Abstract
Structural analysis of sulfate complexes of two different azacryptands crystallized under identical conditions in the presence of methanol and water reveals that one methanol is selectively trapped in each cavity, assisted by the specific arrangements of three external sulfates close to one tren unit.
Methanol is ubiquitous in nature and is known to bind to an active site of corrinoid protein through the interaction between methanolic hydroxyl group and zinc ion.1 It is extensively used as a solvent in laboratory and industry and also as a supplement to gasoline.2 Due to its poor ability to interact with a synthetic receptor, very little is known about methanol binding.3
Although azacryptands have been shown to coordinate with cations and anions,4 their binding to neutral species remains relatively unexplored. Protonated azacryptands have the potential to coordinate with H-bond acceptor atoms in neutral molecules. However, such complexation can be hampered due to the presence of counter anions that preferentially bind to the host macrocycles.5 If a counterion is chosen that shows only weak interaction with the macrocycle, then such a cryptand can also act as a host to a neutral molecule. This approach has been applied successfully to bind water in a p-xylyl-based cryptand using iodide,6 perchlorate,7 picrate,8 or phosphate10 as a counter anion. In this communication, we report two novel and remarkable crystal structures of trapped methanol within bicyclic cryptands L1 and L2, each grown from its sulfate salt in water–methanol mixtures under similar conditions. The specific H-bonding interactions of one tren unit with three sulfates located at the outside lead to the complete inclusion of one methanol molecule in the cavity.
The ligands L1 and L2 were synthesized following the literature procedure.10 The sulfate complexes were obtained by titrating the ligands with sulfuric acid in water. Crystals suitable for X-ray analysis were grown by vapor diffusion of methanol to the salts dissolved in a mixture of water and methanol. Although the ligands consist of two different linking spacers, both of them were found to be hexaprotonated, each with several water molecules and one methanol. However, the methanol is trapped within the bicyclic cavity in both the cryptands (Fig. 1). ‡
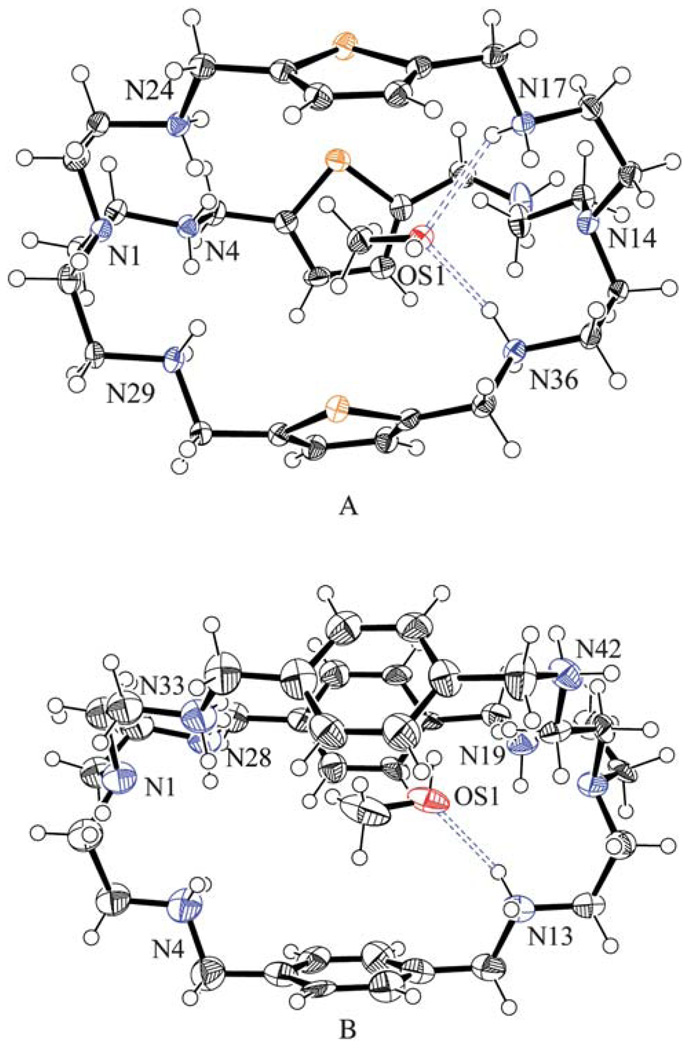
Fig. 1.
ORTEP views of [H6L1(CH3OH)]6+ (A) and [H6L2(CH3OH)]6+ (B) motifs showing encapsulated methanol inside the cavity. Thermal ellipsoids are drawn at the 40% probability level.
