Abstract
Tumor suppressor p53 plays an essential role in protecting cells from malignant transformation by inducing cell cycle arrest and apoptosis. Mutant p53 that is detected in over 50% cases of cancers not only loses its role in suppressing of tumor but also gains oncogenic function. Strategies to convert mutant p53 into wild-type of p53 have been suggested for cancer prevention and treatment, but they face a variety of challenges. Here we report an alternate approach that involves suppression of glucosylceramide synthase (GCS), an enzyme that glycosylates ceramide and blunts its pro-apoptotic activity in cancer cells. Human ovarian cancer cells expressing mutant p53 displayed resistance to apoptosis induced by DNA damage. We found that GCS silencing sensitized these mutant p53 cells to doxorubicin, but did not affect the sensitivity of cells with wild-type p53. GCS silencing increased the levels of phosphorylated p53 and p53-responsive genes including p21Waf1/Cip1, Bax and Puma, consistent with a redirection of the mutant p53 cells to apoptosis. Reactivated p53-dependent apoptosis was similarly verified in p53-mutant tumors where GCS was silenced. Inhibition of ceramide synthase with fumonisin B1 prevented p53 reactivation induced by GCS silencing, whereas addition of exogenous C6-ceramide reactivated p53 function in p53-mutant cells. Our findings indicate that restoring active ceramide to cells can resuscitate wild-type p53 function in p53 mutant cells, offering preclinical support for a novel type of mechanism-based therapy in the many human cancers harboring p53 mutations.
Keywords: mutant p53, restoration, glucosylceramide synthase, ceramide, apoptosis
Introduction
p53 protein is a crucial tumor suppressor in preventing tumorigenesis and tumor progression (1–2). As an essential transcription factor, p53 activates the expressions of p21Waf1/Cip1, Bax, Puma, FAS and others, and consequently regulates cell cycle arrest and apoptosis to remove abnormal cells (1–2). The transcription activity of p53 on p53-responsive genes is sequence-specific, and mainly relies on its DNA-binding domain (DBD, residues 102 to 292) encoded by exon 5 to exon 8 (1–2). In normal cells, the p53 is tightly controlled and kept at low level, but DNA damage, oncogenic stress, hypoxia and cellular distress activate p53 by increasing its expression and protein phosphorylation through various kinases (3–4).
p53 mutants that have been found in approximately 50% cases of solid cancers turn this tumor suppressor into an oncogenic factor (5–6). p53 mutants are mainly located in exon 5–8 (residue 126–306) of the core domain (6). Mutant p53 thus loses sequence specific binding to its responsive genes involved in cycle arrest, senescence and apoptosis. Mutant p53 is unable to activate MDM2 and degrades protein through ubiquitination, resulting in mutant protein accumulated in cell nucleus (7). Mutation of other tumor suppressors often produces truncated proteins or unable to synthesize any protein; however, more than 80% of p53 mutants are missense and synthesized as stable and full-length proteins (8). These p53 mutants confer a dominant-negative or oncogenic gain-of-function in cells (9). p53 mutant that promotes tumor progression and resistance to therapy becomes the most common prognostic indicator for both cancer recurrence and death (10). Restoration of p53 function has been succeeded in regression of lymphomas, sarcomas and hepatocellular carcinoma and represents a more effective means to cancer treatment (11–13). It has been reported that gene therapy and small molecules (e.g. Prima-1, Ellipticin, peptides) restores p53 function by addition of wild-type or alteration of protein conformation (13–15); however, little is known whether p53 expression can be restored in 53-mutant tumors. Gene therapy could not bring an adequate benefit to ovarian cancers with standard chemotherapy (16), that calls for exploration of new approach, restoring the expression of wild-type p53.
Drug resistance is an outcome of multi-gene interactions, and p53 mutant impedes induced-apoptosis when cancer cells are under chemotherapy. Ceramide is an active sphingolipid and plays critical roles in processing of apoptosis and other cellular functions (17–20). Both ceramide and p53 are implicated in DNA-damage induced apoptosis (17–18). Loss of ceramide production confers resistance to radiation-induced apoptosis in B-lymphoma and myeloid leukemia cells that express wild-type p53 (21–22). Glucosylceramide synthase (GCS) catalyzes ceramide glycosylation, converting ceramide to glucosylceramide. GCS is a limiting enzyme controlling intracellular ceramide and glycosphingolipids (23). Overexpression of GCS that confers cell resistance to apoptosis (24–25) becomes a potential marker predicting tumor response to chemotherapy and clinical progression (26–27). Inhibition of GCS by gene silencing or tamoxifen leads p53-mutant cancer cells to apoptosis (25, 28–29). In the present study, we investigate whether disruption of ceramide glycosylation restores p53-dependent apoptosis in p5-mutant cancer cells.
Materials and Methods
Cell culture and treatments
Drug-resistant human NCI/ADR-RES ovary cancer cells (30) and MCF-7 breast adenocarcinoma cells were kindly provided by Dr. Kenneth Cowan (UNMC Eppley Cancer Center, Omaha, NE) and Dr. Merrill Goldsmith(National Cancer Institute, Bethesda, MD) (31). The OVCAR-8 and A2780 human ovarian cancer cell lines were kindly provided by Dr. M. Hollingshead (Division of Cancer Treatment and Diagnosis Tumor Repository at National Cancer Institute). Drug-resistant human ovarian cancer A2780ADR (also named A2780-DX3) was purchased from Sigma-Aldrich (St. Louis, MO) (32). MCF-12A, a normal human mammary epithelial cell line was purchased from ATCC (Manassas, VA). MCF-7, NCI/ADR-RES, and OVCAR-8 cells were cultured in RPMI-1640 medium containing 10% fetal bovine serum (FBS), 100 units/ml penicillin, 100 μg/ml streptomycin, 584 mg/liter L-glutamine. A2780ADR cells were cultured in medium containing 100 nM doxorubicin in addition to the above components. MCF-12A cells were cultured in DMEM/F12 medium containing 5% FBS and 0.01 mg/ml bovine insulin. Cells were maintained in an incubator humidified with 95% air and 5 % CO2 at 37 oC. Cell lines were authenticated in November 2010 at the John Hopkins University Fragment Analysis Facility (Baltimore, MD) using Applied Biosystem Identifiler System to test for 16 STR markers and amelogenin for gender determination. Authenticity was confirmed against the ATCC database (http://bioinformatics.istge.it/clima/) and NCI-60 database published (33).
