Abstract
The influenza RNA-dependent RNA polymerase (RdRp) both replicates the flu's RNA genome and transcribes its mRNA. Replication occurs de novo; however, initiation of transcription requires a 7-methylguanosine 5’ capped primer that is “snatched” from host mRNA via endonuclease and cap binding functions of the influenza polymerase. A key question is how the virus regulates the relative amounts of transcription and replication. We found that the concentration of a capped cellular mRNA, the concentration of the 5’-end of the viral RNA, and the concentration of RdRp all regulate the relative amounts of replication versus transcription. The host mRNA, from which the RdRp snatches its capped primer, acts to upregulate transcription and repress replication. Elevated concentrations of the RdRp itself switch the influenza polymerase towards replication, likely through an oligomerization of the polymerase. The 5’-end of the vRNA template both activates replication and inhibits transcription of the vRNA template, thereby indicating that RdRp contains an allosteric binding site for the 5’ end of the vRNA template. These data provides insights into the regulation of RdRp throughout the viral life cycle and how it synthesizes the appropriate amounts of viral mRNA and replication products (vRNA and cRNA).
Influenza encodes a heterotrimeric RNA-dependent RNA polymerase complex (RdRp1) that both replicates the influenza's RNA genome as well as transcribes its mRNA. The heterotrimeric complex consists of three subunits: PA, PB1, and PB2. The PA subunit is likely responsible for the endonuclease activities of the polymerase (1, 2). The polymerase active site most likely resides in the PB1 subunit while PB2 contains the 7-methylguanosine 5’ cap binding site (3-7).
Replication occurs de novo and begins with vRNA acting as a template to produce a cRNA intermediate which in turn becomes the template for producing more vRNA strands (vRNA → cRNA → vRNA) (8-10). In its transcriptional capacity the polymerase also uses vRNA as a template (vRNA → mRNA). However, the influenza virus cannot produce its own 7-methylguanosine 5’ cap and, therefore, requires a capped primer for transcription. To solve this problem, the influenza polymerase contains cap binding and endonuclease functions that enable it to cleave capped primers from host mRNA (7, 11). Cleavage occurs 9-17 nucleotides from the 5’ end of the host mRNA, typically after a purine. Following cleavage, the polymerase begins synthesizing its own mRNA onto the 3’ end of the stolen, capped primer. When the polymerase encounters an oligo-U tract near the 5’ end of the vRNA template it reiteratively stutters over the sequence, thereby producing a mRNA containing a 3’ polyadenylated tail (12-14). The polymerase, therefore, has two quite distinct functions and presumably must regulate the frequency with which it generates mRNA versus replication products. However, the mechanism by which the polymerase switches between these activities has yet to be fully determined.
Previous studies have shown that at early stages of infection viral mRNA can be readily detected while very little if any cRNA can be detected (15-18). At later stages of infection both cRNA and mRNA are observed, along with increasing amounts of vRNA. This suggests that at early stages of infection the polymerase acts in primarily a transcriptional mode and that at later stages it switches to a replicational mode. One switching model suggests that early in infection both mRNA and cRNA are produced, but the unprotected cRNA is exposed to host nucleases and is degraded while the capped and polyadenylated mRNA is protected (19, 20). At later stages of infection substantial amounts of nascent RdRp and nucleoprotein (NP) have been produced, and these stabilize and protect the cRNA from degradation. It has also been suggested that the influenza's non-structural protein 2 (NS2) plays a more direct role in switching the polymerase from transcription to replication, likely via an interaction with the polymerase complex (18).
In this study we show that the concentration of RdRp, viral template, and host cap source can regulate the polymerase's switch between transcription and replication. In vitro RdRp reconstitution assays indicate that high concentrations of either RdRp or viral template initiate a switch from transcription to replication and that high concentrations of cap source switch the polymerase from replication to transcription.
Experimental Procedures
Plasmids
pcDNA constructs coding for the PA, PB1, and PB2 subunits were used to transfect 293T cells (described below.) The PB2 construct contains a C-terminal TAP tag. The pcDNAPA, pcDNA-PB1, and pcDNA-PB2tap plasmids were a generous gift of Dr. George G. Brownlee, Sir William Dunn School of Pathology, University of Oxford.
RNA
The vRNA 3’ end segment 5’-GGCCUGCUUUUGCU-3’, vRNA 5’ end segment 5’-AGUAGAAACAAGGCC-3’, cRNA 3’ end segment 5’- GGCCUUGUUUCUACU - 3’, cRNA 5’ end segment 5’- AGCAAAAGCAGGCC - 3’, and xRNA segment 5’- AGGGGGUUCCCC - 3’ were purchased from Dharmacon. Lyophilized rabbit globin mRNA was purchased from Sigma.
Preparation and partial purification of influenza polymerase
The RdRp was expressed and purified essentially as previously described (21, 22). The pcDNA-PA, pcDNA-PB1, and pcDNAPB2-TAP plasmids were transfected into 293T cells using Lipofectamine 2000 (Invitrogen) transfection reagent to prepare WT RdRp, while only pcDNA-PB1 and pcDNA-PB2-TAP were transfected to prepare –PA RdRp. 48 hours post transfection the cells were harvested and lysed in lysis buffer (50mM HEPES pH 8.0, 25% glycerol, 0.5% NP40, 40mM NaCl, and 10mM 2-mercaptoethanol) containing Complete-Mini, EDTA-Free protease inhibitor cocktail tablet (Roche). Following centrifugation of the lysate the cell supernatant was added to IgG Sepharose 6 Fast Flow beads (GE Healthcare) in binding buffer (10 mM HEPES pH 8.0, 10% glycerol, 0.1% NP40, 150 mM NaCl, and 50 μM PMSF) and the polymerase was partially purified by taking advantage of the Protein-A binding site genetically encoded in the TAP tag of the PB2 construct. The polymerase was eluted in elution buffer (10 mM HEPES pH 8.0, 10% glycerol, 0.1% NP40, 150 mM NaCl, 50 μM PMSF, and 1mM DTT) from the beads with tobacco etch virus protease that takes advantage of a TEV cleavage site separating the Protein-A tag from the PB2 subunit (21, 22). The partially purified polymerase was analyzed via silver stain and Bradford assay to assess purity and concentration, respectively, and stored at -20°C in 45% glycerol.
