.05 (0.00) | 9.05 (0.00) |
| S2D ⋀ S7D | 2.63 (0.000) | 6.61 (0.00) | 9.16 (0.00) |
| S3D ⋀ S6D | 1.79 (0.000) | 6.57 (0.00) | 9.80 (0.00) |
| S3D ⋀ S8D | 0.97 (0.001) | 6.56 (0.00) | 7.84 (0.00) |
| 0.84 (0.043) | 0.92 (0.02) | 0.92 (0.02) |
The identification of different two-way interactions consisting of SNPs from the two LD-blocks containing the truly associated SNPs led to a reduced value of the respective VIMTrio. Employing VIMLD however resulted in a substantially increased importance (see Table 4). If also three-way interactions composed of one of these two-way interactions and another SNP were found, the importance of this two-way interaction was further increased when using VIMAdj. For example, several three-way interactions containing either S2D ⋀ S7D or S3D ⋀ S7D were identified in the analysis which led to the results presented in Table 4, but no higher-order interaction consisting of S3D ⋀ S7D. Thus, the importances of the former interactions, but not of the latter, were increased when using VIMAdj.
Case Study
In this section, we consider 461 autistic children and their parents from 289 families recruited by the Autism Genetic Resource Exchange (AGRE; http://www.agre.org), a collaborative gene bank created by Cure Autism Now (CAN) and the Human Biological Data Exchange (HBD) to advance genetic research in autism spectrum disorders by consolidating large numbers of families into one collection. Genetic biomaterials and clinical data were obtained for families with at least one offspring diagnosed with an Autism Spectrum Disorder based on evaluation by the Autism Diagnostic Interview-Revised (ADI-R) and the Autism Diagnostic Observational Schedule (Geschwind et al., 2001). Cases were included if they had an ADI-R diagnosis of Autism, and data for both parents were available.
Two of the available 331 SNPs were excluded from the analysis since they were almost monomor-phic. Further, ten of the 461 trios were removed as more than 2% of the SNPs in each of these trios exhibited Mendelian errors. The haplotype-based procedure proposed by Li et al. (2010a) was used to impute the missing genotypes, and to transform the case-pseudo-control data into a format suitable for trio logic regression. We then applied trioFS with B = 20 iterations to these data.
The most important interaction detected by trioFS was a three-way interaction of rs11017112, rs7082126, and rs11017128, all showing the homozygous reference genotypes (Table 5). When considering the adjusted importance measure, a three-way interaction of the latter two SNPs and rs11017114 (also represented by the binary variable using dominant coding, i.e., the variable indicating at least one variant allele) exhibits the second largest importance. All these SNPs are from the gene Glutaredoxin 3 (GLRX3) on chromosome 10.
Table 5.
The five interactions with the largest value for VIMTrio derived from the application of trioFS to the autism data set. For conciseness, the SNP rs-IDs are abbreviated as follows: S180: rs11017112; S185: rs7082126; S192: rs11017128; S183: rs11017114; S193: rs4751178; S143: rs553822; S148: rs502862
| Interaction | VIMTrio | VIMLD | VIMAdj | ORTrio | 95% CI |
|---|---|---|---|---|---|
| 8.18 | 15.46 | 15.46 | 4.44 | (3.26, 6.05) | |
| 5.12 | 5.12 | 15.01 | 2.84 | (2.10, 3.85) | |
| S183D ⋀ S185D ⋀ S192D | 5.06 | 9.94 | 15.17 | 2.58 | (1.88, 3.53) |
| S183D ⋀ S185D ⋀ S192D ⋀ S193D | 4.95 | 4.95 | 4.95 | 3.43 | (2.45, 4.80) |
| 3.95 | 3.95 | 4.40 | 2.41 | (1.81, 3.22) |
Since the haplotype-based imputation of Li et al. (2010a) is probabilistic and data sets created by this procedure can differ, we generated ten data sets with this method and applied trioFS to each of those. All these applications led to the detection of at least one of the three-way interactions mentioned above, with a permutation-based p-value of zero throughout, where sometimes rs11017114D was replaced by rs11017112D, or by (and r2 = 0.985 for these two SNPs). Typically, one of these three-way interactions was also identified as the most important one. Other interactions between SNPs from the same gene and/or SNPs from different genes or chromosomes were also picked up occasionally, but usually not for multiple of these data sets, hinting at spurious associations.
