Abstract
Tumor cell rolling on the endothelium plays a key role in the initial steps of cancer metastasis, i.e. extravasation of circulating tumor cells (CTCs). Identification of the ligands that induce the rolling of cells is thus critical to understand how cancers metastasize. We have previously demonstrated that MCF-7 cells, human breast cancer cells, exhibit the rolling response selectively on E-selectin-immobilized surfaces. However, the ligand that induces rolling of MCF-7 cells on E-selectin has not yet been identified, as these cells lack commonly known E-selectin ligands. Here we report, for the first time to our knowledge, a set of quantitative and direct evidence demonstrating that CD24 expressed on MCF-7 cell membranes is responsible for rolling of the cells on E-selectin. The binding kinetics between CD24 and E-selectin was directly measured using surface plasmon resonance (SPR), which revealed that CD24 has a binding affinity against E-selectin (KD = 3.4 ± 0.7 nM). The involvement of CD24 in MCF-7 cell rolling was confirmed by the rolling behavior that was completely blocked when cells were treated with anti-CD24. A simulated study by flowing microspheres coated with CD24 onto E-selectin-immobilized surfaces further revealed that the binding is Ca2+ dependent. Additionally, we have found that actin filaments are involved in the CD24-mediated cell rolling, as observed by the decreased rolling velocities of the MCF-7 cells upon treatment with cytochalasin D (an inhibitor of actin-filament dynamics) and the stationary binding of CD24-coated microspheres (the lack of actins) on the E-selectin-immobilized slides. Given that CD24 is known to be directly related to enhanced invasiveness of cancer cells, our results imply that CD24-based cell rolling on E-selectin mediates, at least partially, cancer cell extravasation, resulting in metastasis.
INTRODUCTION
Adhesive interactions between tumor cells and the vascular endothelium are necessary to promote cancer metastasis induced by circulating tumor cells (CTCs).1-3 The transient adhesion of cells to the endothelium in blood circulation, known as cell rolling, is the early event of the adhesive interactions that are mediated by adhesive proteins such as selectins on the endothelium and specific ligands on the cancer cell surface.4-6 To date, three types of selectins have been identified, E-selectin (CD62E), L-selectin (CD62L), and P-selectin (CD62P).7 Among them, E-selectin, an inducible endothelial cell-surface glycoprotein,8 has been suggested as one of the major receptors responsible for causing the metastasis of primary tumors such as breast,9 colon,4 and prostate cancers.10,11 E-selectin has been also shown to mediate the adhesion of leukocytes or carcinoma cells to cytokine-activated human endothelial cells.12-14 Information regarding the ligands for E-selectin on the cancer cells would thus be crucial to understand the mechanism of metastasis and to potentially find a way to prohibit the spreading of primary cancers in patients.
Earlier reports have shown high-affinity binding between E-selectin and its ligands such as E-selectin ligand-1 (ESL-1),15 P-selectin glycoprotein ligand-1 (PSGL-1),16,17 and sialyl Lewis X (sLex) and related sulfated structures including sialyl Lewisa (sLea).18-20 In addition to these known ligands for E-selectin, it is reported that the following molecules also have binding affinity to E-selectin; CD43,21 hematopoietic cell E- and L-selectin ligand (HCELL; a specialized glycoform of CD44),12 β2 integrins,22 and CD44v4.23 In our previous report, MCF-7 cells exhibited the rolling behavior only on E-selectin-immobilized surfaces under flow but no interactions with P-selectin.24 However, to the best of our knowledge, a ligand against E-selectin present on MCF-7 cells has not been identified since MCF-7 cells lack all the known E-selectin ligands listed above.23,25,26
CD24, which is highly expressed by MCF-7 cells, is a mucin-like glycosylphosphatidyl-inositol-linked cell surface protein consisting of 27 amino acids.27 Expression of CD24 from MCF-7 cells was assessed using a fluorescence activated cell sorter (FACS) as shown in Supplementary Figure 1. This protein is also expressed by early stage B-cells and neutrophils, whereas it is not expressed by normal T-cells and monocytes.27 Previous reports have also shown that it is highly expressed on several non-hematopoietic tumors such as breast carcinomas, epithelial ovarian cancer, small and non-small cell lung carcinoma, prostate cancer, pancreatic cancer, and renal cell carcinoma.11,28,29 A higher expression level of CD24 on cells in cancer patients has been correlated with a decreased survival rate, and thus CD24 has demonstrated potential to be used as a marker for more aggressive cancers.28,30 In cancer cell lines, the expression of CD24 molecules is known to enhance the metastatic properties of carcinoma cells, e.g. the adhesiveness29,30 and invasion in vitro31 and in vivo.32,33 Moreover, expedited cell growth and metastatic properties of MCF-7 cells were inhibited by cross-linking of CD24 using anti-human CD24 rabbit polyclonal antibodies on the cell surface.34 Based on the in vitro and in vivo connection between CD24 and the cancer cell properties as well as our previous observation of MCF-7 cell rolling, the possibility that CD24 may be a ligand for E-selectin has emerged.
