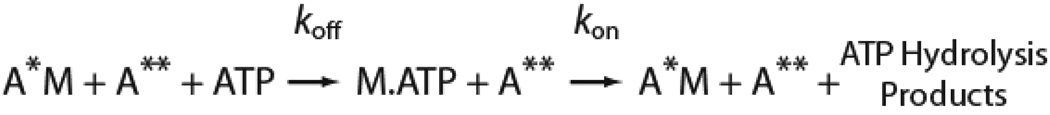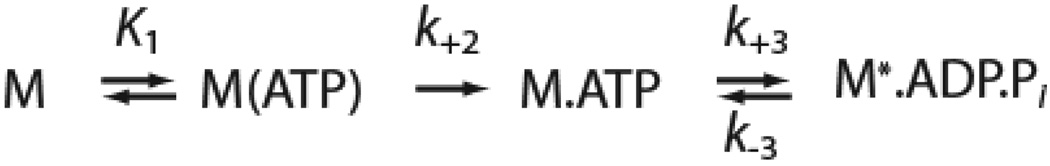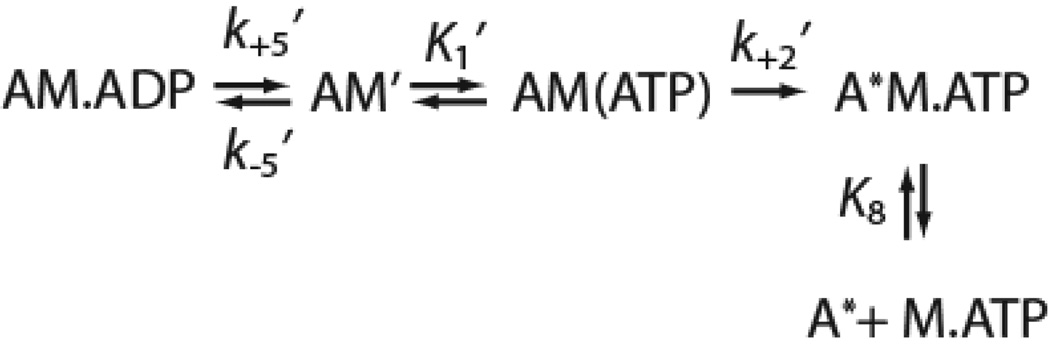Abstract
Myo1c is a member of the myosin superfamily that has been proposed to function as the adaptation motor in vestibular and auditory hair cells. A recent study identified a myo1c point mutation (R156W) in a person with bilateral sensorineural hearing loss. This mutated residue is located at the start of the highly conserved switch-1 region, which is a crucial element for the binding of nucleotide. We characterized the key steps on the ATPase pathway at 37 °C using recombinant wild-type (myo1c3IQ) and mutant myo1c (R156W-myo1c3IQ) constructs that consist of the motor domain and three IQ motifs. The R156W mutation only moderately affects the rates of ATP binding, ATP-induced actomyosin dissociation, and ADP release. The actin-activated ATPase rate of the mutant is inhibited > 4-fold, which is likely due to a decrease in the rate of phosphate release. The rate of actin gliding, as measured by the in vitro motility assay, is unaffected by the mutation at high myosin surface densities, but actin gliding is substantially reduced at low surface densities of R156W-myo1c3IQ. We used a frictional-loading assay to measure the affect of resisting forces on the rate of actin gliding and found that R156W-myo1c3IQ is less force sensitive than myo1c3IQ. Taken together, these results indicate that myo1c with the R156W mutation has a lower duty ratio than the wild-type protein and motile properties that are less sensitive to resisting forces.
Myo1c is the single-headed member of the myosin superfamily that plays roles in trafficking of GLUT4-containing vesicles to the plasma membrane in response to insulin stimulation (1, 2) and in compensatory endocytosis following regulated exocytosis (3). Myo1c has also been proposed to play a key role in the process of adaptation in specialized sensory cells, where it is thought to dynamically adjust tension on mechanosensitive ion channels via its interaction with actin (4, 5).
Cell biological studies have shown that myo1c localizes and fractionates with cell membranes, and biochemical experiments have shown myosin-I isoforms bind directly to phosphoinositides (6, 7). Additionally, mechanical experiments have shown that a related myosin isoform (myo1b) acts as a tension sensor by responding to small resisting loads by increasing its actin-attachment lifetime, allowing it to generate and sustain tension for extended time periods (8). Thus, evidence points to myo1c acting as a tension-sensing motor protein that links cellular membranes to the underlying actin cytoskeleton.
Consistent with the proposed role of myo1c acting as a tension-sensing adaptation motor in sensory cells, a myo1c point-mutation (R156W) has recently been identified in an individual with bilateral sensorineural hearing loss (Fig. 1; (9)). R156 is highly conserved among the members of the myosin superfamily. It is located at the start of the switch-1 region in the motor domain, which is a crucial structural element involved in nucleotide binding, although R156 is not proposed to interact directly with the nucleotide (10). Point mutations in switch-1 of myosins-II and -V affect nucleotide binding, ATP hydrolysis, phosphate release, and ADP release (11–14). Thus, the R156W mutation likely affects the motile and tension-sensing properties of myo1c. Indeed, the role of myo1c in adaptation may be particularly sensitive to even minor alterations in the ATPase kinetics and load dependent mechanochemistry, since the processes of hearing and balance have stringent force and kinetic requirements (15, 16).
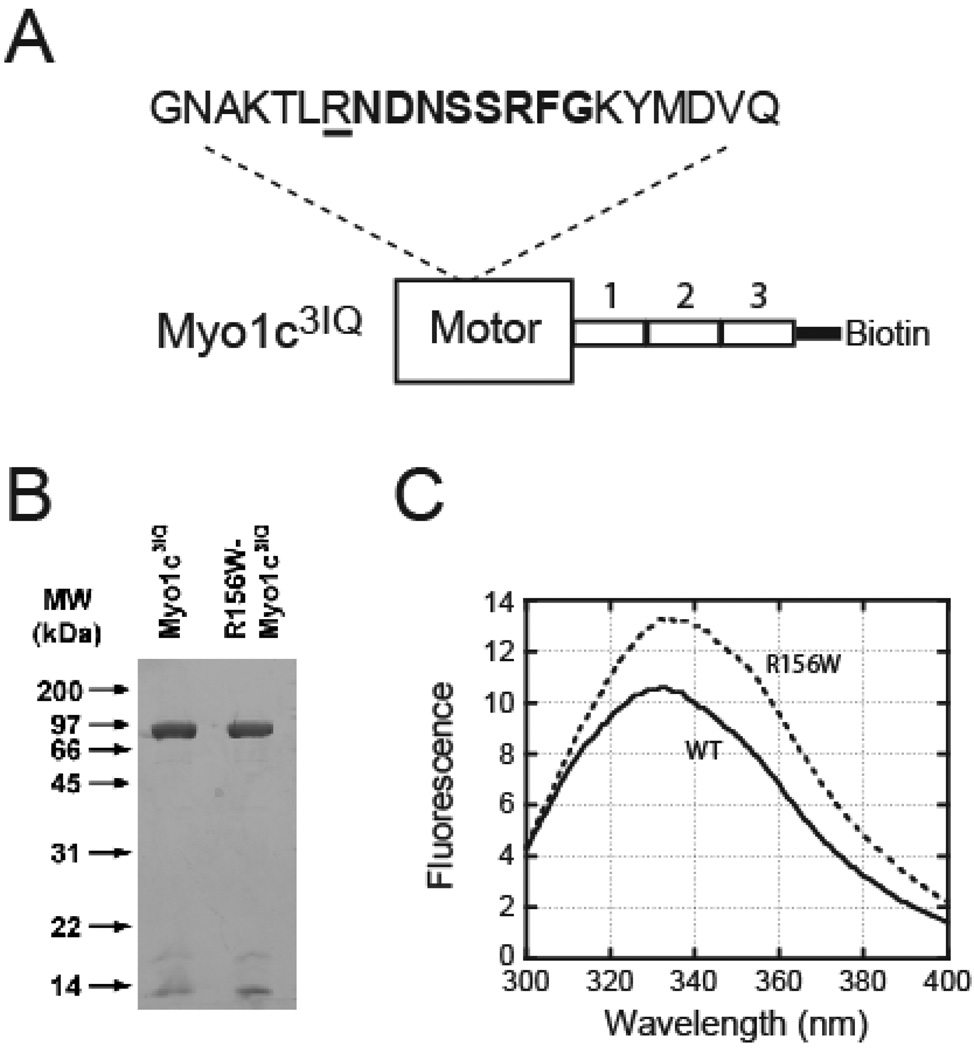
Fig. 1.