Since epistasis is usually defined as interactions between SNPs in different genes and/or genomic regions, we also analyzed a subset of the 329 SNPs that was previously considered elsewhere (Bowers et al., in preparation) Briefly, this subset consists of 138 independent SNPs showing pair-wise r2-values smaller than or equal to 0.2, and each glutathione-related gene is represented by the marker that has the largest estimated marginal effect size. In addition, all SNPs with a marginal p-value less than 0.1 were also included in the analysis. As before, we generated 10 case-pseudo-control data sets by applying the procedure of Li et al. (2010a) to the subset of 138 SNPs (the procedure is also applicable for “degenerate” haplotypes of size 1, i.e. individual SNPs), and analyzed these data sets with trioFS. Even though we strongly biased the selection of SNPs, the application of trioFS did not reveal interesting interactions.
Discussion
One of the main objectives in SNP association studies is the detection of interacting SNPs that explain some of the variability in the response of interest. In this manuscript, we have adapted a method suitable for finding such interactions in population-based case-control studies to case-parent trio designs. We have further proposed and motivated an adjustment of previous measures of interaction importance that corrects for linkage disequilibrium, and potentially over-fitted interactions.
Similar to importance measures from other approaches such as Random Forests, the here proposed importance measures are used to rank the interactions detected by trioFS by their importance for the disease risk, and to assess which of the found interactions are relevant risk factors. This is an appealing feature, however, should be considered a hypothesis generating rather than a hypothesis testing procedure, and ideally, interactions of interest should be validated on an independent data set if such data are available. This is obviously true for other methods as well, and the task to devise statistical methods that help to characterize interactions after discovery, and to quantify their contribution to the variation in the phenotype, is an active research area in the community (Edwards et al., 2010; Greene et al., 2010; Nicodemus et al., 2010, www.epistasis.org). We also note that for our procedure, it is possible to compute permutation-based p-values for the importances of the detected interactions, but we recommend that these p-values should be considered foremost as descriptive statistics.
The value of the proposed importance measure that adjusts for LD is computed for each neighboring interaction individually, although the SNPs forming these interactions are interchangeable. Another idea might be to jointly consider interactions that differ from each other only by SNPs in tight LD, and compute one importance for these interactions between blocks of SNPs. It is an open question whether the proposed importance measures for SNP interactions can also be used for this purpose, or if a more sophisticated measure is required.
Since not all SNPs composing a disease-associated interaction are necessarily of equal importance – some SNPs might be responsible for most of the effect, others might only lead to a marginal improvement in predicting disease risk – it might be beneficial to also develop methods to quantify how much each of the SNPs contribute individually to any particular interaction, and thus to the disease risk. Univariate testing is certainly not the ultimate solution for this problem, as some of the SNPs might not have a main effect at all, and only show an effect when interacting with other SNPs. Employing variable importance measures such as the ones of Random Forests in (trio) logic expression might lead to more reliable results, as such measures take the multivariate data structure into account.
In applications to simulated data, trioFS is almost always able to detect two- and three-way interactions even for small sample sizes when the effect sizes are large. From our experience with logic regression for population-based data and logicFS, we initially expected that trioFS should also be able to detect interactions with odds ratios considerably smaller than 2. However, in our simulations we have frequently seen “spurious” signal, i.e. interactions of null SNPs with large (estimated) odds ratios. A reason for this is due to the trio design, that is, the comparison of observed proband and the pseudo-controls. If, for example, only a single marker is considered, a trio with parents of the same homozygous genotype does not contribute to the likelihood, as all Mendelian children have the same genotype. Thus, in simulations with small data sets, large odds ratios can easily arise, although their actual significance (if assessed individually by a hypothesis test) would be low.