In this study, we hypothesize that CD24 is a ligand for E-selectin, which mediates cell rolling. To test the hypothesis, the binding kinetics of CD24 with E-selectin were measured directly and quantitatively using surface plasmon resonance (SPR) provided by BIAcore® technology. We have also found that the rolling response of the CD24-positive MCF-7 cells was completely blocked by anti-CD24 pre-treatment to the cells. Additionally, we have tested the rolling velocities of MCF-7 cells treated with cytochalasin D (cytoD), an actin-disrupting agent, revealing that the cellular mechanics, such as actin-filament dynamics, play a role in the rolling mechanism mediated by CD24 and E-selectin. The involvement of the actin filament was further supported by the observed no rolling but stationary binding of CD24-coated microspheres on E-selectin. Our results presented here support our hypothesis that CD24 is a ligand for E-selectin and induces the rolling of the MCF-7 cells.
EXPERIMENTAL
Materials
Recombinant human E-selectin/Fc chimera (E-selectin) and anti-human epithelial-cell-adhesion-molecule (EpCAM)/TROP1 antibody (anti-EpCAM) were purchased from R&D systems (Minneapolis, MN). Recombinant human CD24 and sLex–PAA-Biotin were purchased from Santa Cruz Biotechnology, Inc (Santa Cruz, CA) and Glycotech (Gaithersburg, MD), respectively. Unconjugated goat anti-Human IgG (H + L) was acquired from Pierce Biotechnology, Inc (Rockford, IL). The epoxyfunctionalized glass surfaces (SuperEpoxy2®) were purchased from TeleChem International, Inc (Sunnyvale, CA). PE mouse IgG2a (k isotype control), PE mouse anti-human CD24, and purified mouse anti-human CD24 were obtained from BD Pharmingen™ (BD biosciences, San Jose, CA). All other chemicals were obtained from Sigma-Aldrich (St. Louis, MO), unless otherwise specified, and used without further purification.
Surface plasmon resonance measurements
BIAcore® X instrument (GE Healthcare, Pittsburgh, PA) was used for quantitative and direct measurements of the binding kinetics of CD24 and sLex with E-selectin at 25 °C. The E-selectin was immobilized on CM5 sensor chips (GE Healthcare) using the amine coupling kit (GE Healthcare) as described earlier (Supplementary Figure 3).35 Briefly, 6 μL of E-selectin solution (1 mg/mL in PBS buffer) was diluted in 144 μL of sodium acetate buffer (immobilization buffer, 10 mM, pH 5.0). The protein solution (70 μL) was then injected into the channel 2 of a sensor chip CM5 that was pre-activated by 70 μL of a mixture of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) and N-hydroxylsuccinimide (NHS). After protein immobilization, the remaining reactive ester residues were blocked by the injection of 70 μL ethanolamine (pH 8.5). The whole protein immobilization process was carried out at a flow rate of 5 μL/min using HBS-EP (GE Healthcare) as a running buffer.
CM5 sensor chips with an immobilized shift of more than 8,000 resonance units (RU, approximately 8 μg/mm2 of surface presentation) were used for subsequent protein binding analyses (Supplementary Figure 3). The protein-immobilized sensor chips were rinsed with PBS buffer (10 mM, running buffer) until a stable baseline (equilibrium) was obtained, prior to the injection of the analytes. To perform binding assays, 50 μL of CD24 or sLex in PBS buffer at a series of concentrations was injected into the two channels (channel 2 with E-selectin and channel 1 as reference) at a flow rate of 50 μL/min, which allowed the analytes to interact with the chip surface for 1 min, followed by flowing the running buffer for 5 min. After obtaining the binding curves, the sensor chip surface was regenerated using Glycin-HCl (10 mM, pH 2.5 or pH 3.0) buffer for further experiments. To confirm that the response is a result of the specific interactions, the responses (in RU) from the channel 2 were subtracted by those from the channel 1, represented as differences in responses. All sensorgrams were recorded from the start of the injection to the end of the dissociation phase, and were shifted to overlay at the same baseline resonance level. All binding kinetics parameters were obtained by fitting a 1:1 binding model provided by the BIAevaluation software and represented as the mean value ± standard deviation from three independent measurements.