Transcription/ vRNA → cRNA replication assay
Assays (6 μl) were performed essentially as previously described (8) and contained 16 nM partially purified polymerase, 200 μM DTT, 5mM MgCl2, 1 unit/μl RNase OUT (Invitrogen), 1mM ATP, 500 μM CTP, 500 μM UTP, 1 μM GTP, 0.15 μM [α-32P]GTP (3000 Ci/mmol), 1.8 ng/μl (14nM) globin mRNA (Invitrogen), 650 nM vRNA 3’ end segment, and 650 nM vRNA 5’ end segment unless stated otherwise. They were incubated at 30°C and quenched with gel loading buffer (90% formamide). Products were analyzed on a 30% polyacrylamide gel and quantitated on a Typhoon Phosphorimager using Image Quant software (Molecular Dynamics).
cRNA → vRNA replication assay
Assays (6 μl) were performed essentially as previously described ((21)) and contained 16 nM partially purified polymerase, 200 μM DTT, 5mM MgCl2, 1 unit/μl RNase OUT (Invitrogen), 500 μM ATP, 500 μM GTP, 500 μM UTP, 1 μM CTP, 0.15 μM [α-32P]CTP (3000 Ci/mmol), 650 nM cRNA 3’ end segment, and 650 nM cRNA 5’ end segment unless stated otherwise. They were incubated at 30° C and quenched with gel loading buffer (90% formamide). Products were analyzed on a 30% polyacrylamide gel and quantitated on a Typhoon Phosphorimager using Image Quant software.
Results
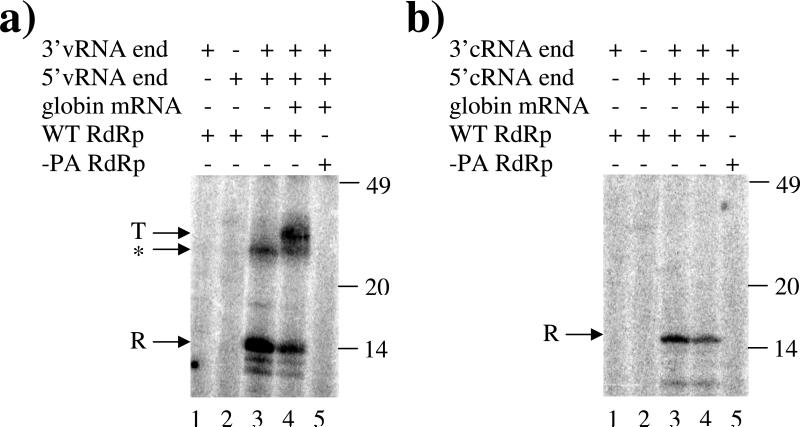
The influenza polymerase is tasked with both replication of the flu's RNA genome and transcription of its mRNA; however the question of how the polymerase regulates these two functions remains largely unanswered. Thus, we examined the effects of the concentrations of cap source, polymerase, and vRNA on replication and transcription using a transcription/replication assay that could simultaneously detect both transcription (vRNA → mRNA) and replication (vRNA → cRNA) from a vRNA template and a cRNA → vRNA replication assay. The assays contained the 3 subunit polymerase produced from recombinant PA, PB1, and PB2 subunits, 14 and 15 nucleotide-long RNA segments that served as the conserved 3’ and 5’ ends of the vRNA or cRNA template, [α-32P]NTPs to body label the various products, and if present, rabbit globin mRNA was used as a 7-methylguanasine-5’-cap source. Importantly, the transcription and replication products can be easily differentiated due to their very different lengths. Replication products will be around 14 or 15 nucleotides long, the length of the 3’ end of the supplied vRNA or cRNA respectively, and their synthesis requires both the 3’ end and the 5’ end of their respective template and all three RdRp subunits (Figures 1a and 1b). Omitting CTP, the third nucleotide required for replication, results in production of a dinucleotide (pppApG) but no longer replication products (data not shown). Transcription products will be around 28 nucleotides, the length of the 3’ end of the vRNA plus the capped primer snatched from the globin mRNA, and their synthesis requires both ends of the vRNA template, globin mRNA, and all three RdRp subunits (Figures 1a and 1b).
Figure 1.
Identification of replicational and transcriptional products. Panel a: Transcription and vRNA→cRNA replication activity was assayed in the presence of 650 nM 3’vRNA end (lanes 1,3,4,and 5), 650 nM 5’vRNA end (lanes 2-5), and 1.8 ng/ul globin mRNA (lanes 4 and 5) using partially purified WT RdRp containing the PA, PB1, and PB2-TAP subunits (lanes 1-4) or partially purified –PA RdRp containing only the PB1 and PB2-TAP subunits (lane 5) as described in Experimental Procedures. Panel b: cRNA→vRNA replication activity was assayed in the presence of 650 nM 3’cRNA end (lanes 1,3,4,and 5), 650 nM 5’cRNA end (lanes 2-5), and 1.8 ng/ul globin mRNA (lanes 4 and 5) using partially purified WT RdRp containing the PA, PB1, and PB2-TAP subunits (lanes 1-4) or partially purified –PA RdRp containing only the PB1 and PB2-TAP subunits (lane 5) as described under Experimental Procedures. The positions of 14 nt, 20 nt, and 49 nt size markers are shown on the right while transcriptional products (T), replicational products (R), and unknown products (*) are indicated on the left.