In an analysis of genotype data from children with autism and their parents, trioFS detects two three-way interactions each composed of SNPs from the same gene that appears to be associated with autism. After removing SNPs in LD by selecting one representative SNP for each gene, trioFS however does not identify interactions that give rise to large values of importance measures. This is not too surprising - interacting markers without main effects would not have entered this analysis, and markers with strong marginal effects might dominate and mask a potential epistatic effect.
In our computing environment, the application of trioFS to the autism data set took about 6.5 hours, where each iteration of trioFS, i.e. each application of trio logic regression, took about 19 minutes, and the computation of the importance measures a few seconds. In general, the computation time of trio logic regression depends on the number of trios and the number of iterations used in the underlying stochastic search algorithm (simulated annealing). Choosing an appropriate number of iterations typically requires some trial and error, and needs to reflect the size of the search space, i.e. should be a function of the number of markers investigated. We note that the total computing time can be cut substantially, since the applications of trio logic regression to the different subsamples of the data can be parallelized. The updated R package logicFS containing trioFS will provide the appropriate functionality for performing such parallel computations.
Nonetheless, the analysis of hundreds of thousands of SNPs would require a way too vast number of iterations, rendering an application of trio logic regression and thus of trioFS to genome-wide association studies impractical. However, we do not believe that the assessment of potential higher order interactions using hundreds of thousands of SNPs without prioritization is desirable, as the required effect sizes to detect such interactions had to be unrealistically large. Logic regression was initially developed for candidate SNP studies and can handle up to a few thousand marker, and thus, the same applies to trio logic regression and trioFS. In particular, if parallelization is employed, it might also be possible to analyze a few ten-thousand SNPs (as they might for example appear in exome sequencing) with a version of (trio) logic regression adapted to this new situation, as recent first attempts with such a modified logic regression show.
Software for trioFS will be available in an updated release of the R package logicFS. This package is freely available at http://www.bioconductor.org, the webpage of the Bioconductor project (Gentleman et al., 2004).
Acknowledgments
Support was provided by grant SCHW15-08 1/1 of the Deutsche Forschungsgemeinschaft (HS), CDC grant “Centers for Autism and Developmental Disabilities Research and Epidemiology” DD06-003 U10 DD000183 (MDF), and R01 HL090577 from the National Heart, Lung, and Blood Institute (IR). We would also like to acknowledge the families who participated in AGRE. The AGRE collection Principal Investigator is Daniel H. Geschwind (UCLA). The Co-Principal Investigators include Stanley F. Nelson and Rita M. Cantor (UCLA), Christa Lese Martin (Univ. Chicago), T. Conrad Gilliam (Columbia). Co-Investigators include Maricela Alarcon (UCLA), Kenneth Lange (UCLA), Sarah J. Spence (UCLA), David H. Ledbetter (Emory) and Hank Juo (Columbia). Scientific oversight of the AGRE program is provided by a steering committee (Chair: Daniel H. Geschwind; Members: W. Ted Brown, Maja Bucan, Joseph D. Buxbaum, T. Conrad Gilliam, David Greenberg, David H. Ledbetter, Bruce Miller, Stanley F. Nelson, Jonathan Pevsner, Carol Sprouse, Gerard D. Schellenberg and Rudolph Tanzi).