Cell culture and treatments
MCF-7 cells purchased from ATCC (Manassas, VA) were grown in DMEM media supplemented with 10% (v/v) FBS and 1% (v/v) penicillin/streptomycin in a humidified incubator at 37 °C and 5% CO2. Prior to all flow-chamber experiments, MCF-7 cells at a concentration of 2 × 105 cells/mL (5 mL) were pre-treated with 240 μg/mL of anti-IgG solution to prevent potential non-specific binding.36 To block CD24 on the MCF-7 cell surface, 100 μL of anti-CD24 (0.5 mg/mL) was added into the cell suspension and incubated for 15 min. The final suspensions were kept on ice throughout the subsequent cell rolling experiments. To investigate the effect of actin-filament dynamics on the cell rolling behavior of MCF-7 cells, 5 mL of the cells at 2 × 105 cells/mL was treated with 40 μM of cytoD for 60 min and were resuspended in the supplemented DMEM media for the flow chamber experiments.37,38
Preparation of CD24- and sLex-coated microspheres
Recombinant human CD24 was used after dialysis (Slide-A-Lyzer® mini dialysis units with 10,000 MWCO, Thermo scientific, Rockford, IL) to remove impurities for high conjugation efficiency of the subsequent reaction with a biotin reagent. The purified, recombinant CD24 proteins were biotinylated using 10 mM EZ-Link® Sulfo-NHS-LC-Biotin (20-fold molar excess, Thermo scientific) for 2 hr on ice, and the biotinylated CD24 proteins were re-purified by dialysis using a Slide-A-Lyzer® mini dialysis unit. Streptavidin-coated microspheres (104.8 μL, 1 × 107 beads, 9.95 μm O.D., ProActive® microspheres, Bangs Laboratories, Inc, Fishers, IN) in 1% (w/v) bovine serum albumin (BSA) in PBS buffer (10 mM, Cellgro®, with 2 mM Ca2+) (BSA solution) were washed three times using a centrifuge at 10,000 rpm for 2 min. The biotinylated CD24 (100 nM) was conjugated to the microspheres in BSA solution by the spontaneous interaction between streptavidin and biotin after 1 hr of incubation at room temperature. The coating of CD24 molecules on microspheres was confirmed by flow cytometry (Supplementary Figure 4) using the same immunostaining condition for the confirmation of CD24 expression on MCF-7 cell using phycoerythrin (PE)-anti-CD24. In parallel, sLex-coated microspheres were also prepared under the exactly same condition used for preparation of the CD24-coated microspheres.39 Given that the microspheres with the same streptavidin density were used, and that the functionalization conditions were identical, the density of CD24 and sLex on the microspheres should be, if not identical, very similar. For parallel flow chamber experiments, CD24 or sLex-coated microspheres were resuspended in 10 mL BSA solution (1 × 106 microspheres/mL). To investigate potential non-specific interactions of microspheres themselves with E-selectin, a flow-chamber experiment (described in detail below) was performed using non-functionalized microspheres, confirming that there is no non-specific binding (Supplementary Figure 2).
Surface functionalization by immobilization of adhesive proteins
A general scheme of the adhesive protein immobilization, along with the characterization of the surfaces, is described in our previous publication.24 Briefly, 150 μL of each protein (E-selectin or anti-EpCAM) at a concentration of 5 μg/mL in PBS buffer was added on an approximately 1 cm2 area of an epoxy functionalized glass slide defined by a polydimethylsiloxane (PDMS) gasket. After incubation at room temperature for 4 hr with constant gentle shaking, the PDMS gasket was removed, and the whole slide surface was washed with PBS three times. Potential non-specific binding of both protein-coated and uncoated regions was blocked by a final incubation with BSA solution. Experiments using the functionalized surfaces were performed immediately.
Flow chamber experiments
Flow chamber experiments were also performed according to our previous report.24 The biofunctionalized glass slide, a gasket (30 mm (L) × 10mm (W) × 0.25 mm (D), Glycotech), and a rectangular parallel plate flow chamber (Glycotech) were assembled in line under vacuum. To observe cellular interactions, MCF-7 cell suspensions with or without anti-CD24 treatment as well as CD24- or sLex-coated microsphere suspensions were injected into the flow chamber at various flow rates (ranging from 50 to 200 μL/min) using a syringe pump (New Era pump Systems Inc., Farmingdale, NY). Note that, in this flow chamber, 50 μL/min of flow rate is correspondent to 0.08 dyn/cm2 of a wall shear stress, 8 s-1 of a wall shear rate, and 20 μm/sec of near-wall non-adherent cell velocity according to the Goldman equation.40 All cellular responses in the flow chamber were monitored using an Olympus IX70 inverted microscope (IX 70-S1F2, Olympus America, Inc., Center Valley, PA) and images were recorded using a 10× objective and a CCD camera (QImaging Retiga 1300B, Olympus America, Inc.). Rolling velocities of cells and the functionalized microspheres on the immobilized proteins were measured, based on the images taken every second for 1 min in repetitive observations using ImageJ (NIH). Cell rolling was defined when the rolling velocities were less than 50% of the free stream velocity (e.g. slower than 10 μm/sec at a flow rate of 50 μL/min). In addition to the flow-based experiments using the functionalized microspheres, the same number of CD24-coated microspheres (1 × 106 microspheres/mL in BSA solution) was incubated with each protein-immobilized surface for 1 min and the number of protein-immobilized microspheres per 1 mm2 that remained on the surface was counted after flowing PBS buffer for 1 min.
RESULTS AND DISCUSSION
We have previously reported MCF-7 cell rolling on immobilized E-selectin.24 However, as indicated, it was unclear which ligand was responsible for inducing the rolling behavior since the cells do not express any known ligands for E-selectin. The purpose of this study was therefore to identify the ligand of MCF-7 cells. Given that cells that maintain a high level of CD24 expression have enhanced invasive and adhesive properties which can potentially related to cancer metastasis,28,32 we hypothesized that CD24 may mediate the rolling response of MCF-7 cells on E-selectin.