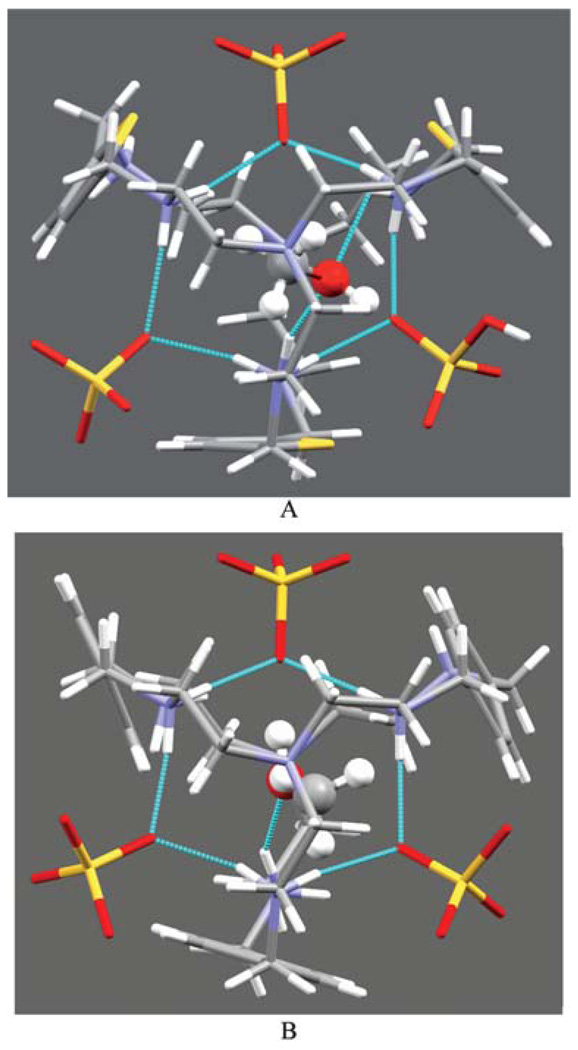
The ligand L1 containing three thiophene spacers crystallizes with sulfate anions in the triclinic space group P1̄. The general shape of this complex is more elongated than similar complexes with different counterions as shown by the distances between the bridgehead nitrogens (9.457 Å this study, 6.096 Å for the chloride complex10b and 5.528 Å for the nitrate11 complex). Despite the presence of several H-bond donors, the cavity does not contain any sulfate or water. However, the single methanol present in the complex is coordinated inside the macrocyclic cavity by two strong hydrogen bonds (N17⋯O1S = 2.947(5) and N36⋯O1S = 2.832(5) Å) with secondary amines of the right side tren unit as shown in Fig. 1A. The other tren unit is involved in coordinating three external sulfate groups (one HSO4− and two SO42−), each being cleft between the arms through two NH⋯O bonds with protonated secondary amines (Fig. 2A). Thus all six protons on the secondary nitrogens of one tren unit are involved in hydrogen bonding with three external anions.
Fig. 2.
Crystal views of [H6L1(CH3OH)(2SO4)(HSO4)]+ (A) and [H6L2(CH3OH)(3SO4)] (B) motifs showing encapsulated methanol (ball and stick) and three cleft bound sulfates.
The ligand L2 with p-xylyl spacers is slightly larger than L1 and its sulfate salt crystallizes in the monoclinic space group P21/c, with six protonated amines. The complex contains one crystalline methanol which is again trapped in the void of the ellipsoid cavity, with one strong N⋯O bond (N13⋯O1S = 2.884(18) Å) via the methanolic oxygen with a protonated amine of one tren unit (Fig. 1B). The trapped molecule lies almost linearly along the pseudo-threefold axis passing through the apical nitrogen atoms, N1 and N16. The distance between the apical atoms is 9.786 Å. The other tren is coordinated to three sulfates in a manner similar to that observed in the structure of the L1 complex as shown in Fig. 2B.
In order to determine the binding energies and stability of L1 and L2 complexes with methanol, we have performed quantum-chemical investigation with the Gaussian 03 program package12 using density functional theory at the M052x/6-31+G(d) level of theory.13 Geometries of water and methanol molecules were fully optimized. Initial geometries of ligands L1 and L2 and their complexes with methanol were derived from the crystallographic coordinates with corrected lengths of bonds that involve hydrogen atoms. Single-point energies were calculated for those structures. The binding energy was computed as ΔE = E (cryptand–methanol) − E (cryptand) − E (methanol). The basis set superposition error was corrected through counterpoise approach.14 The results are −0.59 kcal mol−1 for L1–methanol complex and −5.90 kcal mol−1 for L2–methanol complex. Therefore in both cases formation of the complex is accompanied by release of the energy and stabilization of the system. Stability of the methanol complexes with the cryptands was further compared to the stability of their water complexes. This was done by optimizing the position of the encapsulated small molecule inside the cryptand, whose geometry was frozen. The binding energies for all four complexes are shown in Table 1.