High concentrations of mRNA cap source cause a switch from replication to transcription
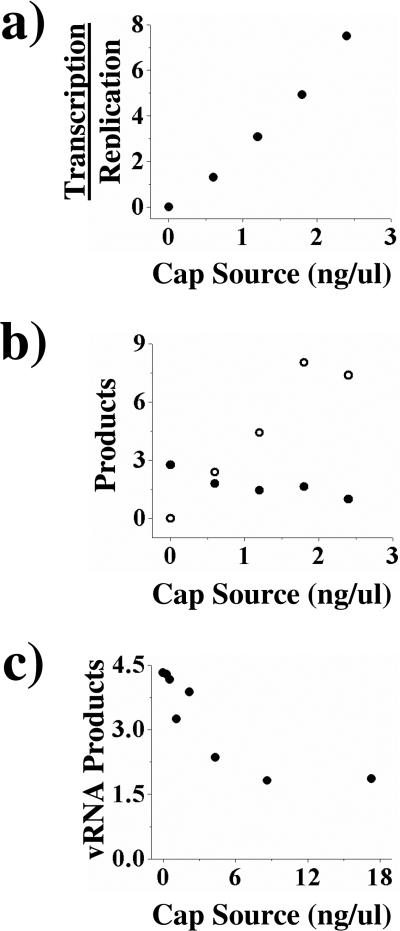
We initially examined how varying the concentration of the mRNA cap source affected the ratio of transcription/replication by titrating globin mRNA into transcription/replication assays, with the globin mRNA acting as the influenza polymerase's 7-methylguanosine 5’ cap source. As the concentration of globin mRNA increased from 0 to 3 ng/ul, the ratio of transcription/replication rose as well (Figure 2a). The increase in the ratio of transcription/replication was due both to an increase in transcription and a decrease in replication, indicating that at higher concentrations of cellular mRNA the polymerase will favor transcription over replication (Figure 2b).
Figure 2.
The effects of cap source concentration on transcription and replication. Cap source was titrated into transcription/replication assays using vRNA (Panels a and b) or cRNA (Panel c) as the template; assays were performed as described under Experimental Procedures. Panel a: The effects on the ratio of transcription to replication. Panel b: The amounts of cRNA (•) or transcriptional products (○) produced (pmol min-1 × 107). Panel c: Effects of cap source on cRNA → vRNA replication on amounts of vRNA products produced (pmol min-1 × 107). The figures shown are single representatives of two separate experiments.
The decrease in vRNA → cRNA replication at higher cap source concentrations raised the possibility that high concentrations of the cap source would also inhibit cRNA → vRNA replication, thereby inhibiting the entire vRNA → cRNA → vRNA replication scheme. To survey the later half of the replicational pathway we titrated globin mRNA into a cRNA → vRNA replication assay. Again, increasing concentrations of globin mRNA inhibited replication (Figure 2c). Thus, higher mRNA concentrations favor transcription over replication by inhibiting both halves of the replication cycle, vRNA → cRNA and cRNA → vRNA, and by increasing transcription.
The 5’ end of the vRNA template helps regulate the ratio of transcription to replication
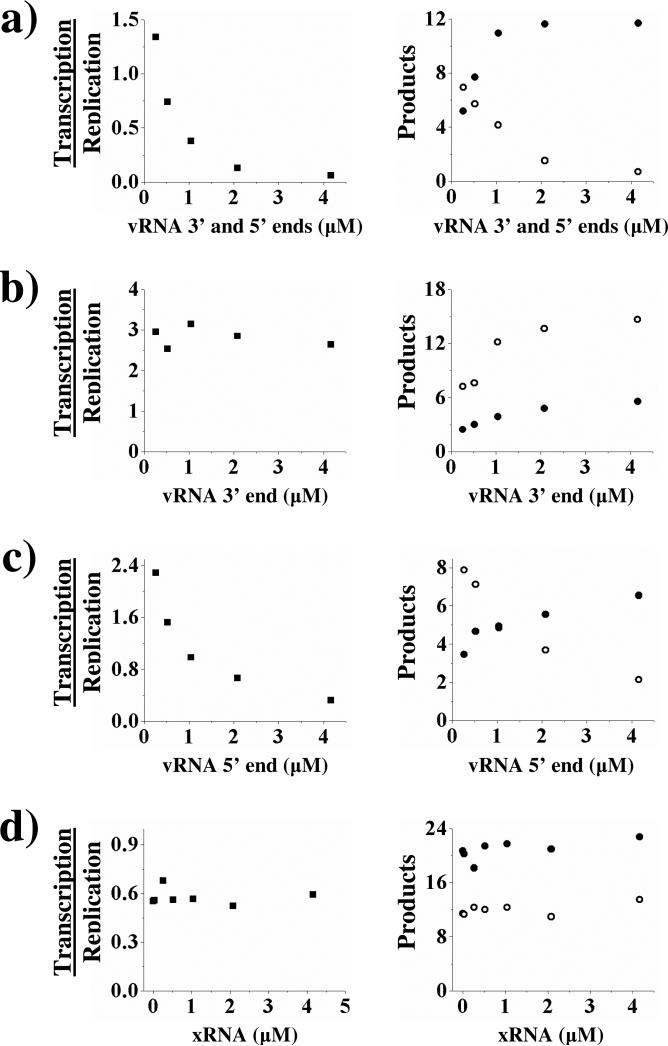
We examined the effect of varying the vRNA concentration on transcription and replication. The vRNA serves as template for both replication and transcription, and its concentration varies greatly during the infection cycle – low upon initial infection, and high at later stages. Increasing the concentration of vRNA in an assay that supports both transcription and replication resulted in decreased mRNA production along with increased vRNA to cRNA replication. Thus, the concentration of vRNA can alter the ratio of transcription/replication such that higher concentrations of vRNA cause a switch from transcription to replication (Figure 3a).
Figure 3.