A. Schematic of the expressed myo1b3IQ construct showing the relationship of the motor domain (large rectangle) to the IQ motifs (smaller numbered rectangles). The inset shows the positional relationship of the mutated residue (underlined) to switch-1 (bold). B. SDS-PAGE showing purified myo1c3IQ and R156W-myo1c3IQ. C. Steady-state fluorescence emission spectra of 1 µM (solid line) myo1c3IQ and (dotted line) R156W-myo1c3IQ (λex = 280 nm). The peak of the R156W-myo1c3IQ spectrum (335 nm) has a 1.3-fold higher intensity and is red-shifted as compared to myo1c3IQ (332 nm).
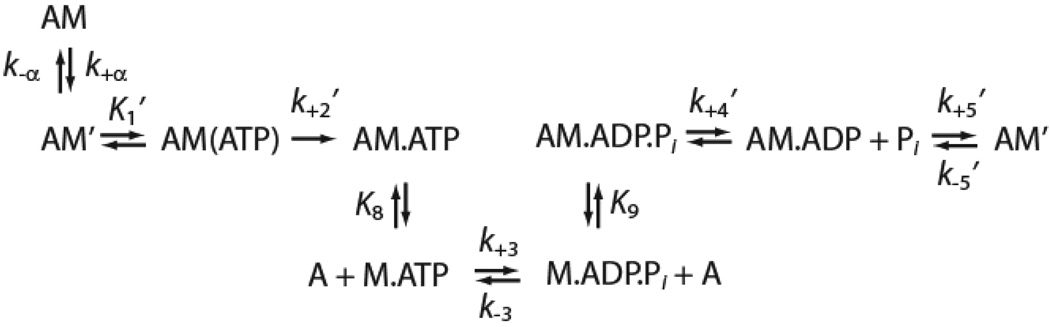
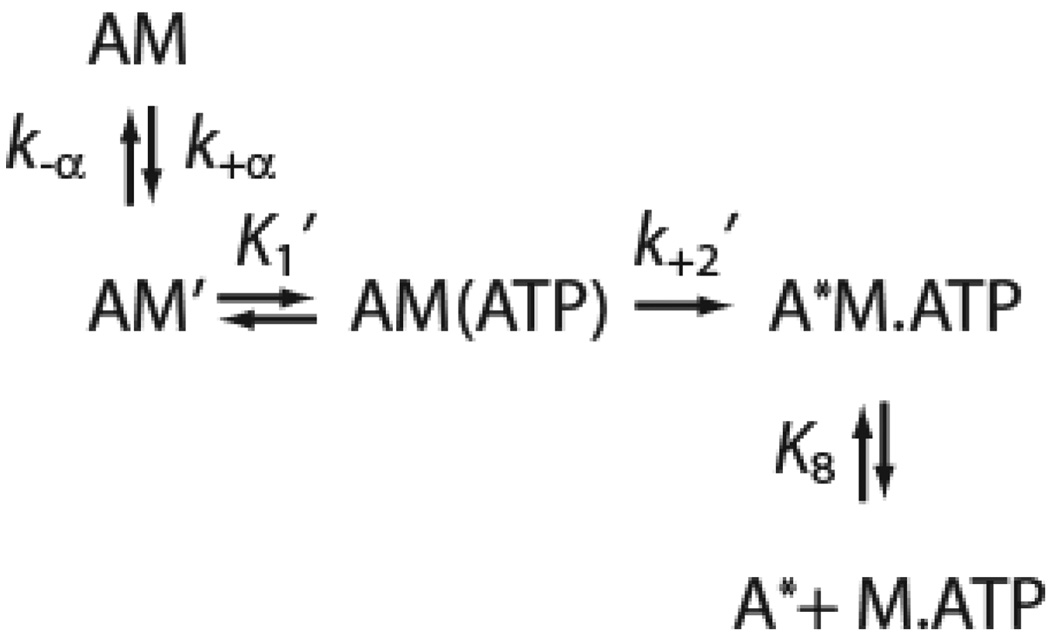
In this study, we determined the effect of the R156W mutation on key steps in the myo1c ATPase cycle (Scheme 1) using transient and steady-state kinetic techniques. We also determined the effect of this mutation on the motile properties of the motor. Experiments were performed with wildtype (myo1c3IQ) and mutated (R156W-myo1c3IQ) myo1c constructs that consist of the motor domain, regulatory domain (contains three calmodulin-binding IQ motifs), and a C-terminal biotinylation tag for site-specific attachment of myosin for motility assays (Fig. 1). We found that the R156W mutation causes a reduction in the myosin duty cycle, likely by reducing the rate of phosphate release. Furthermore, the mutation appears to cause a reduction in the sensitivity of myo1c to resisting loads, possibly explaining the basis of the observed mutant phenotype.
Scheme-1.
EXPERIMENTAL PROCEDURES
Reagents, Proteins, and Buffers
Rabbit skeletal muscle actin was prepared and gel filtered (17). Actin concentrations were determined by absorbance at 290 nm, ε290 = 26,600 M−1cm−1. Actin was labeled with pyrenyl iodoacetamide (pyrene-actin) and gel filtered (18). All actin was stabilized with a molar equivalent of phalloidin (Sigma). Calmodulin was expressed in E. coli and purified as described (19). ADP and ATP concentrations were determined before each experiment by absorbance (ε259 = 15,400 M−1cm−1). Unless stated otherwise, all experiments were performed in KMg25 buffer (10 mM Mops, 25 mM KCl, 1 mM MgCl2, 1 mM EGTA, 1 mM DTT). Unless otherwise stated, all experiments were conducted in the presence of 1 µM free calmodulin (i.e. in excess of the calmodulin already bound to the myosin from the purification).
Myosin-I Expression and Purification
Constructs consisting of the motor domain and three IQ motifs of mouse MYO1C were constructed, expressed in Sf9 cells, and purified as described (20). The R156W-myo1c3IQ construct was made using via QuickChange (Stratagene) and the sequence was verified by sequencing.
Steady-state ATPase measurements
Steady-state ATPase activities were measured in KMg25 at 37 °C using the NADH-coupled assay as described (21, 22). The final protein concentrations after mixing were 100 nM myo1cIQ1-3, 0 – 175 µM actin, and 1 µM free calmodulin. Steady-state fluorescence measurements were made with a Photon Technology International fluorometer.
Transient Kinetic Measurements
Transient kinetic measurements were made with an Applied Photophysics (Surrey, U.K.) SX.18MV stopped-flow (22). A 400 nm long-pass filter was used to monitor pyrene fluorescence (λex = 365 nm), and a 320 nm long-pass filter was used to monitor intrinsic tryptophan fluorescence (λex = 295 nm). Transients were fit to exponential functions using the software supplied with the stopped-flow. All concentrations are given as final after mixing. Single turnover ATPase measurements where were fit to kinetic simulations of Scheme 2 in Berkley Madonna (http://www.berkeleymadonna.com/) to determine the rate constants. For the single turnover measurements, the ratio of myosin to free calmodulin was maintained at 1:1.
Scheme 2.
In vitro motility assays
In vitro motility assays using biotinylated myo1b3IQ and R156W-myo1b3IQ were performed in standard motility chambers at 37° C as described (23). The temperature was controlled using an objective heater (Bioptechs). Nitrocellulose-coated coverslips were exposed to 0.1 mg/mL streptavidin and then blocked with 1 mg/mL BSA. Biotinylated myo1c3IQ (50 – 250 nM) was added to the coverslip and allowed to bind to the immobilized streptavidin. The rate of actin filament gliding was determined using Metamorph (Universal Imaging).
Frictional loading motility assays were performed using an identical procedure to the unloaded motility assays except that the desired amount of α-actinin (Cytoskeleton Inc., Denver, CO) diluted in KMg25 buffer was added to the flow cell and allowed to incubate for 1 minute before the addition of streptavidin. The myosin concentration added to the flow cell was 250 nM. The average gliding rate of 15–35 filaments was drift corrected and measured over 5 frames, captured at rates varying from 10 seconds per frame to 45 seconds per frame, using the freeware tracking software Retrac (http://mc11.mcri.ac.uk/Retrac). Single exponential curves were fit to the data for visualization.