Table 1.
Binding energies (kcal mol−1) of the optimized complexes
| L1 | L2 | ||
|---|---|---|---|
| H2O | CH3OH | H2O | CH3OH |
| −0.61 | −9.94 | −13.24 | −12.78 |
The results clearly show that the formation of methanol complex is more favorable than that of water complex with ligand L1, supporting the trapping of methanol in the presence of water as observed in the crystallographic structure. The bond distances, N36⋯O1S (2.832(5) Å) in L1 and N13⋯O1S (2.884(18) Å) in L2 are very close to the corresponding calculated values, 2.885 and 3.134 Å, respectively, which are also in agreement with the reported values (3.056(6) and 3.099(6) Å) observed in the azametallacrown encapsulating two methanol molecule.3d The difference in binding energies of L2 complexes is insignificantly small, therefore in this case we assume that preferable encapsulation of methanol is due to steric and electronic factors.
In conclusion, we have structurally characterized two azacryptands that are capable of binding a single methanol selectively from sulfate and water species. There are striking similarities between the two complexes with respect to the encapsulated methanol and three cleft-bound sulfate groups. Jiang and coworkers characterized a sulfate complex of L2 grown in an aqueous solution that contained several water molecules in the cavity.9 Bowman-James and coworkers isolated crystals of a sulfate complex with a m-xylyl analogue in which the cavity contained a sulfate group.15 The arrangement of three external sulfates coordinating to one tren unit appears to open the other tren unit making it capable of receiving polar neutral species from the solution. This study outlines a simple strategy for binding neutral molecules that can be extended to other systems.
Supplementary Material
Acknowledgments
Research is sponsored by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences, US Department of Energy via contract with Oak Ridge National Laboratory (MAH). The project described was supported by grant number G12RR013459 from the National Center for Research Resources. This material is based upon work supported by the National Science Foundation under CHE-0821357. This work was also supported by the NSF Nanotoxicity CREST Center Grant No. HRD-0833178. Authors thank Mississippi Center for Supercomputing Research for computational facilities. The authors thank the National Science Foundation (CHE-0130835) and the University of Oklahoma for funds to acquire the diffractometer used in this work.
Footnotes
Electronic supplementary information (ESI) available: Two crystallographic data in CIF format, and the synthetic procedure, crystallographic details and Cartesian coordinates of DFT calculations in pdf format. CCDC reference numbers 755129 and 755130. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c0ce00162g
Notes and references
‡ Crystal data: 2[H6L1(CH3OH)])]·3SO4·6HSO4·11H2O, M = 2378.83, triclinic, a = 11.8698(14) Å, b = 14.1475(18) Å, c = 16.133(2) Å, α = 97.812(7)°, β = 96.193(7)°, γ = 101.013(8)°, V = 2609.5(6) Å3, T = 100(2) K, space group P-1, Z = 1, µ(CuKα) = 3.751 mm°1, 35 685 reflections measured, 9417 independent reflections (Rint = 0.0794). The final R1 value was 0.0573 (I > 2σ(I)).
For H6L2(CH3OH)]·3SO4·12H2O, M = 1141.33, monoclinic, a = 15.303(2) Å, b = 16.583(2) Å, c = 23.124(3) Å, α = 90.00°, β = 104.234(8)°, γ = 90.00°, V = 5688.0(13) Å3, T = 100(2) K, space group P21/c, Z = 4, µ(CuKα) = 1.918 mm°1, 11 543 reflections measured, 1749 independent reflections (Rint = 0.0719). The final R1 value was 0.0777 (I > 2σ(I)).