References
- Andrew AS, Karagas MR, Nelson HH, Guarrera S, Polidoro S, Gamberini S, Sacerdote C, Moore JH, Kelsey KT, Demidenko E, Vineis P, Matullo G. DNA repair polymorphisms modify bladder cancer risk: a multi-factor analytic strategy. Hum. Hered. 2008;65:105–118. doi: 10.1159/000108942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baksh MF, Balding DJ, Vyse TJ, Whittaker JC. A likelihood ratio approach to family-based association studies with covariates. Ann. Hum. Genet. 2006;70:131–139. doi: 10.1111/j.1529-8817.2005.00189.x. [DOI] [PubMed] [Google Scholar]
- Baksh MF, Balding DJ, Vyse TJ, Whittaker JC. Family-based association analysis with ordered categorical phenotypes, covariates and interactions. Genet. Epidemiol. 2007;31:1–8. doi: 10.1002/gepi.20183. [DOI] [PubMed] [Google Scholar]
- Breiman L. Bagging predictors. Mach. Learn. 1996;26:123–140. [Google Scholar]
- Breiman L. Random Forests. Mach. Learn. 2001;45:5–32. [Google Scholar]
- Breiman L, Friedman JH, Olshen RA, Stone CJ. Classification and regression trees. Wadsworth; Belmont, CA: 1984. [Google Scholar]
- Buehlmann P, Yu B. Analyzing bagging. Ann. Statist. 2002;30:927–961. [Google Scholar]
- Bureau A, Dupuis J, Falls K, Lunetta KL, Hayward B, Keith TP, Eerdewegh PV. Identifying SNPs predictive of phenotype using Random Forests. Genet. Epidemiol. 2005;28:171–182. doi: 10.1002/gepi.20041. [DOI] [PubMed] [Google Scholar]
- Chen X, Liu CT, Zhang M, Zhang H. A forest-based approach to identifying gene and gene gene interactions. Proc. Natl. Acad. Sci. USA. 2007;104:19199–19203. doi: 10.1073/pnas.0709868104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark TG, De Iorio M, Griffiths RC. Bayesian logistic regression using a perfect phylogeny. Biostat. 2007;8:32–52. doi: 10.1093/biostatistics/kxj030. [DOI] [PubMed] [Google Scholar]
- Clark TG, De Iorio M, Griffiths RC. An evolutionary algorithm to find associations in dense genetic maps. IEEE Trans. Evol. Comp. 2008;12:297–306. [Google Scholar]
- Clark TG, De Iorio M, Griffiths RC, Farrall M. Finding associations in dense genetic maps: A genetic algorithm approach. Hum. Hered. 2005;60:97–108. doi: 10.1159/000088845. [DOI] [PubMed] [Google Scholar]
- Cordell HJ, Barratt BJ, Clayton DG. Case/pseudocontrol analysis in genetic association studies: A unified framework for detection of genotype and haplotype associations, gene-gene and gene-environment interactions, and parent-of-origin effects. Genet. Epidemiol. 2004;26:167–185. doi: 10.1002/gepi.10307. [DOI] [PubMed] [Google Scholar]
- Cordell HJ, Clayton DG. A unified stepwise regression procedure for evaluating the relative effects of polymorphisms within a gene using case/control or family data: application to HLA in type 1 diabetes. Am. J. Hum. Genet. 2002;70:124–141. doi: 10.1086/338007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culverhouse R, Klein T, Shannon W. Detecting epistatic interactions contributing to quantitative traits. Genet. Epidemiol. 2004;27:141–152. doi: 10.1002/gepi.20006. [DOI] [PubMed] [Google Scholar]
- Culverhouse R, Suarez BK, Lin J, Reich T. A perspective on epistasis: limits of models displaying no main effect. Am. J. Hum. Genet. 2002;70:461–471. doi: 10.1086/338759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards TL, Turner SD, Torstenson ES, Dudek SM, Martin ER, Ritchie MD. A general framework for formal tests of interaction after exhaustive search methods with applications to mdr and mdr-pdt. PLoS One. 2010;5:e9363. doi: 10.1371/journal.pone.0009363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etzioni R, Falcon S, Gann PH, Kooperberg CL, Penson DF, Stampfer MJ. Prostate-specific antigen and free prostate-specific antigen in the early detection of prostate cancer: do combination tests improve detection? Cancer Epidemiol. Biomarkers Prev. 2004;13:1640–1645. [PubMed] [Google Scholar]