The effect of vRNA on transcription and vRNA → cRNA replication. The vRNA 3’ and 5’ ends (Panel a), vRNA 3’ end (Panel b), vRNA 5’ end (Panel c), and a control, xRNA, (Panel d) were titrated into transcription/replication assays that were performed as described under Experimental Procedures. The panels on the left depict the ratio of transcription to replication upon addition of the noted RNA. The panels on the right show the absolute amounts of cRNA (•) and transcriptional products (○) produced (pmol min-1 × 107). The figures shown are single representatives of two separate experiments.
Both the 3’ and 5’ ends of the vRNA template must bind to influenza polymerase before either transcription or replication can begin (23-25). This raises the question of whether the 3’ end, 5’ end, or both ends of vRNA are required to regulate the ratio of transcription to replication. The transcription/replication reconstitution assay uses separate RNA segments to mimic the ends of the vRNA (a 14 nucleotide segment to mimic the 3’-end and a 15 nucleotide segment to mimic the 5’ end). Thus, we can hold the concentration of one end constant while independently varying the concentration of the other. The concentration of the 3’ end of vRNA had no effect on the ratio of transcription/replication, whereas increasing the concentration of the 5’ end of vRNA decreased the ratio of transcription/replication (Figure 3b and 3c). These data establish the 5’ end of vRNA as a regulator of the switch from transcription to replication.
Control RNA has no effect on the switch between replication and transcription
To provide further evidence that the effects of varying the concentrations of vRNA and capped mRNA did not result from non-specific effects, we examined xRNA, a short oligoribonucleotide unrelated to influenza (Table 1). As expected, control experiments showed that the influenza polymerase could not replicate or transcribe the xRNA (data not shown). Titrating xRNA into assays showed that xRNA did not affect transcription of vRNA, vRNA → cRNA replication (Figure 3d), or cRNA→ vRNA replication (data not shown). These data indicate that the effects of vRNA template and mRNA cap source result from specific sequence and/or chemical features of the 5’ end of the vRNA and cap source.
Table 1.
Table of RNA sequences used in Transcription/Replication assays.
| RNA name | RNA sequence |
|---|---|
| vRNA 3’ end | 5’-GGCCUGCUUUUGCU-3’ |
| vRNA 5’ end | 5’-AGUAGAAACAAGGCC-3’ |
| cRNA 3’ end | 5’- GGCCUUGUUUCUACU - 3’ |
| cRNA 5’ end | 5’- AGCAAAAGCAGGCC - 3’ |
| xRNA | 5’- AGGGGGUUCCCC - 3’ |
vRNA has little effect on cRNA to vRNA replication
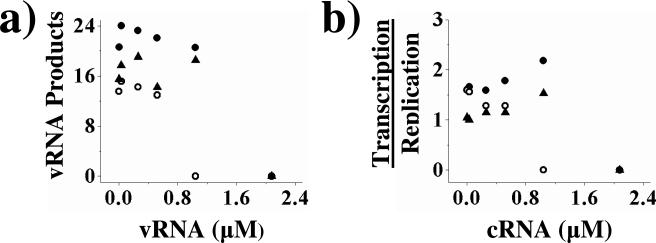
We next examined the effect of vRNA concentration on the cRNA → vRNA replication reaction. In contrast to the effects of increased vRNA concentration on the vRNA → cRNA replication reaction, increasing the concentration of the 3’ end, 5’ end, or both ends of the vRNA template had little effect on the rate of cRNA → vRNA replication until the concentrations of vRNA were equal to or greater than the concentration of cRNA (Figure 4a). At this point replication was strongly inhibited, most likely the result of the 3’ end of the vRNA template hybridizing to the complementary 5’ end of the cRNA template and/or the 5’ end of the vRNA template hybridizing with the complementary 3’ end of the cRNA template (Table 1). Presumably, once the vRNA had annealed with its cRNA counterpart, the cRNA was no longer available for use by the polymerase as a template for replication. We tested this hypothesis by repeating these experiments with one half the initial concentration of cRNA template. The concentration of vRNA needed to give strong inhibition also decreased by one half, consistent with strong inhibition resulting from hybridization of vRNA and cRNA (data not shown). These data suggest that the concentration of vRNA template has little direct effect on the replication of cRNA to vRNA.
Figure 4.
The effects of vRNA on cRNA → vRNA replication and cRNA on the ratio of transcription/replication. Panel a: The 3’ end (•), 5’ end (○), and both 3’ and 5’ ends (▲) of vRNA were titrated into cRNA → vRNA replication assays. The graph indicates the amount of vRNA produced (pmol min-1 × 107). Panel b: The 3’ end (•), 5’ end (○), and both 3’ and 5’ ends (▲) of cRNA were titrated into replication/transcription assays using a vRNA template. The graph depicts the ratio of transcription to replication. Assays were performed as described under Experimental Procedures. The figures shown are single representatives of two separate experiments.
The cRNA template does not affect the ratio of transcription/replication
To determine if the concentration of cRNA affects the relative amounts of transcription and replication on a vRNA template, we titrated cRNA into transcription/replication assays. The concentration of the 3’ end, 5’ end, or both cRNA ends had little effect on the ratio of transcription/replication until the concentration of cRNA was equal to or greater than the concentration of vRNA, at which point both replication and transcription were completely inhibited (Figure 4b). This strong inhibition at equimolar concentrations of vRNA and cRNA most likely resulted from hybridization of the vRNA ends with the cRNA ends due to their complementarity, as described above. Additionally, these data suggest that the concentration of the cRNA template plays no role in controlling the switch between transcription and replication.