RESULTS
Protein expression and steady-state ATPase activity
We expressed wild type and R156W recombinant myo1c protein constructs that contain the motor domain and three IQ motifs, including a C-terminal sequence for site-specific biotinylation (Fig. 1). The presence of the additional tryptophan is apparent in the intrinsic fluorescence of the protein, as the steady-state emission spectrum (λex = 280 nm) of R156W-myo1c3IQ has a peak intensity that is 1.3-fold greater than myo1c3IQ (Fig. 1). There is also a 3 nm red-shift in the emission peak of the R156W-myo1c3IQ protein.
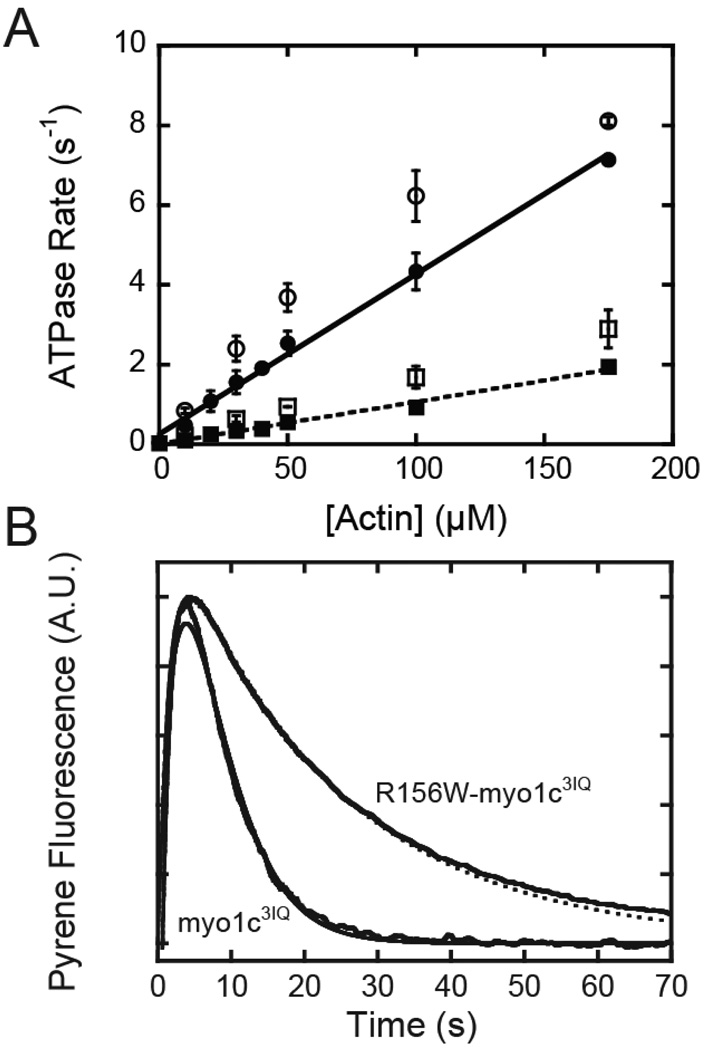
The steady-state ATPase activity of myo1c3IQ is linearly activated by actin with an apparent second order rate constant of va (va = 0.041 (± 0.0015) µM−1s−1; Fig. 2). We were not able to achieve actin concentrations higher than 175 µM due to mixing artifacts resulting from high viscosities. This linear actin-activation suggests that the maximum steady-state ATPase rate (Vmax) is faster than 7 s−1. It also indicates that the affinity of myo1c3IQ for actin in the pre-power-stroke states is very weak, as found for other myosin-I isoforms (24, 25). The ATPase activity of R156W-myo1c3IQ is also linearly related to the actin concentration (va = 0.010 (± 0.0004) µM−1s−1; Fig. 2), but the rates are 4-fold lower than myo1c3IQ. ATPase rates obtained in the absence of added salt were only slightly faster (va = 0.046 (± 0.0052) µM−1s−1 for myo1c3IQ; (va = 0.016 (± 0.0004) µM−1s−1 for R156W-myo1c3IQ), indicating that the affinity of myo1c3IQ for actin is less sensitive to ionic strength than other characterized myosins (26, 27).
Fig. 2.
A. Steady-state ATPase activity of myo1c. Actin concentration dependence of the steady-state ATPase rate of (circles) myo1c3IQ and (squares) R156W-myo1c3IQ measured using the NADH-coupled assay at 37 °C. Closed symbols were acquired in the presence of in KMg25, and open symbols were acquired in the same buffer in the absence of KCl. Solid lines are linear fits to the data acquired in KMg25 with slopes of 0.041 µM−1 s−1 for myo1c3IQ and 0.010 µM−1s−1 for R156W-myo1c3IQ. B. Single turnover measurements of myo1c ATPase activity acquired in KMg25. Pyrene fluorescence transients were obtained by mixing 0.75 µM ATP with a pre-equilibrated mixture of 2.5 µM myosin and 7.5 µM pyrene-actin at 37 °C. Five traces were averaged and normalized. Transients were fit to kinetic simulations of Scheme 2 (smooth lines) to obtain effective rate constants for detachment (koff = 0.1 µM−1 s−1 for myo1c3IQ and koff = 0.28 µM−1s−1 for R156W-myo1c3IQ) and for the strong binding of myosin to pyrene-actin (kon = 0.05 µM−1 s−1 for myo1c3IQ and kon = 0.008 µM−1s−1 for R156W-myo1c3IQ).
To ensure that the measured steady-state ATPase activity of the mutant was not decreased due to the presence of inactive protein, we performed single turnover ATPase measurements (Fig. 2). ATP (0.75 µM) was mixed with a pre-equilibrated complex of 7.5 µM pyrene-labeled actin and 2.5 µM myosin. Upon mixing, the intensity of fluorescence transients increased rapidly due to ATP-induced dissociation of myosin from pyrene actin, which was followed by slow quenching due to repopulation of the strong-binding state. Experiments performed with R156W-myo1c3IQ had a quenching time course that was substantially slower than myo1c3IQ (Fig. 2). Thus, the kinetic step that limits entry into the quenched pyrene state is decreased for R156W-myo1c3IQ. Data were fit via simulation to the following pathway: where A* is the quenched state of actin, A** is the high fluorescence state of actin, koff is the effective rate constant for ATP-induced dissociation of the pyrene-actomyosin complex, and kon is the effective rate constant for myosin binding to actin and formation of a strongly-bound state. Fits of the data yielded kon = 0.05 µM−1s−1 for myo1c3IQ and kon = 0.008 µM−1s−1 for R156W-myo1c3IQ, which are similar to those measured for the steady-state ATPase rates (Fig. 2; Table 1). These results confirm that the observed reduction of ATPase activity of the mutant protein is due to kinetic changes in the myosin and also suggest that the rate limiting step of the ATPase cycle is a transition that precedes entry into the strong binding state.
Table 1.
Rates and equilibrium constants for the myo1c3IQ and R156W-myo1c3IQ ATPasea
| Myo1c3IQ | R156W-myo1c3IQ | |
|---|---|---|
| Steady-State ATPase Activity | ||
| [Actin] dependence of ATPase Rate | 0.041 (± 0.0015) µM−1s−1 | 0.010 (± 0.0004) µM−1s−1 |
| Vmax | > 7 s−1 | > 2 s−1 |
| ATP Binding | ||
| 1 / K1’ | 220 (± 60) µM | 250 (± 40) µM |
| k2’ | 150 (± 15) s−1 | 230 (± 14) s−1 |
| Kα | 1.2 (± 0.31) | 4.2 (± 1.6) |
| k+α | 37 (± 5.4) s−1 | 26 (± 3.1) s−1 |
| k−α | 31 (± 9.1) s−1 | 6.2 (± 2.5) s−1 |
| ATP Hydrolysis | ||
| k3app | 140 (± 9.0) s−1 | 160 (± 14) s−1 |
| ADP release | ||
| K5’ | 1.8 (± 0.54) µM | 1.6 (± 0.27) µM |
| k+5’ | 24 (± 0.50) s−1 | 19 (± 0.087) s−1 |
| k−5’ | 13 (± 4.0) µM−1s−1 | 12 (± 2.0) µM−1s−1 |
Experiments performed in KMg25 (10 mM MOPS (pH 7.0), 25 mM KCl, 1 mM EGTA, 1 mM DTT, 1 mM MgCl2) at 37 °C.