For MBO-asGCS (mixed-backbone oligonucleotide against GCS) pretreatments, experiments were conducted as described previously (29, 34). Briefly, after overnight culture, cells (2 x 106 /100 mm dish) were transfected with MBO-asGCS (50–200 nM; Integrated DNA Technologies, Inc. Coralville, IA) using LipofectamineTM 2000 (Invitrogen, Carlsbad, CA) and cultured in 10% FBS medium. MBO-asGCS were introduced into cells twice in 7 days to observe p53 expression. Cells were exposed to doxorubicin (1 or 2.5 μM, 48 hr) to induce p53.
GCS inhibitor, D-threo-1-phenyl-2-decanoylamino-3-morpholino-1-propanol HCl (D-PDMP), C6-ceramide (N-hexanoyl-D-erythro-sphingosine) and C6-dihydroceramide (N-hexanoyl-D-erythro-sphinganine) were purchased from Matreya (Pleasant Gap, PA). Doxorubicin hydrochloride and acidic sphingomyelinase (100 units/mg) of human placenta was purchased from Sigma-Aldrich (St. Louis, MO). Ceramide synthase inhibitor, fumonisin B1 (FB1) was purchased from Biomol (Plymouth Meeting, PA). Puma siRNA and its scrambled control were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Cell viability assay
Cell viability was determined by quantization of cellular ATP, using the CellTiter-Glo luminescent cell viability assay kit (Promega, Madison, WI) (29, 34–35). Briefly, cells (4,000 cells/well) were grown in 96-well plates with 10% FBS RPMI-1640 medium overnight. After pretreatments, cells were incubated with doxorubicin in 5% FBS medium for additional 72 hr. Cell viability was determined by the measurement of luminescent ATP in a Synergy HT microplate reader (BioTek, Winnooski, VT) following incubation with CellTiter-Glo reagent.
Western blot analysis
After treatments, cells or tissue homogenates were lysed in NP40 cell lysis buffer (Biosource, Camarillo, CA). Equal amount of proteins (50 μg/lane) were resolved using 4–20% gradient PAGE (Invitrogen). After transferring, blots were blocked in 5% fat-free milk in PBST (0.05% tween-20, 20 mM phosphate buffered saline, pH 7.4) and incubated with antibodies against p53 (Invitrogen), phosphporylated p53 at Ser15 (pp53), cleaved PARP (c-PARP), Puma (Cell Signaling Technology, Danvers, MA), p21Waf1/Cip1, Bax (Santa Cruz Biotechnology), active caspase-7 (a-Casp7, EMD Chemicals, Gibbstown, NJ) and GCS (1:500 to 1:5000 dilution), respectively, at 4 oC, overnight. After washing, these blots were incubated with horseradish peroxide-conjugated secondary antibodies, and developed using SuperSignalR West Pico ECL (Thermo scientific, Rockford, IL), as described previously (34–35). Endogenous GAPDH was used as a loading control. The levels of pp53 protein were represented by the ratios of optical densities in pp53 bands normalized against unphosphorylated p53.
Immunocytochemistry
Cells (10,000 cells/chamber) were grown in 4-chamber slides with treatments for 48 hr. After fixation with methanol, cells were blocked with 5% goat serum PBS (block solution) incubated with antibodies against GCS (1:1000) or ceramide (1:500; clone MID 15B4 from Sigma) and pp53 (1:1000) in block solution at 4oC, overnight. Cellular GCS and pp53 tagged with antibodies were recognized by Alexa FluorR488 goat anti-rabbit IgG and Alexa FluorR555 goat anti-mouse IgG (1:1000). Cellular ceramide and pp53 tagged with antibodies were recognized by Alexa FluorR555 goat anti-mouse IgG and Alexa FluorR488 goat anti-rabbit IgG (1:1000), respectively. Cell nuclei were counterstained with DAPI (4′, 6-diamidino-2-phenylindole) in mounting solution. Slides were observed under Olympus IX71 fluorescence microscope coupled with digital camera and images were captured using Olympus DP controller software. Confocal images were captured using LSM Pascal confocal microscope (Carl Zeiss Microimaging Inc., Thornwood, NY).
Tumors were fixed and maintained in paraffin blocks. Microsections of each tumor (5 μm) were stained with Hematoxylin and Eosin (H&E) and identified by pathologist. For immunostaining, antigens were retrieved in steaming sodium citrate buffer (10 mM, 0.05% Tween-20, pH 6.0). After blocking, slides were incubated with anti-GCS rabbit serum (1:100) and anti-pp53 mouse primary antibodies at 4°C overnight. Cell nuclei were counterstained with DAPI.
Flow cytometry assay
The analyses were performed, as described previously (29). Cells (5x105 cells/100-mm dish) were exposed to doxorubicin (2.5 μM) for 48 hr and harvested with trypsinization and centrifugation. Cells resuspended were incubated with 0.01% propidium iodide (PI) in staining solution (0.1% sodium citrate, 0.3% Triton X-100, 2 mg/ml ribonuclease A) at 4°C for 30 min. The cells were analyzed using FACSCalibur with CellQuest Pro program (BD Biosciences, San Jose, CA). For each sample, 10,000 events were counted three times and sub-phase G0/G1 was defined as indicative of apoptotic cells in cell cycle histograms.
Tumor xenografts and treatments
All animal experiments were approved by the IACUC, University of Louisiana at Monroe, and were handled in strict accordance with good animal practice as defined by NIH guidelines. Tumor model was established, as described previously (29, 34). Athymic nude mice (Foxn1nu/Foxn1+, 4–5 weeks, female) were purchased from Harlan (Indianapolis, IN). Cell suspension of NCI/ADR-RES (3–5 passages, 1 x 106 cells in 20 μl/mouse) was subcutaneously injected in the left flank of the mice. Mice were monitored by measuring tumor growth, body weight and clinical observation. Once tumors were visible (~2 mm in diameter), mice were randomly allotted to different treatment groups (10 mice/group). For treatment, MBO-asGCS (1 mg/kg/3-day) or MBO-SC (scrambled control for MBO-asGCS) was administered intratumorally alone or with doxorubicin (2 mg/kg/week, i.p.) for 32 days. The control group received saline and doxorubicin combination. Tumor volume was calculated by the formula L/2 x W2 (L, the length; W, the width).