Direct measurements of binding kinetics using SPR
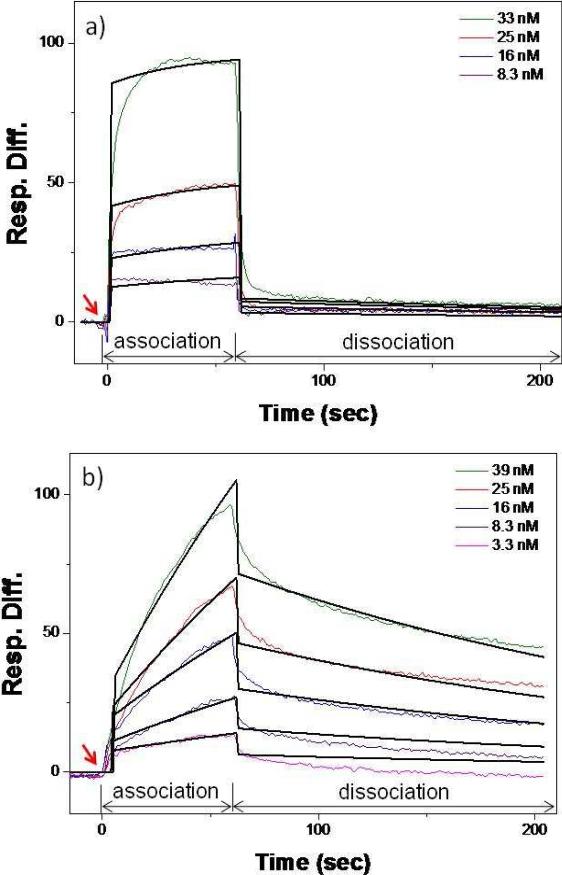
For the first time to our knowledge, the binding kinetics of recombinant CD24 with E-selectin was quantitatively measured using a direct binding assay by BIAcore® X. A well-known ligand for E-selectin, sLex was also employed as a control. The BIAcore® sensorgrams illustrating the binding interactions between the E-selectin-immobilized sensor chips and CD24 or sLex are shown in Figure 1, and all measured kinetic parameters are listed in Table 1. Note that concentrations of the analytes (CD24 and sLex) used for the SPR measurements were optimized to achieve similar levels of resonance unit (RU) (preferably lower than 100) between sensorgrams. The SPR measurements revealed that CD24 indeed bound to E-selectin. The association rate constant (ka) and the dissociation rate constant (kd) were 3.8 × 105 M-1s-1 and 1.6 × 10-3 s-1, respectively, and the dissociation constant (KD) of CD24 was 3.4 nM. The binding parameters ka, kd, and KD of sLex were measured to be 7.9 × 104s-1, 5.6 × 10-3 s-1 and 78.3 nM, respectively. While the kd values of CD24 and sLex were relatively similar, ka of CD24 was higher than those of sLex by an order of magnitude. Based on the ratio of the directly measured binding parameters, especially the dissociation constants, we have found that the binding affinity of CD24 with E-selectin is stronger than that of sLex by approximately 20 fold.
Figure 1.
SPR sensorgrams showing the binding curves of (a) CD24 and (b) sLex with the immobilized E-selectin. The colored lines represent the raw data curves, and the solid black lines are simulated fitting curves using an 1:1 interaction model. The arrows indicate the injection time of the analytes.
Table 1.
Quantitatively measured kinetic parameters of binding of CD24 and sLex with E-selectin using SPR.
| Kinetic Parameters1 | Ligands |
|
|---|---|---|
| CD24 | sLex | |
| ka (× 104 M-1s-1) | 37.9 ± 30.4 | 7.9 ± 2.6 |
| kd (× 10-3 s-1) | 1.6 ± 1.3 | 5.6 ± 0.8 |
| KA (× 107 M-1) | 31.8 ± 6.5 | 1.4 ± 0.3 |
| KD (nM) | 3.4 ± 0.7 | 78.3 ± 15.7 |
ka: association rate constant, kd: dissociation rate constant, KA: association constant, and KD: dissociation constant. All the kinetic values were obtained by averaging three independent runs of SPR measurements, presented as averages ± standard deviations.
Cellular responses on selectin- and anti-EpCAM-functionalized surfaces
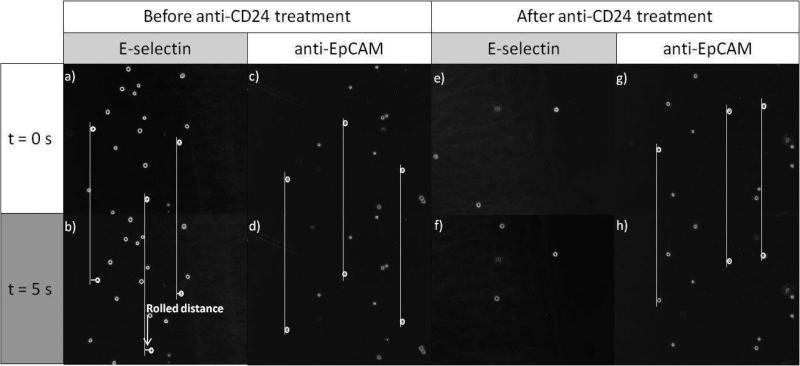
We then investigated how the measured binding parameters correlate with cell interactions with various proteins on surfaces under flow using a parallel plate flow chamber. As shown in our previous report, no interactions were observed between MCF-7 cells and P-selectin.24 Although CD24 is known as a P-selectin ligand, it requires modification by mechanical enzymes to bind to P-selectin, which requires sLex decoration.26,41 This can explain the lack of interaction between sLex-deficient MCF-7 cells and P-selectin.24 Figure 2 shows the surface interactions of MCF-7 cells with E-selectin and anti-EpCAM. The cell interactions with each of the individual proteins in Figure 2a-d were compared to those of the anti-CD24-treated MCF-7 cells in Figure 2e-h. Images of the cells on each surface were taken at t = 0 s (a randomly chosen time to begin recording during the flow experiment) and at t = 5 s to observe any dynamic cell response such as cell rolling. Untreated MCF-7 cells exhibited the rolling response on E-selectin and stationary adhesion on anti-EpCAM. The average rolling velocity of MCF-7 cells was 2.8 ± 0.2 μm/sec on E-selectin-immobilized slides at a wall shear stress of ~0.1 dyn/cm2. Upon anti-CD24 treatment, the rolling behavior of MCF-7 cells on E-selectin was completely interrupted (Figure 2e and f), while the responses of MCF-7 cells on anti-EpCAM remained the same (Figure 2g and h), i.e. stationary binding with anti-EpCAM. The directly measured binding kinetics between CD24 and E-selectin using Biacore, along with the ceased rolling of MCF-7 cells upon anti-CD24 treatment, confirms that CD24 is a mediator for the MCF-7 cell rolling.