ATP-Induced Population of the Weakly-Bound States
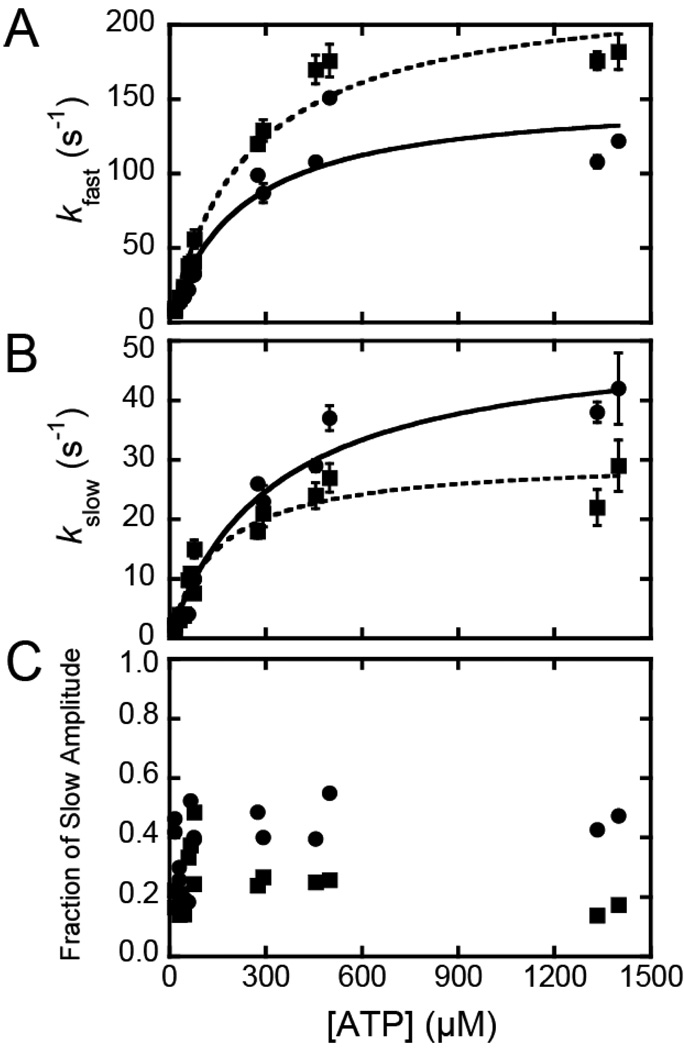
Pyrene-actin fluorescence was used to measure the rate of ATP binding and population of the weakly bound states at 37 °C (22, 28). Mixing actomyo1c with ATP resulted in transient increases in pyrene-actin fluorescence that were best fit to the sum of two exponential rates (Fig. 3). The rates were hyperbolically related to the ATP concentration, with R156W-myo1c3IQ having faster rates than myo1c3IQ. The fast phase (kfast) of the increase in pyrene-actin fluorescence was modeled as ATP binding to the AM state and subsequent population of the AM.ATP state (K1’k+2’), and the slow phase (kslow) was modeled as a transition (k+α) from a nucleotide-insensitive state (AM) to a state that can bind ATP (AM’) as proposed by Geeves (29): where A* represents the unquenched fluorescent state of pyrene-actin. We analyzed the ATP dependence of the fast phase as:
| eq. 1 |
where K1’ is a rapid equilibrium and k+2’ is a rate-limiting isomerization to the high fluorescence AM.ATP state. Values for K1’ and k+2’ for the wildtype and mutant proteins are given in Table 1.
Fig. 3.
ATP-induced population of weakly-bound actomyo1c states. Pyrene fluorescence transients obtained by mixing 1 µM (circles) myo1c3IQ-pyrene-actin or (squares) R156W-myo1c3IQ-pyrene-actin with 16 – 1400 µM ATP at 37 °C were fitted to a double exponential function (kobs= Afast(1−e−kfast·t) + Aslow(1−e−kslow·t)). The rates (A) kfast and (B) kslow are plotted as a function of ATP concentration. The fractional amplitude of the slow phase (Aslow/(Aslow + Afast) is also plotted (C) as a function of ATP concentration. Solid lines are the best fits of the data to equation 1.
At high ATP concentrations, the rate of the slow phase reports the isomerization of AM to AM’ (k+α), and the ratio of the amplitudes of the fast phase to the slow phase reports the equilibrium constant between AM and AM’ (28, 29). Values for Kα, k+α, and k−α were determined by averaging points acquired at ATP concentrations > 300 µM. Interestingly, R156W-myo1c3IQ has a larger Kα than the wildtype protein (Table 1), as can be seen in a plot of the ATP dependence of the amplitude of the slow phase, which reports the mole fraction of the AM state (Fig. 3; Scheme 3).
Scheme 3.
Intrinsic tryptophan fluorescence changes in myo1c3IQ and R156W-myo1c3IQ
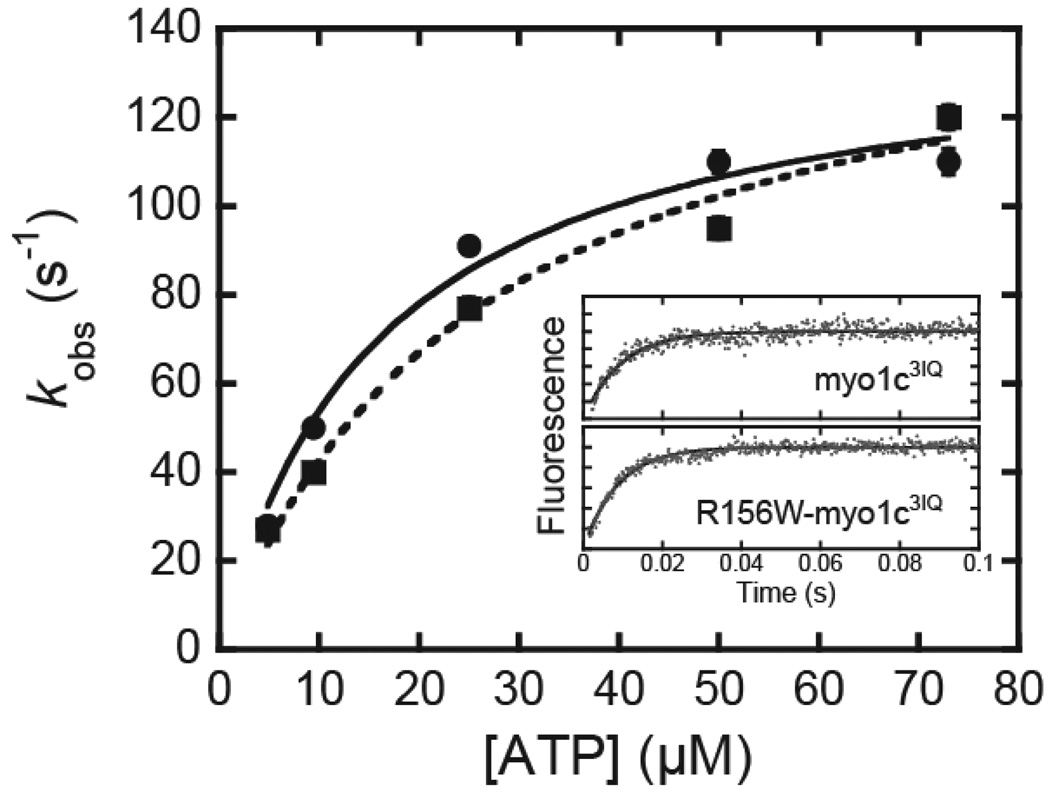
Changes in the intrinsic tryptophan fluorescence of myo1c3IQ and R156W-myo1c3IQ upon mixing with ATP were determined by stopped-flow at 37 °C (Fig. 4). Fluorescence time courses were best fit to single exponential function, and the rates were found to increase hyperbolically with the ATP concentration (Fig. 4). Previous kinetic reports associated this myo1c fluorescence change with the structural transition that accompanies ATP hydrolysis, as shown in Scheme 4 where M* represents the enhanced fluorescence state (30).
Fig. 4.