Terminal-deoxynucleotide-transferase-mediated dUTP nick end labeling (TUNEL) staining
Apoptotic cells of tumor tissues were detected by measurement of nuclear DNA fragmentation using TUNEL system (Promega), following the manufacturer's instruction, as described previously (29). Briefly, after antigen retrieval, slides were put in 0.2 mg/ml proteinase K in 10 mM Tris-HCl, pH 8.0 for 20 min for digestion and labeled for 90 min with fluorescein-12-dUTP terminal deoxynucleotide transferase reaction mixture at 37°C in a humidified chamber. After mounting with DAPI, the sections were observed.
Statistical analysis
Cell experiments in triplicate were repeated twice. All data represent the mean ± SD. Student’s t test was employed to compare mean values, using a Prism 4 program (GraphPad software, San Diego, CA).
Results
Silencing of GCS by MBO-asGCS sensitized mutant p53 cells to doxorubicin
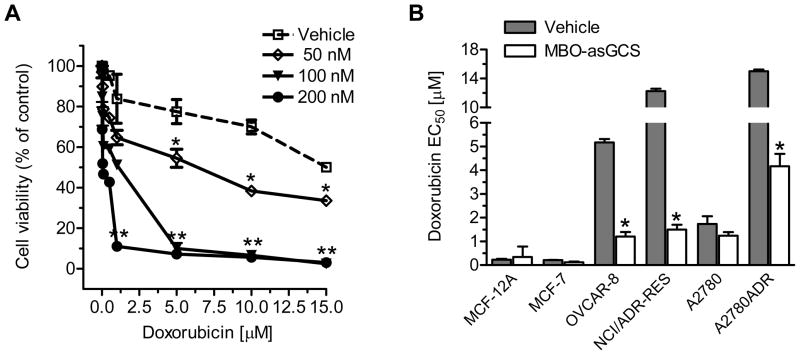
Mutant p53, particularly the deletion is highly associated with poor-response to chemotherapy (10–11). NCI/ADR-RES and OVCAR-8 cells are mutant p53 cell lines that dominantly express the p53 with deleted 21-bp and 18-bp within the DNA-binding domain (36–37). NCI/ADR-RES has an additional point mutation, arginine instead of proline at codon 72 of p53 (36). A2780ADR (also named A2780-DX3) cells do not respond to cisplatin-induced p53 activation, even though the mutation has not been determined (32) (Table 1). NCI/ADR-RES, OVCAR-8 and A2780ADR display considerable resistance to several anticancer drugs including doxorubicin and cisplatin (31, 37) (Table 1). To examine whether disruption of ceramide glycosylation restores p53-dependent apoptosis, we treated NCI/ADR-RES cells with MBO-asGCS to silence GCS and then tested cell response to doxorubicin. As shown in Fig. 1A, MBO-asGCS treatments significantly increased cell response to doxorubicin, as suppressed GCS expression in dose-dependent fashion (Fig. S1A). At 200 nM, MBO-asGCS decreased the EC50 for doxorubicin by 17-fold (12.9 μM vs. 0.8 μM), as compared with vehicle control. To test whether this sensitization is associated with p53 status, we silenced GCS with MBO-asGCS (50 nM, 7 days) in cell lines with variant p53 status (Table 1). OVCAR-8 and NCI/ADR-RES cells sharing mutant p53 displayed doxorubicin-resistance, and their EC50 values for doxorubicin were 22-fold (5.2 μM vs. 0.23 μM) and 53-fold (12.4 μM vs. 0.23 μM) greater than p53 wild-type cells, either MCF-12A or MCF-7 (Fig. 1B). Interestingly, silencing of GCS with MBO-asGCS sensitized p53-mutant cells, but not p53 wild-type cells. With decreases of GCS protein levels (Fig. S1B), MBO-asGCS treatments decreased EC50 values for doxorubicin in OVCAR-8, NCI/ADR-RES and A2780ADR by 4-fold, 8-fold and 4-fold, respectively. However, MBO- asGCS minimally reduced GCS protein (Fig. S1B) and the EC50 values in MCF-12A, MCF-7 and A2780 cells (Fig. 1B).
Table 1.
p53 status and cell response to anticancer drugs.
| Cell line | p53 tumor suppressor | GCS mRNA (copy) | Dox Resistance EC50 (μM) | ||
|---|---|---|---|---|---|
| Mutation | Exon | Codon | |||
| MCF-12A | wild-type | 500 | 0.23 | ||
| MCF-7 | wild-type | 600 | 0.21 | ||
| OVCAR-8 | deletion, 18 bp | 5 | 126–132 | 2400 | 5.17 |
| NCI/ADR-RES | deletion, 21 bp | 5 | 126–133 | 2624 | 12.5 |
| A2780 | wild-type | 488 | 1.74 | ||
| A2780ADR* | undetermined | 1499 | 15.0 | ||
Figure 1.
Silencing of GCS sensitized mutant p53 cancer cells to doxorubicin. A. Cell response to doxorubicin. NCI/ADR-RES cells were pretreated with MBO-asGCS for 7 days and exposed to doxorubicin for additional 72 hr. *, p<0.01 compared with vehicle control; **, p<0.001 compared with vehicle control. B. EC50 values for doxorubicin. Cells were pretreated with MBO-asGCS (50 nM) or vehicle (Lipofectamine 2000) for 7 days, and exposed to doxorubicin in 5% FBS medium for additional 72 hr. EC50 was calculated using Prism software after measurements. *, p<0.001 compared with vehicle control.