- Feng Q, Balasubramanian A, Hawes SE, Toure P, Sow PS, Dem A, Dembele B, Critchlow CW, Xi L, Lu H, McIntosh MW, Young AM, Kiviat NB. Detection of hypermethylated genes in women with and without cervical neoplasia. J. Natl. Cancer Inst. 2005;97:273–282. doi: 10.1093/jnci/dji041. [DOI] [PubMed] [Google Scholar]
- Garte S. Metabolic susceptibility genes as cancer risk factors: Time for a reassessment? Cancer Epidemiol. Biomarkers Prev. 2001;10:1233–1237. [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, D. M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JYH, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, Sowinski J, Lord C, Iversen P, Shestack J, Jones P, Ducat L, Spence SJ, AGRE Steering Committee The autism genetic resource exchange: A resource for the study of autism and related neuropsychiatric conditions. Am. J. Hum. Genet. 2001;69:463–466. doi: 10.1086/321292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman JE, Mechanic LE, Luke BT, Ambs S, Chanock S, Harris CC. Exploring SNP-SNP interactions and colon cancer risk using polymorphism interaction analysis. Int. J. Cancer. 2006;118:1790–1797. doi: 10.1002/ijc.21523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene CS, Himmelstein DS, Nelson HH, Kelsey KT, Williams SM, Andrew AS, Karagas MR, Moore JH. Enabling personal genomics with an explicit test of epistasis. Pac Symp Biocomput. 2010:327–336. doi: 10.1142/9789814295291_0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn LW, Ritchie MD, Moore JH. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics. 2003;19:376–382. doi: 10.1093/bioinformatics/btf869. [DOI] [PubMed] [Google Scholar]
- Harth V, Schaefer M, Abel J, Maintz L, Neuhaus T, Besuden M, Primke R, Wilkesmann A, Thier R, Vetter H, Ko YD, Bruening T, Bolt HM, Ickstadt K. Head and neck squamous-cell cancer and its association with polymorphic enzymes of xenobiotic metabolism and repair. J. Toxicol. Environ. Health A. 2008;71:887–897. doi: 10.1080/15287390801988160. [DOI] [PubMed] [Google Scholar]
- Heidema GA, Boer JMA, Nagelkerke N, Mariman ECM, van de A DL, Feskens EJM. The challenge for genetic epidemiologists: How to analyze large numbers of SNPs in relation to complex diseases. BioMed Genet. 2006;7 doi: 10.1186/1471-2156-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justenhoven C, Hamann U, Schubert F, Zapatka M, Pierl CB, Rabstein S, Selinski S, Mueller T, Ickstadt K, Gilbert M, Ko YD, Baisch C, Pesch B, Harth V, Bolt HM, Vollmert C, Illig T, Eils R, Dippon J, Brauch H. Breast cancer: a candidate gene approach across the estrogen metabolic pathway. Breast Cancer Res. Treat. 2008;108:137–149. doi: 10.1007/s10549-007-9586-8. [DOI] [PubMed] [Google Scholar]
- Keles S, van der Laan MJ, Vulpe C. Regulatory motif finding by logic regression. Bioinformatics. 2004;20:2799–2811. doi: 10.1093/bioinformatics/bth333. [DOI] [PubMed] [Google Scholar]
- Kooperberg C, Ruczinski I. Identifying interacting SNPs using Monte Carlo logic regression. Genet. Epidemiol. 2005;28:157–170. doi: 10.1002/gepi.20042. [DOI] [PubMed] [Google Scholar]
- Kooperberg C, Ruczinski I, LeBlanc M, Hsu L. Sequence analysis using logic regression. Genet Epidemiol. 2001;21:626–631. doi: 10.1002/gepi.2001.21.s1.s626. [DOI] [PubMed] [Google Scholar]