Rate of ATP hydrolysis as measured by intrinsic tryptophan fluorescence. ATP concentration dependence of the rate of change in tryptophan fluorescence of (circles) myo1c3IQ and (squares) R156W- myo1c3IQ at 37 °C. Solid lines are hyperbolic fits, yielding maximum rates k3app = 140 (± 9.0) s−1 for myo1c3IQ and k3app = 160 (± 14) s−1 for R156W-myo1c3IQ. The inset shows time courses of a fluorescence increase after mixing proteins with 73 µM ATP. The smooth lines are the best fits of the data to a single exponential function with a rates kobs = 110 (± 4.1) s−1 for myo1c3IQ and kobs = 120 (± 2.7) s−1 for R156W-myo1c3IQ.
Scheme 4.
Thus, the maximum rate of the fluorescence change reports the effective rate of ATP hydrolysis (k3app = k+3 + k−3) for the wildtype (k3app = 140 (± 9.0) s−1) and mutant proteins (k3app = 160 (± 14) s−1). The amplitudes and rates of the fluorescent transients for the two proteins are similar, so it appears that the fluorescence of the tryptophan introduced via the point mutation does not change upon ATP binding or hydrolysis, despite its proximity to the nucleotide binding site.
ADP Release
The rate of ADP release (k+5’; Scheme 1) was determined by ATP-induced dissociation of the myo1c3IQ constructs from pyrene-actin (22). When the active-sites of myo1c3IQ and R156W-myo1c3IQ are saturated with ADP, ATP binding is rate-limited by the slow dissociation of ADP (k5’; Scheme 5).
Scheme 5.
ADP (10 µM) was incubated with 250 nM pyrene-actomyo1c and mixed with 500 µM ATP, and transients were best fit by a single exponential function (Fig. 5, inset). The rate of ADP release from myo1c3IQ (k5’ = 24 (± 0.50) s−1) is slightly faster than from R156W-myo1c3IQ (k5’ = 19 (± 0.090) s−1; Table 1).
Fig. 5.
A. ADP release from actomyo1c3IQ. The fractional amplitudes of the slow phase were obtained by fitting pyrene-actomyo1c dissociation transients from experiments with (circles) myo1c3IQ or (squares) R156W-myo1c3IQ to a double exponential as a function of ADP concentration at 37 °C. Values were normalized to the total change in amplitude. The lines are fits of the (solid) myo1c3IQ and (dashed) R156W-myo1c3IQ data to a hyperbolic function. The inset shows time courses of pyrene-actin fluorescence after mixing 500 µM ATP with 0.25 µM myo1c3IQ-pyrene-actin and R156W-myo1c3IQ-pyrene-actin equilibrated with 10 µM ADP. The smooth lines are best fits of the data to a single exponential function. B. Calculated duty ratio as a function of actin concentration for (solid) myo1c3IQ and (dashed) R156W-myo1c3IQ. Duty ratios were calculated as defined in equation 2.
ATP-induced pyrene-actin fluorescence transients were acquired as a function of ADP concentration to determine the affinity of ADP (K5’) for actin-bound myo1c3IQ proteins. Time courses were fit to two-exponential functions, with the slow component reporting ADP release (k+5’) and the rate of the fast phase reporting ATP binding (K1’k+2’). The affinity of the actomyo1cIQ for ADP was determined by monitoring the change in the amplitude of the slow phase as a function of ADP concentration (Fig. 5). Hyperbolic fits to the data yield similar affinities for myo1c3IQ (K5’ = 1.8 (± 0.54) µM) and R156W-myo1c3IQ (K5’ = 1.6 (± 0.27) µM; Table 1).
In Vitro Motility Assays
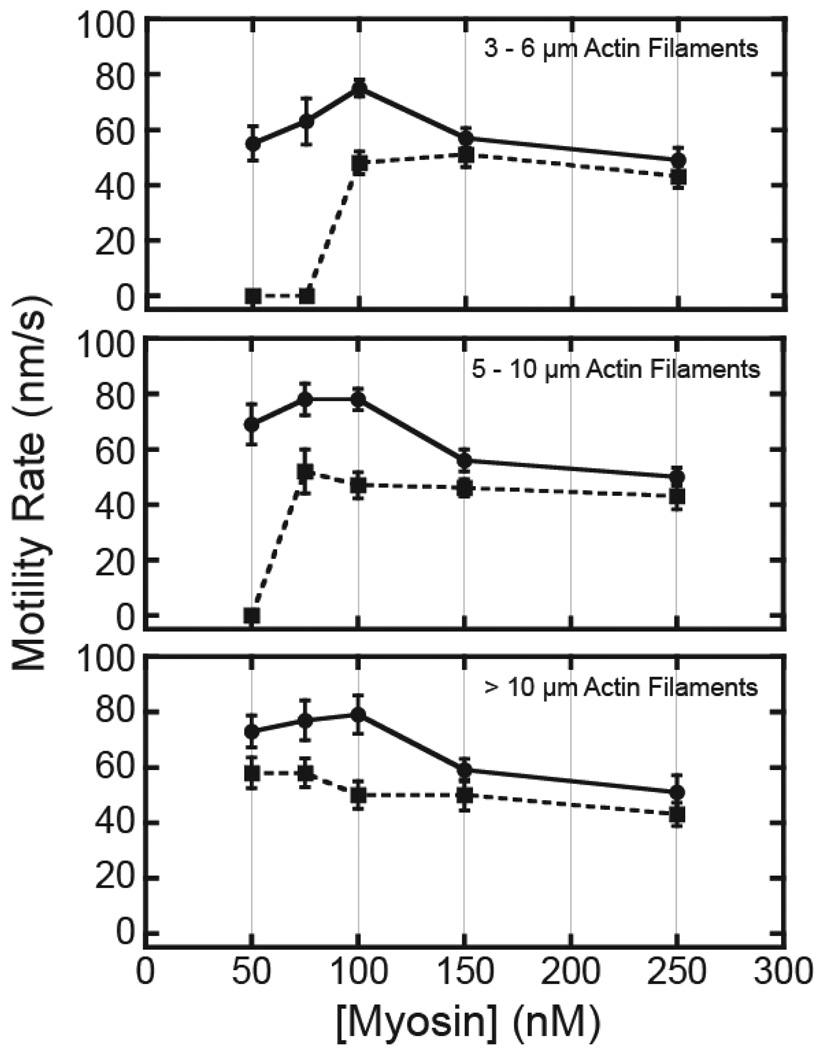
We determined the motile activity of myo1c3IQ and R156W-myo1c3IQ at 37 °C using the in vitro motility assay (23, 31). The myo1c proteins contain a C-terminal biotin that allows site-specific attachment of the myosin to the motility surface (see Methods). Incubation of 250 nM myo1c in streptavidin-coated chambers resulted in actin gliding rates of myo1c3IQ that are the same as R156W-myo1c3IQ (Fig. 6). This finding is consistent with the two proteins having similar rates of ADP release (Fig. 5), as this step is expected to limit the rate of unloaded sliding at high ATP concentrations of several different myosin isoforms (e.g., (8, 32–34)).
Fig. 6.
Velocity of actin filament gliding measured by the in vitro motility assay. Actin gliding rates were measured at five different surface densities of (circles) myo1c3IQ and (squares) R156W-myo1c3IQ at 37 °C. Myosin concentrations indicate the quantity of protein incubated in the flow chamber before washing (see Methods). Gliding rates were determined over a 5 min time period as a function of actin filament length. Actin filaments with rates of 0 nm/s were not immobile, but rather exhibited non-directional diffusion. Points are the averages of 10 filaments and error bars are standard deviations.
Actin gliding velocity increased ~1.5-fold at lower myo1c3IQ concentrations, which is likely due to a reduced drag on actin at lower surface densities. However, R156W-myo1c3IQ did not show increased motility rates at lower surface densities. Interestingly, when the concentration of R156W-myo1c3IQ in the motility chambers was decreased, we found that shorter actin filaments did not undergo directed motility, but rather appeared to be detached from the coverslip. This inability of R156W-myo1c3IQ to power motility of short actin filaments is likely due to a decrease in the motor’s duty ratio (see below).