The polymerase can switch between replication and transcription
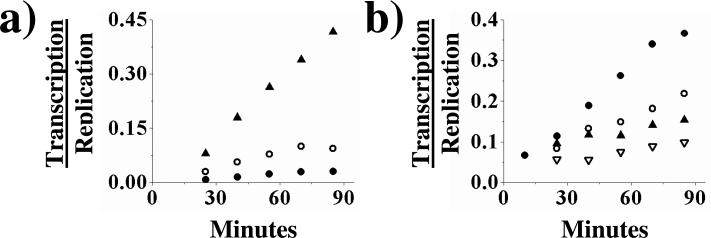
To determine if enzyme that has engaged in replication can switch to transcription, we incubated polymerase with vRNA template and no cap source in a transcription/replication assay, thereby only allowing the polymerase to operate in the replication mode. After 10 minutes, the reaction was split into 3 aliquots that contained different globin mRNA concentration (0.9 ng/ul to 5.4 ng/ul), and the amount of transcription and replication products were determined over the course of an additional 75 minutes. The aliquots containing higher concentrations of globin mRNA had elevated ratios of transcription/replication compared to the aliquots containing lower globin mRNA concentrations (Figure 5a), due both to increased amounts of transcription as well as decreased amounts of replication (data not shown). Thus, RdRp can switch from a purely replicational mode to transcription and higher concentrations of a cap source result in more extensive switching to the transcriptional mode.
Figure 5.
Switching RdRp between transcription and replication. Panel a: A vRNA → cRNA replication assay was incubated for 10 minutes before being split into three aliquots and globin mRNA added to differing final concentrations: 0.9 ng/ul (•) 1.8 ng/ul (○) and 5.4 ng/ul (▲). The assays were allowed to incubate for another 75 minutes and products quantified. Panel b: A transcription/replication assay with an initial concentration of 650 nM vRNA was allowed to incubate for 10 minutes before being split into four aliquots and each with differing amounts of vRNA added to give final concentrations of: 650 nM (•), 1500 nM (○), 2500 nM (▲), and 5000 nM (▽). The assays were allowed to incubate for an additional 75 minutes and products quantified. The graphs depict the ratio of transcription to replication. Assays were performed as described under Experimental Procedures. The figures shown are single representatives of two separate experiments.
vRNA can switch polymerase from transcription to replication
To determine if polymerase that has already committed to transcription can be switched to a replicative mode we incubated vRNA and globin mRNA with polymerase for 10 minutes, conditions where the polymerase is largely engaged in transcription. After 10 minutes the reaction was split into 4 aliquots with varying final concentrations of vRNA (650 nM to 5000 nM). The aliquots containing higher concentrations of vRNA had decreased ratios of transcription/replication (Figure 5b), due both to increased rates of replication and decreased rates of transcription (data not shown). Thus, increasing the concentration of vRNA can switch polymerase that is involved in transcription into the replication mode.
Increased polymerase concentration results in a switch from transcription to replication
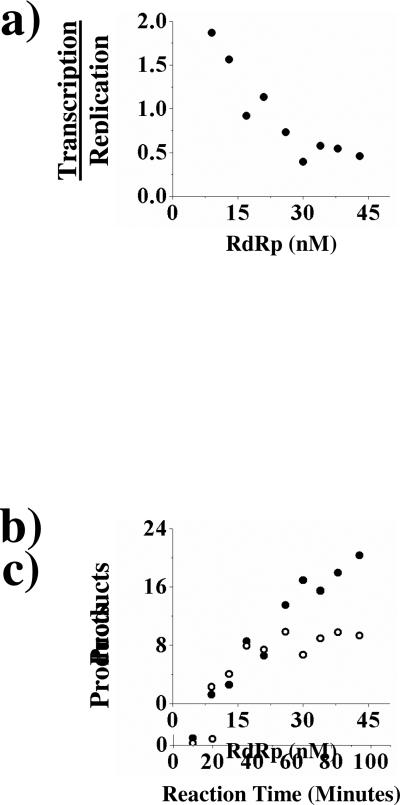
Finally, we examined how varying the concentration of influenza polymerase influenced the ratio of transcription/replication. This was of interest since upon initial infection, the concentration of the polymerase will be very low while at later stages the concentration will be much higher. We titrated influenza polymerase into transcription/replication assays and found that as the concentration of polymerase increases, the amount of replication products increases proportionally. In contrast, the production of transcription products increases only slightly, resulting in a decrease in the ratio of transcription/replication at the higher polymerase concentrations as compared to the lower concentrations (Figure 6a and 6b). This indicates that as the concentration of the polymerase increases, the enzyme synthesizes more replication products compared to transcription products. To ensure that the switch to replication was not due to a decline of globin mRNA or some other substrate over the course of the experiment we examined the formation of replicational products and transcriptional products over the course of 90 minutes at 43 nM RdRp (Figure 6c). The amounts of both replicational products and transcriptional products increased linearly throughout the entire 90 min reaction, indicating that even at the highest concentrations of RdRp the amounts of substrates do not become limiting.
Figure 6.
Titration of RdRp into a transcription/replication assay. RdRp was titrated into a transcription/replication assay that was performed as described under Experimental Procedures. Panel a: The effect of RdRp concentration on the ratio of transcription to replication. Panel b: The effect of RdRp concentration on the amounts of cRNA (•) or transcriptional products (○) produced (pmol min-1 × 105). Panel c: A time course of a transcription/replication assay performed at 43 nM RdRp. Products are the amounts of cRNA (•) or transcriptional products (○) produced (pmol min-1 × 104). The figures shown are single representatives of two separate experiments.
Discussion
The influenza polymerase is responsible for both the replication of influenza's RNA genome and transcription of its mRNA. We found that the concentrations of cap source, vRNA, and influenza polymerase all play a role in the regulation of replication and transcription. We have also shown that populations of polymerase that are replicating RNA can be switched to transcription by increasing the concentration of cap source; similarly, polymerase that is transcribing RNA can be switched to replication by increasing the concentrations of vRNA. These data suggest that the influenza polymerase switches between replication and transcription in a highly controlled manner and responds to multiple effectors.