Figure 2.
Time-course images of MCF-7 cells under shear stress of 0.08 dyn/cm2 on E-selectin and anti-EpCAM-immobilized surfaces. Untreated MCF-7 cells exhibit the rolling behavior on the E-selectin-coated surface at a velocity of 2.76 ± 0.16 μm/sec (a and b) while being stationary bound on the anti-EpCAM-coated surface (c and d). Upon treatment with anti-CD24, the interaction of MCF-7 cells with E-selectin is disappeared (e and f) whereas the binding with anti-EpCAM is not affected (g and h). Note that the cells are not tracked in the images e and f due to no interaction between the anti-CD24-treated cells and the surface, resulting in fast free flow of the cells. Flow direction is from left to right.
Interactions of microspheres on the surfaces functionalized with E-selectin
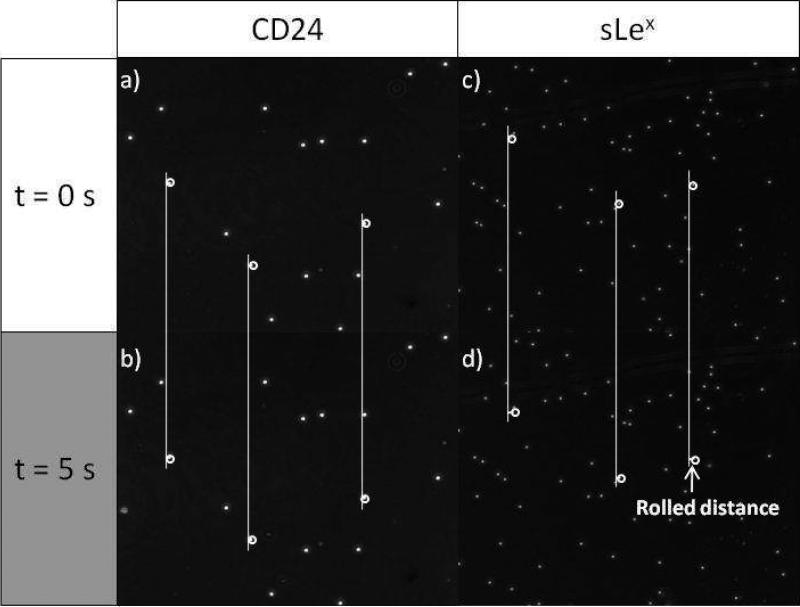
To assess if CD24 is solely responsible for binding and rolling of the MCF-7 cells, CD24-coated microspheres were prepared for a flow-chamber experiment (Figure 3). Microspheres functionalized with sLex were also prepared according to the previous report and used as a control group.39 Note that the streptavidin-immobilized microspheres (without CD24 or sLex) had no non-specific binding with immobilized E-selectin (Supplementary Figure 2a and b). CD24- and sLex-coated microspheres exhibited different behaviors on the E-selectin-immobilized slides. As shown in Figure 4, CD24-coated microspheres exhibited stationary binding to E-selectin, whereas sLex-coated microspheres exhibited the stable rolling response on E-selectin at a velocity of 0.4 m/sec, which is consistent with the previous report.39 The stationary adhesion of CD24 microspheres as opposed to the dynamic rolling of sLex microspheres can be explained by the significantly higher affinity between CD24 and E-selectin than sLex-based binding by ~20 fold, as measured by SPR. Additionally, in a Ca2+-deficient PBS buffer, the CD24 and sLex-coated microspheres did not bind to E-selectin (Supplementary Figure 2c and d), indicating that the interaction of both CD24 and sLex with E-selectin is Ca2+ dependent.
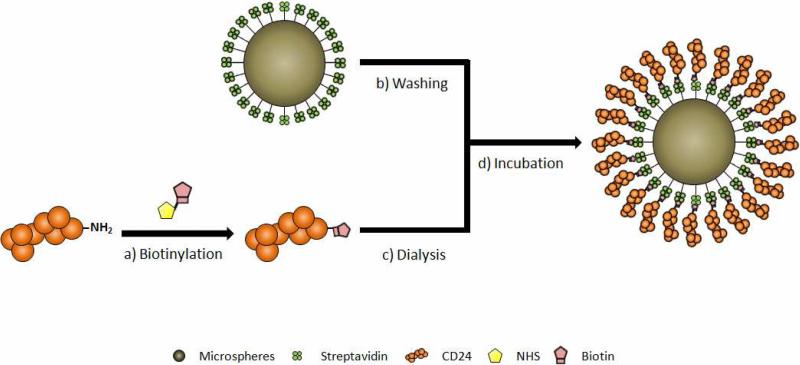
Figure 3.