- 1.(a) Hagemeier CH, Kruer M, Thauer RK, Warkentin E, Ermler U. Proc. Natl. Acad. Sci. U. S. A. 2006;103:18917–18922. doi: 10.1073/pnas.0603650103. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Das A, Fu ZQ, Tempel W, Liu ZJ, Chang J, Chen L, Lee D, Zhou W, Xu H, Shaw N, Rose JP, Ljungdahl LG, Wang BC. Proteins. 2007;67:167–176. doi: 10.1002/prot.21094. [DOI] [PubMed] [Google Scholar]
- 2.Olah GA. Angew. Chem., Int. Ed. 2005;44:2636–2639. doi: 10.1002/anie.200462121. [DOI] [PubMed] [Google Scholar]
- 3.(a) Zhao N, Bullinger JC, Van Stipdonk MJ, Stern CL, Eichhorn DM. Inorg. Chem. 2008;47:5945–5950. doi: 10.1021/ic8003307. [DOI] [PubMed] [Google Scholar]; (b) Rachlewicz K, Latos-Grazyski L, Gebauer A, Vivian A, Sessler JL. J. Chem. Soc., Perkin Trans. 1999;2:2189–2195. [Google Scholar]; (c) Ben-Haida A, Colquhoun HM, Hodge P, Williams DJ. J. Mater. Chem. 2000;10:2011–2016. doi: 10.1002/1521-3765(20001201)6:23<4285::aid-chem4285>3.3.co;2-4. [DOI] [PubMed] [Google Scholar]; (d) Lin S, Liu SX, Huang JQ. J. Chem. Soc., Dalton Trans. 2002:1595–1601. [Google Scholar]; (e) Nakazawa J, Nagiwara J, Mizuki M, Shimazaki Y, Tani F, Naruta Y. Angew. Chem., Int. Ed. 2005;44:3744–3746. doi: 10.1002/anie.200500732. [DOI] [PubMed] [Google Scholar]; (f) Muniappan S, Lipstman S, Goldberg I. Acta Crystallogr., Sect. C: Cryst. Struct. Commun. 2006;62:m477–m479. doi: 10.1107/S0108270106034986. [DOI] [PubMed] [Google Scholar]; (g) Nakazawa J, Mizuki M, Hagiwara J, Shimazaki Y, Tani F, Naruta Y. Bull. Chem. Soc. Jpn. 2006;79:1431–1443. [Google Scholar]; (h) Tolman WB, Rardin RL, Lippard SJ. J. Am. Chem. Soc. 1989;111:4532–4533. [Google Scholar]
- 4.(a) Nelson J, McKee V, Morgan G. Prog. Inorg. Chem. 1998;47:167–316. [Google Scholar]; (b) McKee V, Nelson J, Town RM. Chem. Soc. Rev. 2003;32:309–325. doi: 10.1039/b200672n. [DOI] [PubMed] [Google Scholar]; (c) Hossain MA. Curr. Org. Chem. 2008;12:1231–1256. [Google Scholar]
- 5.Bowman-James K. Acc. Chem. Res., 2005;38:671–678. doi: 10.1021/ar040071t. [DOI] [PubMed] [Google Scholar]
- 6.Lakshminarayanan PS, Kumar DK, Ghosh P. Inorg. Chem. 2005;44:7540–7546. doi: 10.1021/ic051191f. [DOI] [PubMed] [Google Scholar]
- 7.Yang L-Z, Jiang L, Feng X-L, Lu T-B. CrystEngComm. 2008;10:649–651. [Google Scholar]
- 8.Ravikumar I, Lakshminarayanan PS, Suresh E, Ghosh P. Cryst. Growth Des. 2006;6:2630–2633. [Google Scholar]
- 9.Li Y, Jiang L, Feng XL, Lu TB. Cryst. Growth Des. 2008;8:3689–3694. [Google Scholar]
- 10.(a) Hossain MA, Llinares JM, Mason S, Morehouse P, Powell D, Bowman- James K. Angew. Chem., Int. Ed. 2002;41:2335–2338. doi: 10.1002/1521-3773(20020703)41:13<2335::AID-ANIE2335>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]; (b) Saeed MA, Fronczek FR, Hossain MA. Chem. Commun. 2009:6409–6411. doi: 10.1039/b916099j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saeed MA, Fronczek FR, Huang MJ, Hossain MA. Chem. Commun. 2010;46:404–406. doi: 10.1039/b923013k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frisch MJ, et al. Gaussian 03, Revision E.01. Wallingford, CT: Gaussian, Inc.; 2004. [Google Scholar]
- 13.Zhao Y, Schultz NE, Truhlar DG. J. Chem. Theor. Comput. 2006;2:364. doi: 10.1021/ct0502763. [DOI] [PubMed] [Google Scholar]
- 14.Boys SF, Bernardi F. Mol. Phys. 1970;19:553. [Google Scholar]
- 15.Kang SO, Hossain MA, Powell D, Bowman-James K. Chem. Commun. 2005:328–330. doi: 10.1039/b411904e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.