High concentrations of cap source could shut down the entire replication pathway since both vRNA → cRNA as well as cRNA → vRNA replication were inhibited while transcription was stimulated in response to increasing cap source concentrations. While it is possible that there is an allosteric site for cap source separate from the substrate binding site, this is not required to account for the data. Rather, RdRp may contain a single binding site for cap source. Once the polymerase binds a vRNA template there is a simple competition for the initiation of transcription via cap snatching versus the initiation of replication via binding of the 2 NTPs needed to initiate cRNA synthesis. For example, if the two NTPs required to initiate vRNA replication bind prior to the capped cellular RNA, the RdRp replicates the vRNA. However, if a capped cellular mRNA binds the polymerase prior to the two NTPs, this may direct the polymerase to initiate transcription by cleaving the capped mRNA and polymerizing NTPs onto the primer. Alternatively, commitment to transcription may require both binding of the capped mRNA followed by cap snatching.
High concentrations of vRNA, specifically the 5’ end of the vRNA, cause RdRp to switch towards a replicational mode. We propose that since the influenza polymerase must already be bound to a vRNA template for either vRNA → cRNA replication or transcription to occur, RdRp contains an additional binding site for vRNA that is distinct from the substrate binding site. The 3’ vRNA end, the 3’ cRNA end, the 5’ cRNA end, nor xRNA could induce the RdRp to switch towards replication, indicating that this site has sequence specificity for the 5’ terminus of the vRNA template. The binding of the 5’ end of vRNA to an allosteric site might induce a conformational change in the polymerase that inhibits the binding of cap source, which would lock the polymerase in a replicational mode for as long as the 5’ end of vRNA is bound. Alternatively, the 5’ end of vRNA might bind to the RdRp cap binding site and directly prevent the polymerase from binding the cap source required for transcription.
Recently, Perez et al reported the existence of influenza derived small viral RNAs (svRNA) with sequences corresponding to the 5’ ends of the vRNA segments. Infected cells will contain two sources of vRNA 5’ ends – the vRNA itself and the svRNA. Introduction of an antisense locked nucleic acid complimentary to the svRNA into influenza-infected cells caused a disregulation of mRNA, cRNA, and vRNA synthesis (26), consistent with the 5’ end of the vRNA template playing an important role in regulating the influenza polymerase. Thus, these svRNA may be the actual regulator of the switch from transcription to replication. (Note: In these whole cell studies, the antisense locked nucleic acid will have bound to both the vRNA segments as well as to the svRNA, hence it is not strictly possible to determine if the effects of the antisense nucleic acid resulted from binding to svRNA, vRNA or both svRNA and vRNA.)
Increasing the RdRp concentration pushes the polymerase towards synthesizing more replication products relative to transcription products, suggesting that the RdRp can self associate into oligomers. Recently, Huet et al have observed the formation of RdRp oligomers in live cells with florescence cross-correlation spectroscopy and Jorba et al have also detected oligomers of RdRp via gel-filtration analysis (27, 28). Formation of these oligomers, therefore, likely plays a key role in regulating whether RdRp produces viral mRNA or replicates the viral genome. While it is likely that an oligomerization of the three subunit RdRp is involved in this regulation, we cannot discount the possibility that only one of the three polymerase subunits is responsible for the switch.
As influenza begins its infection of a cell it would be advantageous for the virus to quickly produce the viral machinery required to fend off immune responses from the host, replicate its own genome, and package nascent virons. This would require that the viral polymerase primarily produce mRNA early in infection. During the later stages of infection, once the viral components have been produced, the virus can then focus on the replication of its genome and the polymerase can switch towards replication of both cRNA and vRNA.
Consistent with these ideas, previous studies have indicated that during early stages of infection there is an accumulation of influenza mRNA and very little cRNA or vRNA, implying that the influenza polymerase is in a primarily transcriptional mode during the beginning of infection. At latter stages of infection, the concentrations of cRNA and vRNA increase (15-17), suggesting that the RdRp has switched to a replicational mode. Importantly, the results described above accurately predict such a switch.
We have found that high concentrations of cap source switch the influenza polymerase to a transcriptional mode. RdRp has been shown to associate with RNA polymerase II when the eukaryotic polymerase's carboxy terminal domain is hyperphosphorylated, which would give it access to locally high concentrations of cap source (29). Thus, during the early stages of infection the RdRp would have access to these locally high concentrations of cap source while the concentrations of vRNA and RdRp would be low compared to the levels found later in infection; these conditions would result in RdRp primarily transcribing viral mRNA. Vreede et al have provided evidence that influenza polymerase mediates the ubiquitination of RNA polymerase II, thereby leading to the degradation of the host polymerase via the proteasome (30, 31). This degradation should significantly lower the concentration of host cap source available to RdRp, thus decreasing the amount of transcription and increasing the amount of replication. As the infection progresses, the concentrations of all viral proteins increase in response to the accumulation of viral mRNA, with the increase in RdRp and NP concentrations subsequently protecting nascent cRNA from degradation. The newly protected cRNA can now be used as a template for producing more vRNA and/or svRNA. Importantly, this increased vRNA concentration would result in RdRp switching into the replicational mode. The svRNAs consisting of 5’-end of the vRNA segments would also likely contribute to this effect. While it is unknown how the concentration of svRNAs varies during the infection cycle, one would expect them to increase since their production is presumably dependent upon the presence of cRNA. Thus, the increasing levels of vRNA and svRNA serve as a positive feedback loop for even greater vRNA production. The increase in RdRp concentration during the infection cycle would also lead to self association of the polymerase, further enhancing the switch towards replication.
Regulation of the relative amounts of transcription versus replication almost certainly involves processes other than those described above. Vreede et al. have proposed a stabilization model whereby at early stages of infection the polymerase is synthesizing both cRNA and mRNA. However, the product of replication, cRNA, is unprotected by proteins and quickly degraded by host nucleases while the mRNA's 7-methylguanosine 5’ cap and polyadenylated tail protect it from degradation and allow it to be exported from the nucleus. At later stages of infection the mRNA will have been translated to generate large amounts of RdRp and nucleoproteins, both of which bind to and stabilize the cRNA against degradation (19, 20). This stabilization model would also help account for the early accumulation of mRNA followed by a build up of cRNA (and vRNA) later in infection.