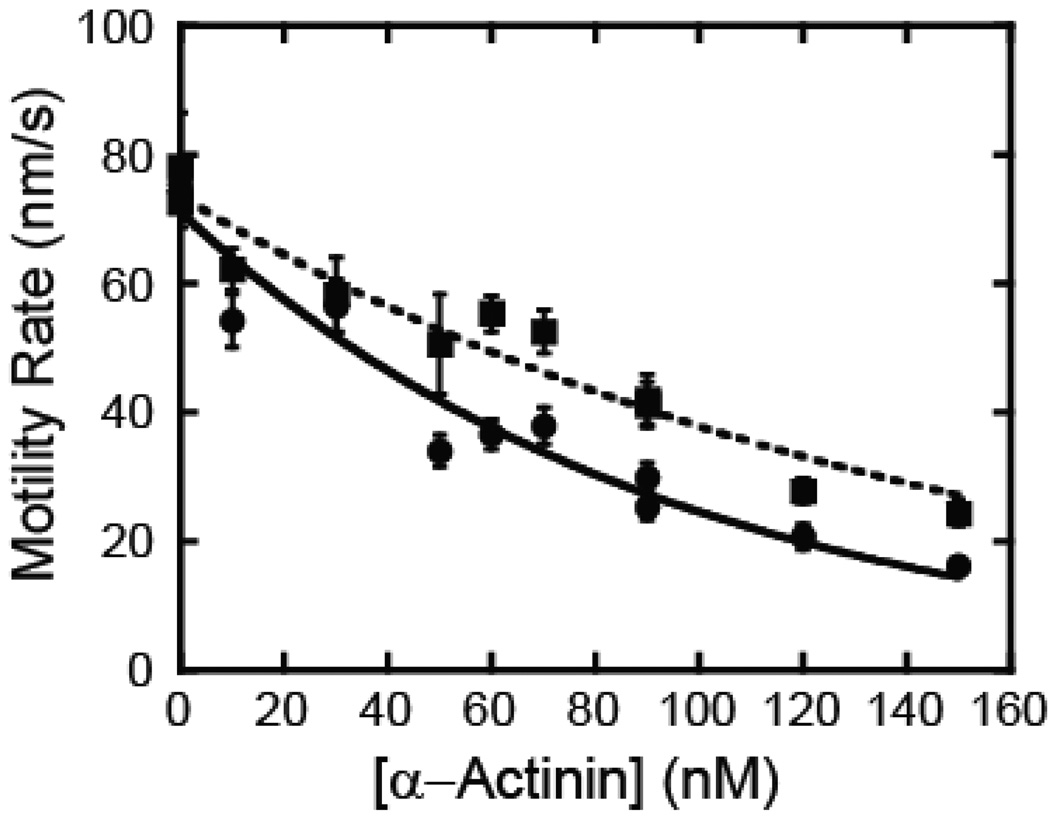
We tested the ability of myo1c to propel actin filaments in the presence of a load by performing a frictional loading assay (35, 36). α-actinin transiently binds to sliding actin filaments, providing a frictional load that opposes the driving force of the bed of myosin. The reduction in actin sliding velocity with load is due to both the drag force of the α-actinin and the load-dependent kinetics of the myosin. The rate of actin gliding was measured in the presence of a range of α-actinin surface densities (Fig. 7). The motility rates of myo1c3IQ decreased by 2-fold with the addition of 60 nM α-actinin, while R156W-myo1c3IQ required approximately 100 nM of α-actinin to cause the same decrease in velocity. Thus, although sensitive to resisting loads, the sliding velocity of R156W-myo1c3IQ appears to be substantially less force-sensitive than the wild-type protein.
Fig. 7.
Frictional loading assays. Actin filament sliding velocity was measured as a function of α-actinin concentration for both (circles) myo1c3IQ and (squares) R156W-myo1c3IQ at 37 °C. Two-parameter exponential decays were fit to the data for visualization. Points are the average sliding velocity of 15–35 filaments averaged over 5 frames and the error bars are standard errors of the mean.
DISCUSSION
ATP-induced dissociation of actomyo1c3IQ and ATP hydrolysis
The R156W mutation increases the rate of ATP-induced dissociation of myo1c3IQ from actin (k2’) ~1.5-fold. This elevated rate is not expected to affect the population of the actin-bound states, since it is substantially faster than the rates that limit the entry and exit from the force-bearing states (below). Consistent with this notion, the unloaded motility of R156W mutant myosin is unchanged compared to the wild type.
It has been proposed that the time course of ATP-induced changes in the intrinsic tryptophan fluorescence of myo1c reports the rate of ATP hydrolysis (30), as shown for other myosins (e.g., (37, 38)). We found the rates of the fluorescence transients from the wildtype and mutant proteins to be fast and not significantly different (Table 1). Thus, it appears that the R156W mutation does not alter ATP hydrolysis kinetics, in contrast to other switch-1 mutations (11–13). It is also interesting to note that, in the absence of actin, conformation changes that have been reported to occur near switch-1 (39) do not affect the fluorescence emission of W156 in the mutant protein.
Steady-state ATP hydrolysis and phosphate release
The R156W point mutation inhibits the actin-activated ATPase activity of myo1c3IQ ~4-fold. This inhibition most likely occurs at the phosphate release step (k4’; Scheme 1) for the following reasons: (a) the other key steps on the ATPase pathway (k1’, k2’, k3, k5’) are substantially faster than the rate that limits the steady-state ATPase activity (Table 1); (b) single turnover experiments (Fig. 2) show that the rate of isomerization into a strongly bound state (a transition associated with phosphate release) has the same rate as the rate that limits the steady-state ATPase activity; (c) phosphate release has been shown to be the rate limiting step for other myosin-I isoforms (24, 25, 28, 40); and (d) the lack of an effect of the mutation on the velocity of actin gliding indicates that the inhibited step occurs while myosin is in a weak binding state (33).
We were not able to determine the rate constant for phosphate release (k4’) directly, so it is possible that the R156W mutation weakens actin affinity, which would lead to a decreased KATPase. Switch-1 is not at the actin-binding interface (41), so the mutation is unlikely to affect actin binding via a direct disruption of the actin-binding site. However, the conformational state of switch-1 impacts actin affinity via modulation of the conformation of myosin’s actin-binding cleft (14, 42). Therefore, this mutation may affect affinity by modulating the ability of the actin-binding cleft to close, which ultimately affects phosphate release. Nevertheless, the functional consequence of the R156W mutation is a decreased myosin duty ratio, which results in decreased force from groups of myosins (below).
ADP release, in vitro motility, and duty ratio
The R156W mutation only slightly affects the rate of ADP release (k+5’) from actomyo1c (Table 1). Since ADP release limits the rate of detachment from the strong-binding states, it is not surprising that the maximum actin gliding velocities are only slightly affected by the R156W mutation. This is true despite the fact that the steady-state ATPase activity of the mutant is inhibited 4-fold. This result is a strong indication that the steady-state ATPase rate is inhibited at a kinetic step that occurs when myosin is detached from (or weakly-bound to) actin filaments (33).
The duty ratio is the fraction of the ATPase cycle that myosin is bound to actin in the strong-binding (force-bearing) states. We can calculate the duty ratios of the wild-type and mutant myosins by assuming that the steady-state ATPase assay reports the rate that limits entry into the strong binding states (K9k4’), and ADP release (k5’) limits exit from these states:
| eq. 2 |
where vA is actin-dependent ATPase rate and [A] is the actin concentration (Fig. 5; (43)). At the maximum experimental actin concentration (175 µM), the duty ratio of R156W-myo1c3IQ is ~2.4-fold less than the wildtype protein. This finding is consistent with the in vitro motility assays which show that lower surface-densities of mutant protein are less likely to propel short actin filaments (Fig. 6), i.e., short actin filaments have fewer myosin binding sites, further reducing the probability of actomyosin interactions necessary for movement.
The frictional loading assays (Fig. 7) indicate that load-induced inhibition of actin gliding is less pronounced in the mutant myosin. ADP release step (k5’) has been shown to be the most load-dependent step for other myosin isoforms (8, 44–46), so it is likely that the load-dependence of this step is affected by the R156W mutation. It is interesting to note that the R156W mutation increases the equilibrium constant (Kα) that defines the transition between the nucleotide sensitive (AM’) and nucleotide insensitive (AM) states (Fig. 3 & Table 1). It has been proposed that myosins that undergo the AM-to-AM’ transition are highly force-sensitive (29), and that this transition may be similar to the force-sensitive transition (AM.ADP-to-AM’) during ATP cycling. Thus, an increase the stabilization in Kα (i.e., a stabilization of the nucleotide accessible AM’ state) may be an indication of decreased tension sensitivity. Single molecule measurements are required to address this speculation directly (8, 44).