- Kotti S, Bickeboeller H, Clerget-Darpoux F. Strategy for detecting susceptibility genes with weak or no marginal effect. Hum. Hered. 2007;63:85–92. doi: 10.1159/000099180. [DOI] [PubMed] [Google Scholar]
- Li Q, Fallin MD, Louis TA, Lasseter VK, McGrath JA, Avramopoulos D, Wolyniec PS, Valle D, Liang KY, Pulver AE, Ruczinski I. Detection of SNP-SNP interactions in trios of parents with schizophrenic children. Genet. Epidemiol. 2010a;34:396–406. doi: 10.1002/gepi.20488. [DOI] [PubMed] [Google Scholar]
- Li Q, Louis TA, Fallin MD, Ruczinski I. Detection of SNP-SNP interactions in case-parent trios. 2010b in revision. [Google Scholar]
- Lucek PR, Ott J. Neural network analysis of complex traits. Genet. Epidemiol. 1997;14:1101–1106. doi: 10.1002/(SICI)1098-2272(1997)14:6<1101::AID-GEPI90>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Lunetta KL, Faraone SV, Biederman J, Laird NM. Family-based tests of association and linkage that use unaffected sibs, covariates, and interactions. Am. J. Hum. Genet. 2000;66:605–614. doi: 10.1086/302782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunetta KL, Hayward LB, Segal J, van Eerdewegh P. Screening large-scale association study data: exploiting interactions using random forests. BMC Genet. 2004;10 doi: 10.1186/1471-2156-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Donnely P, Cardon RC. Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat. Genet. 2005;37:413–416. doi: 10.1038/ng1537. [DOI] [PubMed] [Google Scholar]
- Martin ER, Ritchie MD, Hahn L, Kang S, Moore JH. A novel method to identify gene-gene effects in nuclear families: the MDR-PDT. Genet. Epidemiol. 2006;30:111123. doi: 10.1002/gepi.20128. [DOI] [PubMed] [Google Scholar]
- McKinney BA, Reif DM, Ritchie MD, H MJ. Machine learning for detecting gene-gene interactions: a review. Appl. Bioinformatics. 2006;5:77–88. doi: 10.2165/00822942-200605020-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musani SK, Shriner D, Liu N, Feng R, Coffey CS, Yi N, Tiwari HK, Allison DB. Detection of gene × gene interactions in genome-wide association studies of human population data. Hum. Hered. 2007;63:67–84. doi: 10.1159/000099179. [DOI] [PubMed] [Google Scholar]
- Nicodemus KK, Malley JD. Predictor correlation impacts machine learning algorithms: Implications for genomic studies. Bioinformatics. 2009;25:1884–1890. doi: 10.1093/bioinformatics/btp331. [DOI] [PubMed] [Google Scholar]
- Nicodemus KK, Callicott JH, Higier RG, Luna A, Nixon DC, Lipska BK, Vakkalanka R, Giegling I, Rujescu D, Clair DS, Muglia P, Shugart YY, Weinberger DR. Evidence of statistical epistasis between disc1, cit and ndel1 impacting risk for schizophrenia: biological validation with functional neuroimaging. Hum Genet . 2010 doi: 10.1007/s00439-009-0782-y. [DOI] [PubMed] [Google Scholar]
- North BV, Curtis D, Cassell PG, Hitman GA, Sham PC. Assessing optimal neural network architecture for identifying disease-associated multi-marker genotypes using a permutation test, and application to calpain 10 polymorphisms associated with diabetes. Ann. Hum. Genet. 2003;67:348–356. doi: 10.1046/j.1469-1809.2003.00030.x. [DOI] [PubMed] [Google Scholar]
- Nunkesser R, Bernholt T, Schwender H, Ickstadt K, Wegener I. Detecting high-order interactions of single nucleotide polymorphisms using genetic programming. Bioinformatics. 2007;23:3280–3288. doi: 10.1093/bioinformatics/btm522. [DOI] [PubMed] [Google Scholar]