Robb et al reported that the influenza NS2 protein plays a role in regulating the influenza polymerase by decreasing the ratio of transcriptional/replicational products (18). It seems that influenza NS2 and RdRp are both regulators in a negative feedback loop where influenza polymerase will transcribe viral mRNA which will then be used to translate viral proteins including NS2 and RdRp; once levels of NS2 and RdRp are high enough, they convert RdRp from a transcriptional mode to a replicative mode.
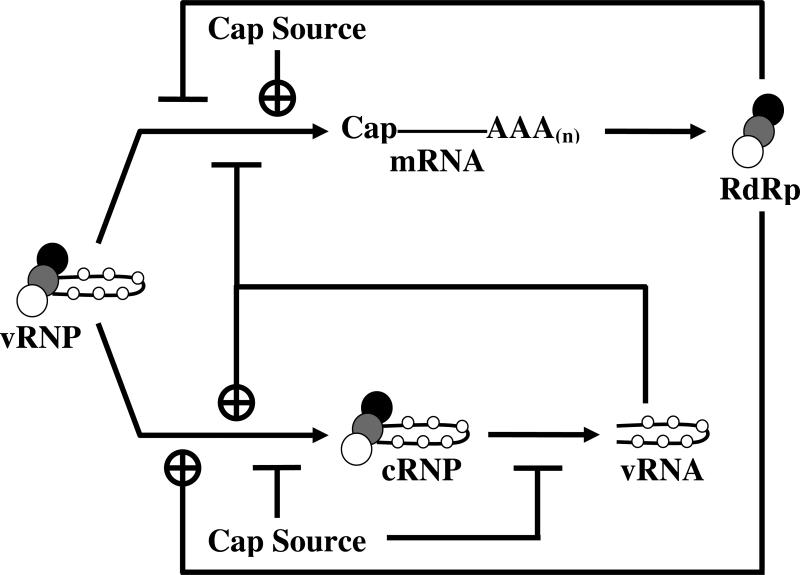
Figure 7 summarizes the effects of varying vRNA, capped cellular mRNA and RdRp on the relative rates of transcription and replication. During early stages of infection there are low levels of vRNA and RdRp and locally high concentrations of capped cellular mRNA. The elevated cap source concentration signals an increase in the production of transcriptional products while inhibiting both vRNA → cRNA and cRNA → vRNA replication, thereby increasing the rate of transcription and, therefore, the rate at which viral proteins accumulate. Later stages of infection feature the decline of the concentration of capped cellular mRNA and an increase in the levels of both vRNA and RdRp, both of which signal a decrease in the rate of transcription and an increase in replication. In combination with other reported regulatory mechanisms described above, it is clear that influenza has developed a remarkably complex regulatory network for ensuring the appropriate levels of viral mRNA needed for protein production and vRNA that will both serve as a substrate and be packaged into new viral particles. Indeed, it would not be surprising if future studies report the existence of additional viral and host factors that regulate the rates and balance between transcription and replication.
Figure 7.
The viral ribonucleoprotein complex (vRNP) can be influenced to adopt a transcriptional mode by increased concentrations of cap source, whereas increased concentrations of vRNA and viral polymerase will effect a switch towards replication.
Footnotes
This work was supported by NIH grants GM54195 and AI071338.
Abbreviations used: Complementary RNA (cRNA), Viral Ribonucleoprotein packaging cRNA (cRNP), Dithiothreitol (DTT), Ethylenediaminetetraacetic acid (EDTA), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), Phenylmethanesulfonylfluoride (PMSF), RNA-dependent RNA polymerase (RdRp), Tandem Affinity Purification (TAP), Viral RNA (vRNA), Viral Ribonucleoprotein packaging vRNA (vRNP)
References
- 1.Dias A, Bouvier D, Crepin T, McCarthy AA, Hart DJ, Baudin F, Cusack S, Ruigrok RW. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature. 2009;458:914–918. doi: 10.1038/nature07745. [DOI] [PubMed] [Google Scholar]
- 2.Yuan P, Bartlam M, Lou Z, Chen S, Zhou J, He X, Lv Z, Ge R, Li X, Deng T, Fodor E, Rao Z, Liu Y. Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature. 2009;458:909–913. doi: 10.1038/nature07720. [DOI] [PubMed] [Google Scholar]
- 3.Guilligay D, Tarendeau F, Resa-Infante P, Coloma R, Crepin T, Sehr P, Lewis J, Ruigrok RW, Ortin J, Hart DJ, Cusack S. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nature structural & molecular biology. 2008;15:500–506. doi: 10.1038/nsmb.1421. [DOI] [PubMed] [Google Scholar]
- 4.Braam J, Ulmanen I, Krug RM. Molecular-Model of a Eukaryotic Transcription Complex - Functions and Movements of Influenza-P Proteins During Capped Rna-Primed Transcription. Cell. 1983;34:609–618. doi: 10.1016/0092-8674(83)90393-8. [DOI] [PubMed] [Google Scholar]
- 5.Blaas D, Patzelt E, Kuechler E. Cap-recognizing protein of influenza virus. Virology. 1982;116:339–348. doi: 10.1016/0042-6822(82)90425-1. [DOI] [PubMed] [Google Scholar]
- 6.Blaas D, Patzelt E, Kuechler E. Identification of the cap binding protein of influenza virus. Nucleic acids research. 1982;10:4803–4812. doi: 10.1093/nar/10.15.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ulmanen I, Broni BA, Krug RM. Role of two of the influenza virus core P proteins in recognizing cap 1 structures (m7GpppNm) on RNAs and in initiating viral RNA transcription. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:7355–7359. doi: 10.1073/pnas.78.12.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng T, Vreede FT, Brownlee GG. Different de novo initiation strategies are used by influenza virus RNA polymerase on its cRNA and viral RNA promoters during viral RNA replication. Journal of virology. 2006;80:2337–2348. doi: 10.1128/JVI.80.5.2337-2348.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hay AJ, Lomniczi B, Bellamy AR, Skehel JJ. Transcription of the influenza virus genome. Virology. 1977;83:337–355. doi: 10.1016/0042-6822(77)90179-9. [DOI] [PubMed] [Google Scholar]
- 10.Hay AJ, Skehel JJ, McCauley J. Characterization of influenza virus RNA complete transcripts. Virology. 1982;116:517–522. doi: 10.1016/0042-6822(82)90144-1. [DOI] [PubMed] [Google Scholar]
- 11.Plotch SJ, Bouloy M, Ulmanen I, Krug RM. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell. 1981;23:847–858. doi: 10.1016/0092-8674(81)90449-9. [DOI] [PubMed] [Google Scholar]