Disruption of ceramide glycosylation increased phosphorylated p53, and induced the expressions of p53-responsive genes in mutant p53 cells
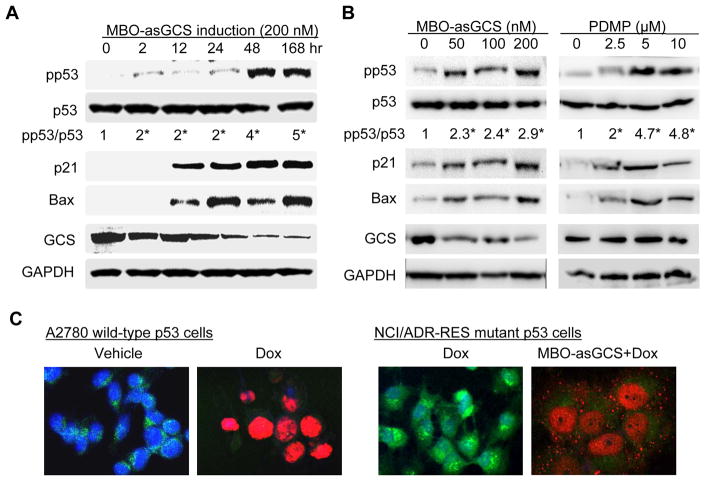
To examine whether disruption of ceramide glycosylation alters p53, we used NCI/ADR-RES cells that dominantly express mutant p53 and high level of GCS (25, 36). It was found that suppression of GCS by MBO-asGCS increased the expression levels of wild-type p53 and p53-resposive genes. After 48 hr treatments, MBO-asGCS increased the levels of phosphorylated p53 (pp53, at Ser15 in DBD) greater than 4-fold with p21Waf1/Cip1 and Bax, as GCS was significantly suppressed in NCI/ADR-RES cells (Fig. 2A). Silencing of GCS by MBO-asGCS reactivated p53 response to doxorubicin-induced DNA damage, as pp53 levels were increased with decrease of GCS protein (Fig. 2B). As expected, GCS protein levels were suppressed by MBO-asGCS in dose-dependent manner; the pp53 levels were significantly increased greater than 2-fold, even though at 50 nM MBO-asGCS, while unphosphorylated p53 protein levels were constant in all treatments (Fig. 2B). Consequently, the levels of p21Waf1/Cip1 and Bax were enhanced with pp53 augmenting. The association of pp53 with GCS suppression was confirmed in NCI/ADR-RES cells treated with D-PDMP, a GCS inhibitor. Disruption of ceramide glycosylation by D-PDMP significantly increased the levels of pp53, p21Waf1/Cip1 and Bax in dose-dependent fashion (Fig. 2B). Furthermore, immunofluorescent staining revealed that suppression of GCS restored p53 response to doxorubicin-induced DNA damage. MBO-asGCS treatment dramatically decreased GCS protein in Golgi apparatus where ceramide glycosylation occurred, consequently that increased nuclear pp53 in more than 90% of NCI/ADR-RES cells exposed to doxorubicin (Fig. 2C). Different from wild-type p53 A2780 cells (left panel), pp53 particles were also detected in cytoplasm even though most pp53 were concentrated in nucleus of NCI/ADR-RES.
Figure 2.
Disruption of ceramide glycosylation enhanced phosphorylated p53 and induces p21Waf1/Cip1 and Bax expressions in p53 mutant NCI/ADR-RES cells. A. GCS suppression in time-course. Cells were treated with MBO-asGCS (200 nM) and then exposed to doxorubicin (2.5 μM, 48 hr). Equal amount of detergent-soluble proteins (50 μg/lane) was resolved by 4–20% PAGE and immunoblotted with antibodies. *, p<0.001 compared with vehicle control; pp53, phosphorylated p53 (at Ser15 in DBD). B. GCS suppression in dose-dependent. Cells were treated with MBO-asGCS (7 days) or D-PDMP (48 hr) and then exposed to doxorubicin (2.5 μM, 48 hr). *, p<0.001 compared with vehicle control. C. Active pp53 and GCS in cells. A2780 cells were exposed to doxorubicin (Dox, 1 μM, 48 hr); NCI/ADR-RES cells were exposed to doxorubicin (Dox, 2.5 μM, 48 hr) following MBO-asGCS pretreatment (50 nM, 7 days). Merged fluorescence microphotographs (x200) was captured by confocal microscopy. Cells were recognized by anti-GCS (green) and anti-pp53 (red) antibodies with Alexa FluorR488- and Alexa FluorR555-conjugated goat antibodies. Nucleus was counterstained with DAPI (blue).
Disruption of ceramide glycosylation induced p53-dependent cell growth arrest and apoptosis in mutant p53 cells
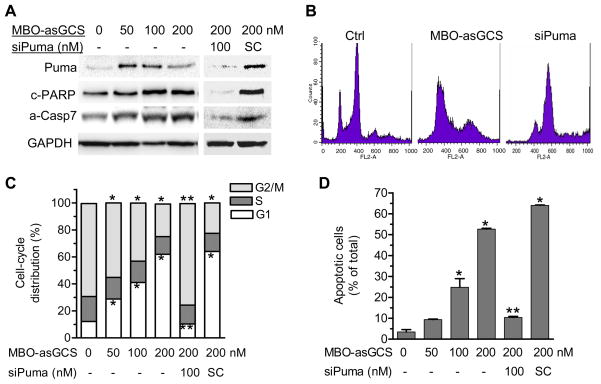
To examine the effects of GCS silencing on p53-responsive genes, we further assessed cell division and apoptosis. Puma is a major p53-responsive gene involved in induced-apoptosis in response to DNA-damage and cleaved PARP (c-PARP) as well as active caspase-7 (a-Casp7) are effectors in p53-dependent apoptosis processing (38-39). We found that the protein levels of Puma were significantly increased in NCI/ADR-RES cells exposed to doxorubicin after MBO-asGCS (Fig. 3A). The levels of c-PARP and a-Casp7 were increased by 2- to 3-fold, respectively. Silencing of Puma with siRNA eliminated the effects of reactivated p53 on Puma, c-PARP and a-Casp7 (Fig. 3A). Flow cytometry assays revealed that MBO-asGCS pretreatments significantly affected cell division and apoptotic cells. The numbers of G2/M phase cells that were under dividing and proliferation in histogram of flow cytometry significantly decreased with increasing concentrations of MBO-asGCS (50–200 nM); however, G1 phase cell numbers were increased by 2-fold, 3-fold and 5-fold, respectively (Fig. 3B, 3C). Furthermore, flow cytometry detected that the numbers of G0/G1 phase cells that are under apoptosis were 3-fold (9.4% vs. 3.4% of total cells), 7-fold (24.8 % vs. 3.4% of total cells), and 15-fold (52.6 vs. 3.4% of total cells) at 50–200 nM MBO-asGCS pretreatment, as compared with vehicle control respectively. Silencing of Puma eliminated the apoptotic effects of restored p53 (Fig. 3B, 3D). These data indicates that silencing of GCS resulted in cell growth arrest and apoptosis through p53-responsive genes.