Preparation of CD24-coated microspheres. a) Biotinylation of recombinant human CD24, b) pre-washing of streptavidin-coated microspheres, c) purification of the biotinylated CD24 using dialysis, and d) incubation of the biotinylated CD24 with streptavidin-coated microspheres.
Figure 4.
Time-course images of microspheres that are coated with (a and b) CD24 and (c and d) sLex under shear stress of 0.08 dyn/cm2 on E-selectin-immobilized surfaces. CD24-coated microspheres are statically bound on E-selectin-immobilized surface, but sLex-coated microspheres exhibit the rolling behavior on E-selectin (0.44 ± 0.02 μm/sec). Flow direction is from left through right.
Actin filament dynamics for the cell rolling behavior
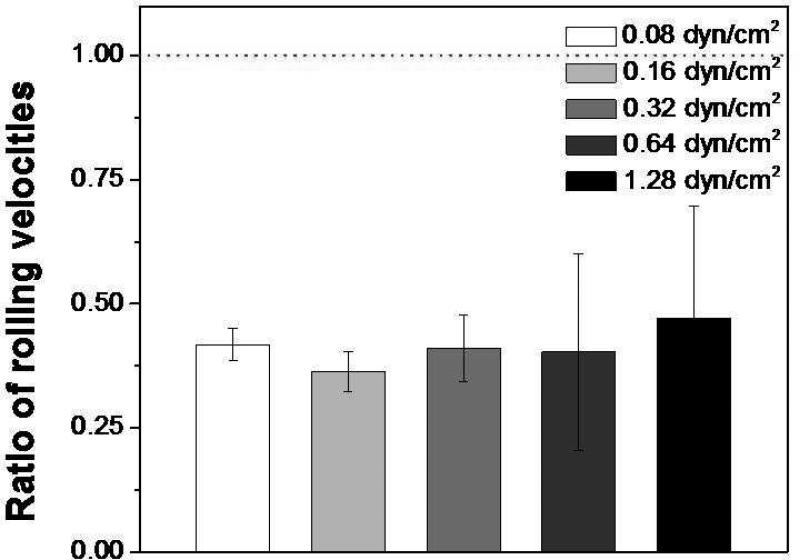
Our results so far still remain a question why CD24-coated microspheres bind statically (Figure 4) unlike MCF-7 cells that exhibit rolling (Figure 2) on the E-selectin-immobilized surface. This may be explained by the role of intracellular dynamics of the live cells. Given that actin filament dynamics are known to be involved in P-selectin-mediated rolling,42 we investigated the effect of the actin filaments on MCF-7 cell rolling. CytoD is a cell permeable inhibitor of actin polymerization, resulting in disruption of actin network organization,43 which alters the rolling response of neutrophiles on P-selectin to the stationary adhesion.42 MCF-7 cells were incubated with various concentrations of cytoD ranging from 10 to 40 μM for 30 min and 60 min, and the rolling velocity ratios (the velocity of treated cells to that of untreated cells on E-selectin) as a function of shear stress were measured. After treating the cells with cytoD at a concentration of 40 μM for 60 min, the rolling velocity was significantly reduced to less than half of that of untreated cells (Figure 5), suggesting that actin filament dynamics play a role in the rolling process. Intracellular dynamics are known to be involved in various cellular interactions with adhesive molecules. Actin filament polymerization is one of the factors that affect cell morphology37 and cellular behaviors (e.g. cell migration44 and intercellular interactions between cancer cells and endothelial cells45). In our experiments, cytoD-treated MCF-7 cells exhibited an increased adhesion, i.e. the decreased rolling velocities (Figure 5). Therefore, actin filament dynamics could be one factor that can control the intercellular interactions, which could explain the different behavior of the CD24-coated microspheres from MCF-7 cells with E-selectin. This can be further supported by the case of human colorectal carcinoma cells (HT-29), where it was reported that the adhesion between cells and the adhesive molecules became tight under dynamic condition after disruption of actin filaments.45
Figure 5.
Ratios of rolling velocities of cytoD-treated MCF-7 cells to those of untreated cells on E-selectin-immobilized surfaces. MCF-7 cells were treated with 40 μM of cytoD for 30 min. The dotted red line (ratio 1) indicates the rolling velocity of the untreated MCF-7 cells. Note that the rolling velocities of the treated MCF-7 cells are significantly reduced in a shear stress independent manner. Error bars: standard error (n=120).
In summary, this study reports the specific interaction between CD24 and E-selectin, which is confirmed by: i) the direct measurements of the binding kinetics of CD24 by SPR (Figure 1); ii) the observed interruption of rolling of MCF-7 cells upon treatment with anti-CD24 (Figure 2); and iii) the specific binding of CD24-coated microspheres on E-selectin (Figures 4 and 5). The identification of CD24 as a ligand for E-selectin will help to better understand the adhesion and invasion mechanisms of potentially metastatic cancer cells. For example, using CD24-mediated cell rolling on E-selectin-immobilized devices, CD24-positive and negative cancer cells may be separated without staining based on different binding and rolling behaviors that are reflected by CD24 expression. Further, in a similar way to several publications,46-48 CD24- and sLex-coated microspheres may be utilized to inhibit E-selectin-mediated cell adhesion (cell migration antagonists), which can be potentially used as treatment of inflammatory diseases and metastatic cancer.