Comparison with previous studies
A kinetic analysis comparing wild type and R156W myo1c was recently published by Adamek et al. (47) using myosin constructs in which the native light chain binding domain was replaced with a single alpha helix domain from myo10. The ATPase rate constants obtained in the Adamek et al. study were substantially slower than those measured in this study, and they did not provide information about the effect of R156W on the myo1c duty ratio. Furthermore, the maximal steady state ATPase rate reassured with the construct containing the single alpha helix domain (0.66 s−1) is over 10-fold slower than the maximal rate measured here (>7 s−1). It is possible that the proteins used in the Adamek et al. study had altered kinetics due to the absence of an intact regulatory domain (20). In a separate study by Adamek et al., utilizing myo1c with a single IQ motif, the maximal steady state ATPase rate measured for the wild type myosin (1.8 s−1) was only 25% of the rate measured here (>7 s−1) (30). The reason for this discrepancy is unclear.
Physiological impact of the R156W mutation
Despite the normal sliding velocity of R156W-myo1c3IQ at low loads and high myosin densities, we propose that ensemble force generation by mutant myosins is impaired. The amount of force generated by a group of myosins is proportional to the number of motors that are strongly bound to actin. Since R156W-myo1c3IQ has a decreased duty ratio, an ensemble of mutant motors will generate less force than wild-type motors. We also propose that the rate at which R156W-myo1c3IQ detaches from actin is less sensitive to resisting forces (Fig. 7), resulting in an impaired ability to alter the duty ratio in response to tension (8). If myo1c is in fact the “adaptation motor,” this altered tension sensing may result in improper gating of mechanosensitive channels. Future mechanical experiments will better address the effect of force on ensembles and single myo1c molecules.
A missense mutation associated with human deafness (E385D) has been identified in the switch-II region of the myosin-I family member myo1a (48). Like the R156W mutation in myo1c, this mutation has a decreased actin-activated ATPase rate (49). However, this protein is unable to propel actin in gliding filament assays. Interestingly, this switch-II mutation also results in the improper localization of the protein. Given the importance of the motor domain in myosin-I localization (50, 51), it will be important to determine if the R156W mutation in myo1c also has an effect on the subcellular localization of this molecular motor.
Abbreviations
- myo1c3IQ
recombinant myosin-IC consisting of the motor domain and three IQ motifs
- R156W-myo1c3IQ
myo1c3IQ containing the R156W mutation
- pyrene-actin
actin labeled at cys-374 with pyrenyl iodoacetamide
Footnotes
This work was supported by grants from the National Institutes of Health to E.M.O (GM57247) and J.M. (HL077280) and by the American Heart Association to M.G. (0815704D).
REFERENCES
- 1.Bose A, Robida S, Furcinitti PS, Chawla A, Fogarty K, Corvera S, Czech MP. Unconventional myosin Myo1c promotes membrane fusion in a regulated exocytic pathway. Mol Cell Biol. 2004;24:5447–5458. doi: 10.1128/MCB.24.12.5447-5458.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bose A, Guilherme A, Robida SI, Nicoloro SM, Zhou QL, Jiang ZY, Pomerleau DP, Czech MP. Glucose transporter recycling in response to insulin is facilitated by myosin Myo1c. Nature. 2002;420:821–824. doi: 10.1038/nature01246. [DOI] [PubMed] [Google Scholar]
- 3.Sokac AM, Schietroma C, Gundersen CB, Bement WM. Myosin-1c couples assembling actin to membranes to drive compensatory endocytosis. Dev Cell. 2006;11:629–640. doi: 10.1016/j.devcel.2006.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holt JR, Gillespie SK, Provance DW, Shah K, Shokat KM, Corey DP, Mercer JA, Gillespie PG. A chemical-genetic strategy implicates myosin-1c in adaptation by hair cells. Cell. 2002;108:371–381. doi: 10.1016/s0092-8674(02)00629-3. [DOI] [PubMed] [Google Scholar]
- 5.Stauffer EA, Scarborough JD, Hirono M, Miller ED, Shah K, Mercer JA, Holt JR, Gillespie PG. Fast adaptation in vestibular hair cells requires myosin-1c activity. Neuron. 2005;47:541–553. doi: 10.1016/j.neuron.2005.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hokanson DE, Ostap EM. Myo1c binds tightly and specifically to phosphatidylinositol 4,5-bisphosphate and inositol 1,4,5-trisphosphate. Proc Natl Acad Sci U S A. 2006;103:3118–3123. doi: 10.1073/pnas.0505685103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hokanson DE, Laakso JM, Lin T, Sept D, Ostap EM. Myo1c Binds Phosphoinositides through a Putative Pleckstrin Homology Domain. Mol Biol Cell. 2006;17:4856–4865. doi: 10.1091/mbc.E06-05-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laakso JM, Lewis JH, Shuman H, Ostap EM. Myosin I can act as a molecular force sensor. Science. 2008;321:133–136. doi: 10.1126/science.1159419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zadro C, Alemanno MS, Bellacchio E, Ficarella R, Donaudy F, Melchionda S, Zelante L, Rabionet R, Hilgert N, Estivill X, Van Camp G, Gasparini P, Carella M. Are MYO1C and MYO1F associated with hearing loss? Biochim Biophys Acta. 2009;1792:27–32. doi: 10.1016/j.bbadis.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 10.Smith CA, Rayment I. X-ray structure of the magnesium(II).ADP.vanadate complex of the Dictyostelium discoideum myosin motor domain to 1.9 A resolution. Biochemistry. 1996;35:5404–5417. doi: 10.1021/bi952633+. [DOI] [PubMed] [Google Scholar]
- 11.Shimada T, Sasaki N, Ohkura R, Sutoh K. Alanine scanning mutagenesis of the switch I region in the ATPase site of Dictyostelium discoideum myosin II. Biochemistry. 1997;36:14037–14043. doi: 10.1021/bi971837i. [DOI] [PubMed] [Google Scholar]
- 12.Li XD, Rhodes TE, Ikebe R, Kambara T, White HD, Ikebe M. Effects of mutations in the gamma-phosphate binding site of myosin on its motor function. J Biol Chem. 1998;273:27404–27411. doi: 10.1074/jbc.273.42.27404. [DOI] [PubMed] [Google Scholar]
- 13.Forgacs E, Sakamoto T, Cartwright S, Belknap B, Kovacs M, Toth J, Webb MR, Sellers JR, White HD. Switch 1 mutation S217A converts myosin V into a low duty ratio motor. J Biol Chem. 2009;284:2138–2149. doi: 10.1074/jbc.M805530200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kintses B, Gyimesi M, Pearson DS, Geeves MA, Zeng W, Bagshaw CR, Malnasi-Csizmadia A. Reversible movement of switch 1 loop of myosin determines actin interaction. Embo J. 2007;26:265–274. doi: 10.1038/sj.emboj.7601482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gillespie PG. Myosin I and adaptation of mechanical transduction by the inner ear. Philos Trans R Soc Lond B Biol Sci. 2004;359:1945–1951. doi: 10.1098/rstb.2004.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Batters C, Wallace MI, Coluccio LM, Molloy JE. A model of stereocilia adaptation based on single molecule mechanical studies of myosin I. Philos Trans R Soc Lond B Biol Sci. 2004;359:1895–1905. doi: 10.1098/rstb.2004.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 18.Kouyama T, Mihashi K. Fluorimetry study of N-(1-pyrenyl)iodoacetamide-labelled F-actin. Local structural change of actin protomer both on polymerization and on binding of heavy meromyosin. Eur J Biochem. 1981;114:33–38. [PubMed] [Google Scholar]
- 19.Putkey JA, Slaughter GR, Means AR. Bacterial expression and characterization of proteins derived from the chicken calmodulin cDNA and a calmodulin processed gene. J Biol Chem. 1985;260:4704–4712. [PubMed] [Google Scholar]