Figure 3.
GCS suppression promoted mutant p53 cells to cell-cycle arrest and apoptosis. NCI/ADR-RES cells were exposed to doxorubicin (2.5 μM, for 48 hr) following MBO-asGCS pretreatments (50 nM, 7 days). Puma siRNA (siPuma, 100 nM) and its scrambled control (iSC, 100 nM) were introduced to cells in last 54 hr. A. Puma, a-Casp7 and c-PARP. Equal amount of proteins (50 μg/lane) was subjected to resolved and immunoblotted. B. Flow cytometry. PI stained cells were analyzed by flow cytometry. C. Cell division. D. Apoptotic cells. *, p<0.001 compared with vehicle control.
Restoration of functional p53 expression induced apoptosis in mutant p53 tumors
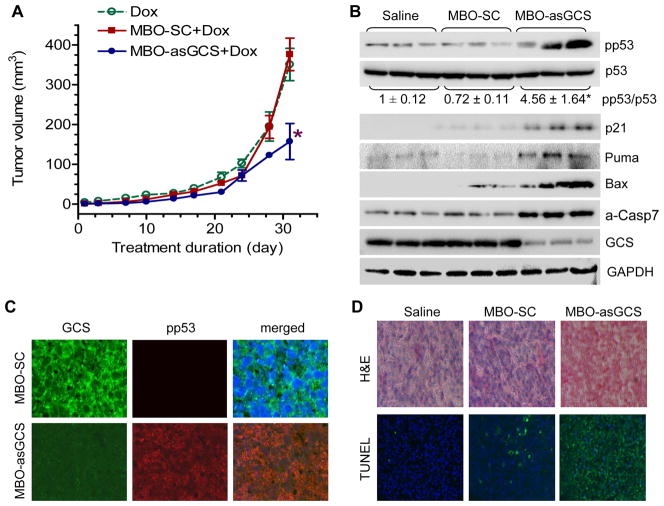
We further investigated whether silencing of GCS restores p53 expression and eliminates mutant p53 tumor growth in vivo. After tumors of NCI/ADR-RES were apparent, mice were treated with MBO-asGCS (1 mg/kg twice a week, intratumoral injection) combined with doxorubicin (2 mg/kg once a week, i.p.) for 32 days. MBO-asGCS combined with doxorubicin decreased tumor volume to 42 % (157 mm3 vs. 376 mm3, p<0.01), as compared with MBO-SC (scrambled control) combined with doxorubicin or doxorubicin alone treatment (Fig 4A). Western blotting detected that silencing of GCS substantially decreased GCS, which in turn increased pp53 and p53-responsive genes in tumors exposed to doxorubicin (Fig. 4B). MBO-asGCS treatments increased wild-type p53, as represented by the ratio of pp53/p53 greater than 4-fold (p<0.001), as compared to vehicle control or MBO-SC. Consequently, the reactivated p53 substantially induced the expression of p21Waf1/Cip1, Bax, Puma and a-Casp7 (Fig. 4B). Immunostaining confirmed these findings and showed that MBO-asGCS, but not MBO-SC, significantly increased nuclear pp53 in tumors while decreased GCS protein in cytoplasm (Fig. 4C). TUNEL analysis indicated that approximately 80% of tumor cells underwent apoptosis in tumors treated with MBO-asGCS combined with doxorubicin, compared to only 3% and 9% of apoptotic cells in doxorubicin alone and doxorubicin combined with MBO-SC (Fig. 4D). Thus, these data indicate that GCS suppression restores wild-type p53 expression and leads mutant p53 tumors to induced-apoptosis.
Figure 4.
Silencing GCS by MBO-asGCS inhibited tumor growth through functional p53 restoration. A. Tumor growth curve. Athymic mice bearing NCI/ADR-RES tumors were treated with doxorubicin (Dox, 2 mg/kg/week) and combinations with MBO-asGCS (1 mg/kg/3-day) or MBO-SC (scrambled control) for 32 days. *, p<0.001 compared with the combination of MBO-SC and Dox. Data represents mean ± SEM (10 cases/group). B. Tumor proteins. Equal amount proteins extracted from tumors (100 μg/lane, 3 cases/group) were resolved by 4–20% PAGE and immunoblotted with antibodies, respectively. *, p<0.001 compared with MBO-SC. C. Immunostaining of pp53 and GCS (x 200). anti-GCS and anti-pp53 antibodies with Alexa FluorR488- and Alexa FluorR555-conjugated goat antibodies. Nucleus was counterstained with DAPI. D. TUNEL staining. Apoptotic cells (TUNEL+) exhibit green fluorescence (x200).