Supplementary Material
ACKNOWLEDGEMENT
This work was supported by the National Science Foundation (NSF) under grant # CBET-0931472. This investigation was conducted in a facility constructed with support from grant C06RR15482 from the NCRR NIH. The authors thank Prof. Richard A. Gemeinhart and his group members for optical/fluorescence microscopy measurements and helpful discussion throughout the work.
REFERENCES
- 1.Psaila B, Lyden D. Nat. Rev. Cancer. 2009;9:285–293. doi: 10.1038/nrc2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nguyen DX, Bos PD, Massague J. Nat. Rev. Cancer. 2009;9:274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 3.Steeg PS. Nat. Med. 2006;12:895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 4.Kohler S, Ullrich S, Richter U, Schumacher U. Br. J. Cancer. 2009;102:602–609. doi: 10.1038/sj.bjc.6605492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Makrilia N, Kollias A, Manolopoulos L, Syrigos K. Cancer Invest. 2009;27:1023–1037. doi: 10.3109/07357900902769749. [DOI] [PubMed] [Google Scholar]
- 6.Miles FL, Pruitt FL, van Golen KL, Cooper CR. Clin. Exp. Metastasis. 2008;25:305–324. doi: 10.1007/s10585-007-9098-2. [DOI] [PubMed] [Google Scholar]
- 7.McEver RP. Thromb. Haemost. 1991;65:223–228. [PubMed] [Google Scholar]
- 8.Kansas GS. Blood. 1996;88:3259–3287. [PubMed] [Google Scholar]
- 9.Shaker OG, Ay El-Deen MA, Abd El-Rahim MT, Talaat RM. Tumori. 2006;92:524–530. doi: 10.1177/030089160609200610. [DOI] [PubMed] [Google Scholar]
- 10.Dimitroff CJ, Lechpammer M, Long-Woodward D, Kutok JL. Cancer Res. 2004;64:5261–5269. doi: 10.1158/0008-5472.CAN-04-0691. [DOI] [PubMed] [Google Scholar]
- 11.Fang X, Zheng P, Tang J, Liu Y. Cell Mol. Immunol. 2010;7:100–103. doi: 10.1038/cmi.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barthel SR, Gavino JD, Descheny L, Dimitroff CJ. Expert Opin. Ther. Targets. 2007;11:1473–1491. doi: 10.1517/14728222.11.11.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brodt P, Fallavollita L, Bresalier RS, Meterissian S, Norton CR, Wolitzky BA. Int. J. Cancer. 1997;71:612–619. doi: 10.1002/(sici)1097-0215(19970516)71:4<612::aid-ijc17>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 14.Tozeren A, Kleinman HK, Grant DS, Morales D, Mercurio AM, Byers SW. Int. J. Cancer. 1995;60:426–431. doi: 10.1002/ijc.2910600326. [DOI] [PubMed] [Google Scholar]
- 15.Steegmaier M, Levinovitz A, Isenmann S, Borges E, Lenter M, Kocher HP, Kleuser B, Vestweber D. Nature. 1995;373:615–620. doi: 10.1038/373615a0. [DOI] [PubMed] [Google Scholar]
- 16.Aeed PA, Geng JG, Asa D, Raycroft L, Ma L, Elhammer AP. Cell Res. 2001;11:28–36. doi: 10.1038/sj.cr.7290063. [DOI] [PubMed] [Google Scholar]
- 17.Zou X, Shinde Patil VR, Dagia NM, Smith LA, Wargo MJ, Interliggi KA, Lloyd CM, Tees DF, Walcheck B, Lawrence MB, Goetz DJ. Am. J. Physiol., Cell Physiol. 2005;289:C415–424. doi: 10.1152/ajpcell.00289.2004. [DOI] [PubMed] [Google Scholar]
- 18.Varki A. Nature. 2007;446:1023–1029. doi: 10.1038/nature05816. [DOI] [PubMed] [Google Scholar]
- 19.Fuster MM, Esko JD. Nat. Rev. Cancer. 2005;5:526–542. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 20.Thurin M, Kieber-Emmons T. Hybrid. Hybridomics. 2002;21:111–116. doi: 10.1089/153685902317401708. [DOI] [PubMed] [Google Scholar]
- 21.Matsumoto M, Shigeta A, Furukawa Y, Tanaka T, Miyasaka M, Hirata T. J. Immunol. 2007;178:2499–2506. doi: 10.4049/jimmunol.178.4.2499. [DOI] [PubMed] [Google Scholar]
- 22.Moser M, Bauer M, Schmid S, Ruppert R, Schmidt S, Sixt M, Wang HV, Sperandio M, Fassler R. Nat. Med. 2009;15:300–305. doi: 10.1038/nm.1921. [DOI] [PubMed] [Google Scholar]