- 20.Manceva S, Lin T, Pham H, Lewis JH, Goldman YE, Ostap EM. Calcium regulation of calmodulin binding to and dissociation from the myo1c regulatory domain. Biochemistry. 2007;46:11718–11726. doi: 10.1021/bi700894h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De La Cruz EM, Sweeney HL, Ostap EM. ADP inhibition of myosin V ATPase activity. Biophys J. 2000;79:1524–1529. doi: 10.1016/S0006-3495(00)76403-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De La Cruz EM, Ostap EM. Kinetic and equilibrium analysis of the myosin ATPase. Methods Enzymol. 2009;455:157–192. doi: 10.1016/S0076-6879(08)04206-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin T, Tang N, Ostap EM. Biochemical and motile properties of Myo1b splice isoforms. J Biol Chem. 2005;280:41562–41567. doi: 10.1074/jbc.M508653200. [DOI] [PubMed] [Google Scholar]
- 24.Ostap EM, Pollard TD. Biochemical kinetic characterization of the Acanthamoeba myosin-I ATPase. J Cell Biol. 1996;132:1053–1060. doi: 10.1083/jcb.132.6.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El Mezgueldi M, Tang N, Rosenfeld SS, Ostap EM. The kinetic mechanism of Myo1e (human myosin-IC) J Biol Chem. 2002;277:21514–21521. doi: 10.1074/jbc.M200713200. [DOI] [PubMed] [Google Scholar]
- 26.Furch M, Remmel B, Geeves MA, Manstein DJ. Stabilization of the actomyosin complex by negative charges on myosin. Biochemistry. 2000;39:11602–11608. doi: 10.1021/bi000985x. [DOI] [PubMed] [Google Scholar]
- 27.Furch M, Geeves MA, Manstein DJ. Modulation of actin affinity and actomyosin adenosine triphosphatase by charge changes in the myosin motor domain. Biochemistry. 1998;37:6317–6326. doi: 10.1021/bi972851y. [DOI] [PubMed] [Google Scholar]
- 28.Lewis JH, Lin T, Hokanson DE, Ostap EM. Temperature dependence of nucleotide association and kinetic characterization of myo1b. Biochemistry. 2006;45:11589–11597. doi: 10.1021/bi0611917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geeves MA, Perreault-Micale C, Coluccio LM. Kinetic analyses of a truncated mammalian myosin I suggest a novel isomerization event preceding nucleotide binding. J Biol Chem. 2000;275:21624–21630. doi: 10.1074/jbc.M000342200. [DOI] [PubMed] [Google Scholar]
- 30.Adamek N, Coluccio LM, Geeves MA. Calcium sensitivity of the cross-bridge cycle of Myo1c, the adaptation motor in the inner ear. Proc Natl Acad Sci U S A. 2008;105:5710–5715. doi: 10.1073/pnas.0710520105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kron SJ, Spudich JA. Fluorescent actin filaments move on myosin fixed to a glass surface. Proc Natl Acad Sci U S A. 1986;83:6272–6276. doi: 10.1073/pnas.83.17.6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rock RS, Rice SE, Wells AL, Purcell TJ, Spudich JA, Sweeney HL. Myosin VI is a processive motor with a large step size. Proc Natl Acad Sci U S A. 2001;98:13655–13659. doi: 10.1073/pnas.191512398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siemankowski RF, Wiseman MO, White HD. ADP dissociation from actomyosin subfragment 1 is sufficiently slow to limit the unloaded shortening velocity in vertebrate muscle. Proc Natl Acad Sci U S A. 1985;82:658–662. doi: 10.1073/pnas.82.3.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rief M, Rock RS, Mehta AD, Mooseker MS, Cheney RE, Spudich JA. Myosin-V stepping kinetics: a molecular model for processivity. Proc Natl Acad Sci U S A. 2000;97:9482–9486. doi: 10.1073/pnas.97.17.9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bing W, Knott A, Marston SB. A simple method for measuring the relative force exerted by myosin on actin filaments in the in vitro motility assay: evidence that tropomyosin and troponin increase force in single thin filaments. Biochem J. 2000;350(Pt 3):693–699. [PMC free article] [PubMed] [Google Scholar]
- 36.Greenberg MJ, Moore JR. The molecular basis of frictional loads in the in vitro motility assay with applications to the study of the loaded mechanochemistry of molecular motors. Cytoskeleton (Hoboken) 2010;67:273–285. doi: 10.1002/cm.20441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De La Cruz EM, Wells AL, Rosenfeld SS, Ostap EM, Sweeney HL. The kinetic mechanism of myosin V. Proc Natl Acad Sci U S A. 1999;96:13726–13731. doi: 10.1073/pnas.96.24.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson KA, Taylor EW. Intermediate states of subfragment 1 and actosubfragment 1 ATPase: reevaluation of the mechanism. Biochemistry. 1978;17:3432–3442. doi: 10.1021/bi00610a002. [DOI] [PubMed] [Google Scholar]
- 39.Malnasi-Csizmadia A, Dickens JL, Zeng W, Bagshaw CR. Switch movements and the myosin crossbridge stroke. J Muscle Res Cell Motil. 2005;26:31–37. doi: 10.1007/s10974-005-9004-y. [DOI] [PubMed] [Google Scholar]
- 40.Jontes JD, Milligan RA, Pollard TD, Ostap EM. Kinetic characterization of brush border myosin-I ATPase. Proc Natl Acad Sci U S A. 1997;94:14332–14337. doi: 10.1073/pnas.94.26.14332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rayment I, Holden HM, Whittaker M, Yohn CB, Lorenz M, Holmes KC, Milligan RA. Structure of the actin-myosin complex and its implications for muscle contraction. Science. 1993;261:58–65. doi: 10.1126/science.8316858. [DOI] [PubMed] [Google Scholar]
- 42.Furch M, Fujita-Becker S, Geeves MA, Holmes KC, Manstein DJ. Role of the salt-bridge between switch-1 and switch-2 of Dictyostelium myosin. J Mol Biol. 1999;290:797–809. doi: 10.1006/jmbi.1999.2921. [DOI] [PubMed] [Google Scholar]
- 43.De La Cruz EM, Wells AL, Sweeney HL, Ostap EM. Actin and light chain isoform dependence of myosin V kinetics. Biochemistry. 2000;39:14196–14202. doi: 10.1021/bi001701b. [DOI] [PubMed] [Google Scholar]
- 44.Laakso JM, Lewis JH, Shuman H, Ostap EM. Control of myosin-I force sensing by alternative splicing. Proc Natl Acad Sci U S A. 2010;107:698–702. doi: 10.1073/pnas.0911426107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Veigel C, Schmitz S, Wang F, Sellers JR. Load-dependent kinetics of myosin-V can explain its high processivity. Nat Cell Biol. 2005;7:861–869. doi: 10.1038/ncb1287. [DOI] [PubMed] [Google Scholar]
- 46.Veigel C, Molloy JE, Schmitz S, Kendrick-Jones J. Load-dependent kinetics of force production by smooth muscle myosin measured with optical tweezers. Nat Cell Biol. 2003;5:980–986. doi: 10.1038/ncb1060. [DOI] [PubMed] [Google Scholar]
- 47.Adamek N, Geeves MA, Coluccio LM. Myo1c mutations associated with hearing loss cause defects in the interaction with nucleotide and actin. Cell Mol Life Sci. 2010 doi: 10.1007/s00018-010-0448-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Donaudy F, Ferrara A, Esposito L, Hertzano R, Ben-David O, Bell RE, Melchionda S, Zelante L, Avraham KB, Gasparini P. Multiple mutations of MYO1A, a cochlear-expressed gene, in sensorineural hearing loss. Am J Hum Genet. 2003;72:1571–1577. doi: 10.1086/375654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yengo CM, Ananthanarayanan SK, Brosey CA, Mao S, Tyska MJ. Human deafness mutation E385D disrupts the mechanochemical coupling and subcellular targeting of myosin-1a. Biophys J. 2008;94:L5–L7. doi: 10.1529/biophysj.107.122689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruppert C, Godel J, Muller RT, Kroschewski R, Reinhard J, Bahler M. Localization of the rat myosin I molecules myr 1 and myr 2 and in vivo targeting of their tail domains. J Cell Sci. 1995;108(Pt 12):3775–3786. doi: 10.1242/jcs.108.12.3775. [DOI] [PubMed] [Google Scholar]
- 51.Tang N, Ostap EM. Motor domain-dependent localization of myo1b (myr-1) Curr Biol. 2001;11:1131–1135. doi: 10.1016/s0960-9822(01)00320-7. [DOI] [PubMed] [Google Scholar]