Ceramide mediated the restoration of p53 expression
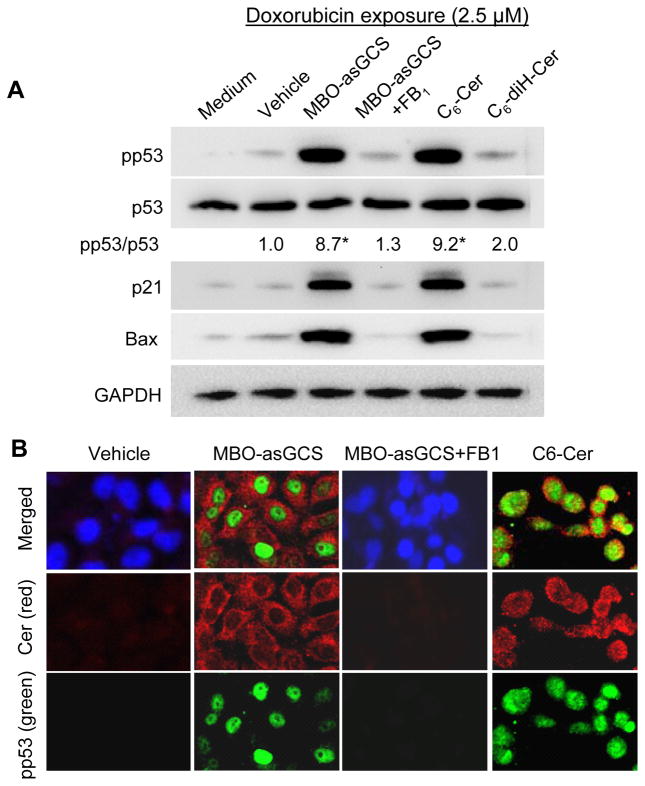
To determine whether ceramide or glucosylceramide after disruption of ceramide glycosylation is the factor restoring p53, we assessed its effects on cellular ceramide and glycosphingolipids. In NCI/ADR-RES cells, MBO-asGCS treatments (50–200 nM) decreased globotriaosylceramide (Gb3) and significantly increased ceramide (Fig. S2A) in dose-depdendent fashion. At 200 nM of MBO-asGCS, endogenous ceramide increased more than 160% (1.01 vs. 1.65 ng/100 μg protein, p<0.001) (Fig. S2C). Additionally, MBO-asGCS significantly enhanced ceramide in other p53 mutant OVCAR-8 and A2780ADR cells, but not p53 wild type cells (Fig. S2D). Further, we examined the expression of pp53 and p53-responsive genes in NCI/ADR-RES cells treated with ceramide synthase inhibitor and cell permeable ceramide, respectively. As shown in Fig. 5A, fumonisin B1 (FB1) treatment (25 μM, 28 hr) that inhibited ceramide synthesis in de novo pathway eliminated the effects of MBO-asGCS on restoration of p53. MBO-asGCS alone increased pp53 protein level by approximately 9-fold, but MBO-asGCS combined with FB1 could not enhance pp53 level or p21Waf1/Cip1 and Bax. Exogenous cell-permeable C6-ceramide (C6-Cer, 25 μM, 48 hr) substantially increased pp53 level by 9-fold; however, C6-dihydroceramide (C6-diH-Cer; 25 μM, 48 hr) that had less bioactivity enhanced pp53 by 2-fold and could not significantly increase p21Waf1/Cip1 or Bax (Fig. 5A). Immunofluorescent staining confirmed that MBO-asGCS dramatically increased cellular ceramide, and pp53 dramatically appeared in nucleus in cells exposed to doxorubicin (Fig. 5B). FB1 that inhibited ceramide synthase prevented cellular ceramide and pp53 reactivated in nucleus, even though MBO-asGCS disrupted ceramide glycosylation. C6-Cer treatment significantly increased pp53 in nucleus, as C6-Cer was detected mostly in cytoplasm and nuclear envelope (Fig. 5B).
Figure 5.
Ceramide restored p53 expression in mutant p53 cells. After pretreatments of MBO-asGCS (50 nM, 7 days), FB1 (25 μM, 48 hr) C6-ceramide (5 μM, 48 hr; C6-Cer,) and C6-dihydroceramide (5 μM, 48 hr; C6-diH-Cer), NCI/ADR-RES/asGCS cells were exposed to doxorubicin (2.5 μM, 48 hr). A. Western blotting. Equal amount proteins extracted from tumors (100 μg/lane) were resolved by 4–20% PAGE and immunoblotted with antibodies. *, p<0.001 compared with Vehicle. B. pp53 and ceramide in cells. Merged fluorescence microphotographs (x200) was captured by confocal microscopy. Green, cells were incubated with anti-pp53 (green) and anti-ceramide (red) following addition of Alexa FluorR488- and Alexa FluorR555-conjugated goat antibodies. Nucleus was counterstained with DAPI.
To understand how silencing of GCS reactivates p53, we examined protein, mRNA and hnRNA of mutant p53 and wild-type p53 in cell lines that dominantly express wild-type 53 or mutant p53 after GCS silencing. As shown in Fig. S3A, MBO-asGCS pretreatment (50 nM, 7 days) did not significantly affect pp53 levels in MCF-12A and A2780 cells, as p53 expression in these wild-type p53 responded to doxorubicin exposure. In contrast, MBO-asGCS considerably increased pp53 by approximately 3-fold in mutant p53 OVCAR-8 and NCI/ADR-RES cells. The mRNA that included p53 exon-5 could not be detected in mutant p53 OVCAR-8 and NCI/ADR-RES cells, even under doxorubicin exposure (Fig. S3B). However, we detected p53 exon-5 in mutant p53 cells after silencing of GCS with MBO-asGCS pretreatment (Fig. S3B). Analysis of p53 hnRNA found that p53 exon-5 was transcribed in both wild-type and mutant p53 cell lines (Fig. S3C). These data indicate that silencing GCS may restore p53 at the level of post-transcriptional processing.
Discussion
Reactivation of mutant p53 can trigger massive cell death and efficiently eliminate abnormal cells. Previous works from those of others have shown that both introduction of wild-type p53 by gene transfection and modulation of the protein conformation by small molecules activate p53-responsive genes and restore p53-dependent apoptosis in mutant p53 cancer cells (11, 14–15). The present study, for the first time, has demonstrated that suppression of GCS restores p53 expression and its functions in mutant p53 ovarian cancer cells. Suppression of GCS expression by MBO-asGCS disrupts ceramide glycosylation and increase ceramide, which in turn leads cell death and eliminates tumor progression through p53-dependent apoptosis.