- 12.Robertson JS, Schubert M, Lazzarini RA. Polyadenylation sites for influenza virus mRNA. Journal of virology. 1981;38:157–163. doi: 10.1128/jvi.38.1.157-163.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poon LL, Pritlove DC, Fodor E, Brownlee GG. Direct evidence that the poly(A) tail of influenza A virus mRNA is synthesized by reiterative copying of a U track in the virion RNA template. Journal of virology. 1999;73:3473–3476. doi: 10.1128/jvi.73.4.3473-3476.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poon LL, Fodor E, Brownlee GG. Polyuridylated mRNA synthesized by a recombinant influenza virus is defective in nuclear export. Journal of virology. 2000;74:418–427. doi: 10.1128/jvi.74.1.418-427.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor JM, Illmensee R, Litwin S, Herring L, Broni B, Krug RM. Use of specific radioactive probes to study transcription and replication of the influenza virus genome. Journal of virology. 1977;21:530–540. doi: 10.1128/jvi.21.2.530-540.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mark GE, Taylor JM, Broni B, Krug RM. Nuclear accumulation of influenza viral RNA transcripts and the effects of cycloheximide, actinomycin D, and alpha-amanitin. Journal of virology. 1979;29:744–752. doi: 10.1128/jvi.29.2.744-752.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engelhardt OG, Fodor E. Functional association between viral and cellular transcription during influenza virus infection. Rev. Med. Virol. 2006;16:329–345. doi: 10.1002/rmv.512. [DOI] [PubMed] [Google Scholar]
- 18.Robb NC, Smith M, Vreede FT, Fodor E. NS2/NEP protein regulates transcription and replication of the influenza virus RNA genome. The Journal of general virology. 2009;90:1398–1407. doi: 10.1099/vir.0.009639-0. [DOI] [PubMed] [Google Scholar]
- 19.Vreede FT, Brownlee GG. Influenza virion-derived viral ribonucleoproteins synthesize both mRNA and cRNA in vitro. Journal of virology. 2007;81:2196–2204. doi: 10.1128/JVI.02187-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vreede FT, Jung TE, Brownlee GG. Model suggesting that replication of influenza virus is regulated by stabilization of replicative intermediates. Journal of virology. 2004;78:9568–9572. doi: 10.1128/JVI.78.17.9568-9572.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kashiwagi T, Leung BW, Deng T, Chen H, Brownlee GG. The N-terminal region of the PA subunit of the RNA polymerase of influenza A/HongKong/156/97 (H5N1) influences promoter binding. PloS one. 2009;4:e5473. doi: 10.1371/journal.pone.0005473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng T, Sharps J, Fodor E, Brownlee GG. In vitro assembly of PB2 with a PB1-PA dimer supports a new model of assembly of influenza A virus polymerase subunits into a functional trimeric complex. Journal of virology. 2005;79:8669–8674. doi: 10.1128/JVI.79.13.8669-8674.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fodor E, Pritlove DC, Brownlee GG. The influenza virus panhandle is involved in the initiation of transcription. Journal of virology. 1994;68:4092–4096. doi: 10.1128/jvi.68.6.4092-4096.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cianci C, Tiley L, Krystal M. Differential activation of the influenza virus polymerase via template RNA binding. Journal of virology. 1995;69:3995–3999. doi: 10.1128/jvi.69.7.3995-3999.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagen M, Chung TD, Butcher JA, Krystal M. Recombinant influenza virus polymerase: requirement of both 5' and 3' viral ends for endonuclease activity. Journal of virology. 1994;68:1509–1515. doi: 10.1128/jvi.68.3.1509-1515.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perez JT, Varble A, Sachidanandam R, Zlatev I, Manoharan M, Garcia-Sastre A, Tenoever BR. Influenza A virus-generated small RNAs regulate the switch from transcription to replication. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11525–11530. doi: 10.1073/pnas.1001984107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huet S, Avilov SV, Ferbitz L, Daigle N, Cusack S, Ellenberg J. Nuclear import and assembly of influenza A virus RNA polymerase studied in live cells by fluorescence cross-correlation spectroscopy. Journal of virology. 2010;84:1254–1264. doi: 10.1128/JVI.01533-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jorba N, Area E, Ortin J. Oligomerization of the influenza virus polymerase complex in vivo. The Journal of general virology. 2008;89:520–524. doi: 10.1099/vir.0.83387-0. [DOI] [PubMed] [Google Scholar]
- 29.Engelhardt OG, Smith M, Fodor E. Association of the influenza A virus RNA-dependent RNA polymerase with cellular RNA polymerase II. Journal of virology. 2005;79:5812–5818. doi: 10.1128/JVI.79.9.5812-5818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vreede FT, Chan AY, Sharps J, Fodor E. Mechanisms and functional implications of the degradation of host RNA polymerase II in influenza virus infected cells. Virology. 396:125–134. doi: 10.1016/j.virol.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan AY, Vreede FT, Smith M, Engelhardt OG, Fodor E. Influenza virus inhibits RNA polymerase II elongation. Virology. 2006;351:210–217. doi: 10.1016/j.virol.2006.03.005. [DOI] [PubMed] [Google Scholar]