- Ritchie MD, Hahn LW, Moore JH. (Power of multifactor dimensionality reduction for detecting gene-gene interactions in the presence of genotyping error, missing data, phenocopy, and genetic heterogeneity. Genet. Epidemiol. 2003;24:150–157. doi: 10.1002/gepi.10218. [DOI] [PubMed] [Google Scholar]
- Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, Moore JH. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am. J. Hum. Genet. 2001;69:138–147. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie MD, White BC, Parker JS, Hahn LW, Moore JH. Optimization of neural network architecture using genetic programming improves detection and modeling of gene-gene interactions in studies of human diseases. BMC Bioinformatics. 2003b;4:28. doi: 10.1186/1471-2105-4-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruczinski I, Kooperberg C, LeBlanc M. Logic regression. J. Comput. Graph. Stat. 2003;12:475–511. [Google Scholar]
- Ruczinski I, Kooperberg C, LeBlanc M. Exploring interactions in high-dimensional genomic data: An overview of logic regression, with applications. J. Mult. Anal. 2004;90:178–195. [Google Scholar]
- Schaid DJ. General score tests for associations of genetic markers with disease using cases and their parents. Genet. Epidemiol. 1996;13:423–449. doi: 10.1002/(SICI)1098-2272(1996)13:5<423::AID-GEPI1>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Schaid DJ. Likelihoods and TDT for the case-parents design. Genet. Epidemiol. 1999;16:250260. doi: 10.1002/(SICI)1098-2272(1999)16:3<250::AID-GEPI2>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Schmid M, Hothorn T. Flexible boosting of accelerated failure time models. BMC Bioinformatics. 2008;9:269. doi: 10.1186/1471-2105-9-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwender H, Ickstadt K. Identification of SNP interactions using logic regression. Biostatistics. 2008;9:187–198. doi: 10.1093/biostatistics/kxm024. [DOI] [PubMed] [Google Scholar]
- Schwender H, Ruczinski I, Ickstadt K. Testing SNPs and sets of SNPs for importance in association studies. Biostatistics. 2010 doi: 10.1093/biostatistics/kxq042. to appear. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal MR, Barbour JD, Grant RM. Relating HIV-1 sequence variation to replication capacity via trees and forests. Stat. Appl. Genet. Mol. Biol. 2004;3:2. doi: 10.2202/1544-6115.1031. [DOI] [PubMed] [Google Scholar]
- Suehiro Y, Wong CW, Chirieac LR, Kondo Y, Shen L, Webb CR, Chan YW, Chan ASY, Chan TL, Wu TT, Rashid A, Hamanaka Y, Hinoda Y, Shannon RL, Wang X, Morris J, Issa JPJ, Yuen ST, Leung SY, Hamilton SR. Epigenetic-genetic interactions in the apc/wnt, ras/raf, and p53 pathways in colorectal carcinoma. Clin. Cancer Res. 2008;14:2560–2569. doi: 10.1158/1078-0432.CCR-07-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita Y, Tomida S, Hasegawa Y, Suzuki Y, Shirakawa T, Kobayashi T, Honda H. Artificial neural network approach for selection of susceptible single nucleotide polymorphisms and construction of prediction model on childhood allergic asthma. BMC Bioinformatics. 2004;5:120. doi: 10.1186/1471-2105-5-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidya VS, Waikar SS, Ferguson MA, Collings FB, Sunderland K, Gioules C, Bradwin G, Matsouaka R, Betensky R, Curhan GC, Bonventre JV. Urinary biomarkers for sensitive and specific detection of acute kidney injury in humans. Clin. Transl. Sci. 2008;3:200–208. doi: 10.1111/j.1752-8062.2008.00053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte JS, Fijal BA. Introduction: Analysis of sequence data and population structure. Genet. Epidemiol. 2001;21:600–601. doi: 10.1002/gepi.2001.21.s1.s600. [DOI] [PubMed] [Google Scholar]