Mutant p53 has been reported as a marker of poor prognosis and a cause of chemotherapy failure in many types of cancers (3, 6). More than 85% of point mutants occur in the DBD encoded by exon-3 to exon-8 (5, 9). Exon-5 mutations are prognostic indicators of shortened survival in non-small-cell lung cancer (40). Small fragment-deletion is less common than point mutant detected in p53 mRNA and protein; however, all deletion mutants in DBD completely lose p53 functions (5). In OVCAR-8 and NCI/ADR-RES cells, the 18-bp and 21-bp deletions in exon-5 confer these cells resistance to anticancer drugs, particularly those inducing DNA-damage including doxorubicin and cisplatin (25, 36). It has been demonstrated that ceramide activates apoptosis processing in response to DAN-damage stress, when cells are exposed to anticancer drugs, cytokines and irradiation (19, 41). Ceramide can independently induce apoptosis in testicular germ-cell tumor cells with null or mutant p53 (42), although ceramide response to DNA-damage stress is p53-dependent in others (18). Enhancing endogenous ceramide by disrupting ceramide glycosylation sensitizes mutant p53 cells to drug-induced apoptosis, even though it does not significantly alter p53 protein levels, as reported previously (25) and detected by Western blotting in the present study (Fig. 2A, 2B, 4B, 5A, S1A). However, further assessments unveil that the levels of pp53, p53-responsive genes (p21Waf1/Cip1, Bax, Puma), and apoptosis are all significantly increased in dose-dependent fashion (Fig. 2–4). These evidences indicate that functional p53, but not mutant p53, is involved in DNA-damage induced-apoptosis after disruption of ceramide glycosylation. In particular type of cells, the direct effects of ceramide-activated mitochondrial alterations may be dominant. It is more likely that ceramide processing apoptosis is associated with p53-dependent pathways, as we have found that ceramide increased after MBO-asGCS treatment (Fig S2) (29) or exogenous C6-ceramide reactivates pp53 in nucleus and enhances p53-responsive genes including p21Waf1/Cip1, Bax and Puma (Fig. 2, 3, 4, 5).
Mutant p53 is a defined target for cancer therapies; however, discovering an effective approach to restore functional p53 is a great challenge. Mutant p53 is often expressed at high levels in cancers, and it affects tumor progression and treatment outcome in terms of loss-of-function in tumor suppression as well as gain-of-function in oncogenic effects (12, 43). Introduction of wild-type p53 by gene therapy can correct the loss-of-function in tumor suppression, but it cannot diminish the oncogenic effects of mutant p53 on tumors. It has been reported that a group of small molecular compounds (Ellipticine, CP-31398, Prima-1) and peptides (C369-382, C361-382) modulate protein conformations of mutant p53, reactivating its transcription function (15, 44). CP-31398, a styrylquinazoline compound, upregulates the expression of p53-responsive genes, and represses tumor growth through binding to DBD and stabilizing the active protein conformation in mutant p53 cells (45–46). Prima-1 reactivates mutant p53 (His175, His273) by covalent binding to and modification of thiol groups in the core domain in the H1299-His175 lung adenocarcinoma or Saos-2-His273 osteosarcoma cells (15). On the other hand, peptide C361-382 restores the transcriptional activity of mutant p53 by an allosteric mechanism of negative regulation (44, 47). In the present study, we have found that ceramide mediates p53 restoration, as silencing of GCS with MBO-asGCS can increase endogenous ceramide (29) and C6-ceramide leads to the same restorative effects (Fig. 2, 4, 5).
It is not clear how ceramide restores functional p53 in deletion mutant of p53. In addition to direct effects on processing of apoptosis, ceramide mediates gene expressions including MDR1, telomerase, c-fos, p21Waf1/Cip1, GCS, caspase-9 and Bcl-x (34–35, 48–49). Ceramide might reactivate functional p53 by modulating either mutant p53 conformation or wild-type p53 expression, but based on this study, it is more likely that ceramide restores wild-type p53 expression at posttranscriptional processing. Suppression of GCS restores p53 reactivation in 12 hr to several days (Fig. 2A), and wild-type p53 is detected in mRNA and protein (Fig. 2, 4, 5, Fig. S1). Additionally, suppression GCS does not have any effect on p53 transactivation (data not shown). These strongly suggest ceramide-mediated post-transcriptional processing may play a critical role in regulating expression of p53 wild-type or mutant.
High levels of GCS and mutant p53 are coincidently detected in several drug-resistant ovarian cancer cells lines, such as OVCAR-8, NCI/ADR-RES and A2780ADR in this study. MBO-asGCS, a specific agent for GCS suppression effectively restores p53-dependent apoptosis in vitro and in vivo. Further investigating the association of GCS and mutant p53 in drug resistance and the molecular mechanism by which ceramide mediates p53 restoration in mutant p53-barrier cancers may lead to discovering effective approaches to improve cancer treatment.
Supplementary Material
Acknowledgments
We thank Dr. Karen Briski and Mr. Amit Gujar (Basic Pharmaceutical Sciences, University of Louisiana at Monroe) for help in confocal microscope. This work was supported by United State Public Health Service/NIH grant P20 RR16456 from the NCRR (Y.Y.L, S.M.J), and Department of Defense Breast Cancer Research Program DAMD17-01-1-0536 (Y.Y.L.).