- 23.Zen K, Liu DQ, Guo YL, Wang C, Shan J, Fang M, Zhang CY, Liu Y. PLoS One. 2008;3:e1826. doi: 10.1371/journal.pone.0001826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myung JH, Launiere CA, Eddington DT, Hong S. Langmuir. 2010;26:8589–8596. doi: 10.1021/la904678p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aigner S, Sthoeger ZM, Fogel M, Weber E, Zarn J, Ruppert M, Zeller Y, Vestweber D, Stahel R, Sammar M, Altevogt P. Blood. 1997;89:3385–3395. [PubMed] [Google Scholar]
- 26.Aigner S, Ramos CL, Hafezi-Moghadam A, Lawrence MB, Friederichs J, Altevogt P, Ley K. Faseb J. 1998;12:1241–1251. doi: 10.1096/fasebj.12.12.1241. [DOI] [PubMed] [Google Scholar]
- 27.Pirruccello SJ, LeBien TW. J. Immunol. 1986;136:3779–3784. [PubMed] [Google Scholar]
- 28.Kristiansen G, Sammar M, Altevogt P. J. Mol. Histol. 2004;35:255–262. doi: 10.1023/b:hijo.0000032357.16261.c5. [DOI] [PubMed] [Google Scholar]
- 29.Lim SC. Biomed. Pharmacother. 2005;59(Suppl 2):S351–354. doi: 10.1016/s0753-3322(05)80076-9. [DOI] [PubMed] [Google Scholar]
- 30.Baumhoer D, Riener MO, Zlobec I, Tornillo L, Vogetseder A, Kristiansen G, Dietmaier W, Hartmann A, Wuensch PH, Sessa F, Ruemmele P, Terracciano LM. Mod. Pathol. 2009;22:306–313. doi: 10.1038/modpathol.2008.192. [DOI] [PubMed] [Google Scholar]
- 31.Kim HJ, Kim JB, Lee KM, Shin I, Han W, Ko E, Bae JY, Noh DY. Cancer Lett. 2007;258:98–108. doi: 10.1016/j.canlet.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 32.Baumann P, Cremers N, Kroese F, Orend G, Chiquet-Ehrismann R, Uede T, Yagita H, Sleeman JP. Cancer Res. 2005;65:10783–10793. doi: 10.1158/0008-5472.CAN-05-0619. [DOI] [PubMed] [Google Scholar]
- 33.Fogel M, Friederichs J, Zeller Y, Husar M, Smirnov A, Roitman L, Altevogt P, Sthoeger ZM. Cancer Lett. 1999;143:87–94. doi: 10.1016/s0304-3835(99)00195-0. [DOI] [PubMed] [Google Scholar]
- 34.Kim JB, Ko E, Han W, Lee JE, Lee KM, Shin I, Kim S, Lee JW, Cho J, Bae JY, Jee HG, Noh DY. BMC Cancer. 2008;8:118. doi: 10.1186/1471-2407-8-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hong S, Leroueil PR, Majoros IJ, Orr BG, Baker JR, Jr., Banaszak Holl MM. Chem. Biol. 2007;14:107–115. doi: 10.1016/j.chembiol.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 36.Nilsson R, Sjogren HO. J. Immunol. Methods. 1984;66:17–25. doi: 10.1016/0022-1759(84)90243-6. [DOI] [PubMed] [Google Scholar]
- 37.Stevenson BR, Begg DA. J. Cell Sci. 1994;107:367–375. doi: 10.1242/jcs.107.3.367. [DOI] [PubMed] [Google Scholar]
- 38.Hong S, Rattan R, Majoros IJ, Mullen DG, Peters JL, Shi X, Bielinska AU, Blanco L, Orr BG, Baker JR, Holl MM. Bioconjug. Chem. 2009;20:1503–1513. doi: 10.1021/bc900029k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hong S, Lee D, Zhang H, Zhang JQ, Resvick JN, Khademhosseini A, King MR, Langer R, Karp JM. Langmuir. 2007;23:12261–12268. doi: 10.1021/la7014397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldman AJ, Cox RG, Brenner H. Chem. Eng. Sci. 1967;22:653–660. [Google Scholar]
- 41.Lawrence MB. Curr. Opin. Chem. Biol. 1999;3:659–664. doi: 10.1016/s1367-5931(99)00023-x. [DOI] [PubMed] [Google Scholar]
- 42.Sheikh S, Nash GB. J. Cell. Physiol. 1998;174:206–216. doi: 10.1002/(SICI)1097-4652(199802)174:2<206::AID-JCP8>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 43.Schliwa M. J. Cell Biol. 1982;92:79–91. doi: 10.1083/jcb.92.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hayot C, Debeir O, Van Ham P, Van Damme M, Kiss R, Decaestecker C. Toxicol. Appl. Pharmacol. 2006;211:30–40. doi: 10.1016/j.taap.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 45.Haier J, Nicolson GL. Clin. Exp. Metastasis. 1999;17:713–721. doi: 10.1023/a:1006754829564. [DOI] [PubMed] [Google Scholar]
- 46.Mackay CR. Nat. Immunol. 2008;9:988–998. doi: 10.1038/ni.f.210. [DOI] [PubMed] [Google Scholar]
- 47.Rossi B, Constantin G. Inflamm. Allergy Drug Targets. 2008;7:85–93. doi: 10.2174/187152808785107633. [DOI] [PubMed] [Google Scholar]
- 48.Stahn R, Schafer H, Kernchen F, Schreiber J. Glycobiology. 1998;8:311–319. doi: 10.1093/glycob/8.4.311. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.