References
- 1.Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15–6. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 2.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 3.Gudkov AV, Komarova EA. The role of p53 in determining sensitivity to radiotherapy. Nat Rev Cancer. 2003;3:117–29. doi: 10.1038/nrc992. [DOI] [PubMed] [Google Scholar]
- 4.Efeyan A, Garcia-Cao I, Herranz D, Velasco-Miguel S, Serrano M. Tumour biology: Policing of oncogene activity by p53. Nature. 2006;443:159. doi: 10.1038/443159a. [DOI] [PubMed] [Google Scholar]
- 5.Sigal A, Rotter V. Oncogenic mutations of the p53 tumor suppressor: the demons of the guardian of the genome. Cancer Res. 2000;60:6788–93. [PubMed] [Google Scholar]
- 6.Soussi T, Beroud C. Assessing TP53 status in human tumours to evaluate clinical outcome. Nature Reviews. 2001;1:233–40. doi: 10.1038/35106009. [DOI] [PubMed] [Google Scholar]
- 7.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–9. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 8.Olivier M, Eeles R, Hollstein M, Khan MA, Harris CC, Hainaut P. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum Mutat. 2002;19:607–14. doi: 10.1002/humu.10081. [DOI] [PubMed] [Google Scholar]
- 9.Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer. 2009;9:701–13. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- 10.Norberg T, Klaar S, Karf G, Nordgren H, Holmberg L, Bergh J. Increased p53 mutation frequency during tumor progression--results from a breast cancer cohort. Cancer Res. 2001;61:8317–21. [PubMed] [Google Scholar]
- 11.Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006;127:1323–34. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 12.Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–5. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 13.Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–60. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vasey PA, Shulman LN, Campos S, Davis J, Gore M, Johnston S, et al. Phase I trial of intraperitoneal injection of the E1B-55-kd-gene-deleted adenovirus ONYX-015 (dl1520) given on days 1 through 5 every 3 weeks in patients with recurrent/refractory epithelial ovarian cancer. J Clin Oncol. 2002;20:1562–9. doi: 10.1200/JCO.2002.20.6.1562. [DOI] [PubMed] [Google Scholar]
- 15.Lambert JM, Gorzov P, Veprintsev DB, Soderqvist M, Segerback D, Bergman J, et al. PRIMA-1 reactivates mutant p53 by covalent binding to the core domain. Cancer Cell. 2009;15:376–88. doi: 10.1016/j.ccr.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Zeimet AG, Marth C. Why did p53 gene therapy fail in ovarian cancer? Lancet Oncol. 2003;4:415–22. doi: 10.1016/s1470-2045(03)01139-2. [DOI] [PubMed] [Google Scholar]
- 17.Santana P, Pena LA, Haimovitz-Friedman A, Martin S, Green D, McLoughlin M, et al. Acid sphingomyelinase-deficient human lymphoblasts and mice are defective in radiation-induced apoptosis. Cell. 1996;86:189–99. doi: 10.1016/s0092-8674(00)80091-4. [DOI] [PubMed] [Google Scholar]
- 18.Dbaibo GS, Pushkareva MY, Rachid RA, Alter N, Smyth MJ, Obeid LM, et al. p53- dependent ceramide response to genotoxic stress. J Clin Invest. 1998;102:329–39. doi: 10.1172/JCI1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9:139–50. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 20.Patwardhan GA, Liu YY. Sphingolipids and expression regulation of genes in cancer. Prog Lipid Res. 2010 doi: 10.1016/j.plipres.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chmura SJ, Nodzenski E, Beckett MA, Kufe DW, Quintans J, Weichselbaum RR. Loss of ceramide production confers resistance to radiation-induced apoptosis. Cancer Res. 1997;57:1270–5. [PubMed] [Google Scholar]
- 22.Bruno AP, Laurent G, Averbeck D, Demur C, Bonnet J, Bettaieb A, et al. Lack of ceramide generation in TF-1 human myeloid leukemic cells resistant to ionizing radiation. Cell Death Differ. 1998;5:172–82. doi: 10.1038/sj.cdd.4400330. [DOI] [PubMed] [Google Scholar]
- 23.Ichikawa S, Sakiyama H, Suzuki G, Hidari KI, Hirabayashi Y. Expression cloning of a cDNA for human ceramide glucosyltransferase that catalyzes the first glycosylation step of glycosphingolipid synthesis. Proc Natl Acad Sci U S A. 1996;93:4638–43. doi: 10.1073/pnas.93.10.4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu YY, Han TY, Giuliano AE, Cabot MC. Expression of glucosylceramide synthase, converting ceramide to glucosylceramide, confers adriamycin resistance in human breast cancer cells. J Biol Chem. 1999;274:1140–6. doi: 10.1074/jbc.274.2.1140. [DOI] [PubMed] [Google Scholar]
- 25.Liu YY, Han TY, Giuliano AE, Cabot MC. Ceramide glycosylation potentiates cellular multidrug resistance. FASEB J. 2001;15:719–30. doi: 10.1096/fj.00-0223com. [DOI] [PubMed] [Google Scholar]
- 26.Itoh M, Kitano T, Watanabe M, Kondo T, Yabu T, Taguchi Y, et al. Possible role of ceramide as an indicator of chemoresistance: decrease of the ceramide content via activation of glucosylceramide synthase and sphingomyelin synthase in chemoresistant leukemia. Clin Cancer Res. 2003;9:415–23. [PubMed] [Google Scholar]
- 27.Juul N, Szallasi Z, Eklund AC, Li Q, Burrell RA, Gerlinger M, et al. Assessment of an RNA interference screen-derived mitotic and ceramide pathway metagene as a predictor of response to neoadjuvant paclitaxel for primary triple-negative breast cancer: a retrospective analysis of five clinical trials. Lancet Oncol. 2010;11:358–65. doi: 10.1016/S1470-2045(10)70018-8. [DOI] [PubMed] [Google Scholar]
- 28.Furlong SJ, Mader JS, Hoskin DW. Lactoferricin-induced apoptosis in estrogen-nonresponsive MDA-MB-435 breast cancer cells is enhanced by C6 ceramide or tamoxifen. Oncol Rep. 2006;15:1385–90. [PubMed] [Google Scholar]
- 29.Patwardhan GA, Zhang QJ, Yin D, Gupta V, Bao J, Senkal CE, et al. A new mixed-backbone oligonucleotide against glucosylceramide synthase Sensitizes multidrug-resistant tumors to apoptosis. PLoS One. 2009;4:e6938. doi: 10.1371/journal.pone.0006938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mehta K, Devarajan E, Chen J, Multani A, Pathak S. Multidrug-resistant MCF-7 cells: an identity crisis? J Natl Cancer Inst. 2002;94:1652–4. doi: 10.1093/jnci/94.21.1652-b. author reply 4. [DOI] [PubMed] [Google Scholar]
- 31.Fairchild CR, Ivy SP, Kao-Shan CS, Whang-Peng J, Rosen N, Israel MA, et al. Isolation of amplified and overexpressed DNA sequences from adriamycin-resistant human breast cancer cells. Cancer Res. 1987;47:5141–8. [PubMed] [Google Scholar]
- 32.Vikhanskaya F, Clerico L, Valenti M, Stanzione MS, Broggini M, Parodi S, et al. Mechanism of resistance to cisplatin in a human ovarian-carcinoma cell line selected for resistance to doxorubicin: possible role of p53. Int J Cancer. 1997;72:155–9. doi: 10.1002/(sici)1097-0215(19970703)72:1<155::aid-ijc22>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 33.Lorenzi PL, Reinhold WC, Varma S, Hutchinson AA, Pommier Y, Chanock SJ, et al. DNA fingerprinting of the NCI-60 cell line panel. Mol Cancer Ther. 2009;8:713–24. doi: 10.1158/1535-7163.MCT-08-0921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu YY, Gupta V, Patwardhan GA, Bhinge K, Zhao Y, Bao J, et al. Glucosylceramide synthase upregulates MDR1 expression in the regulation of cancer drug resistance through cSrc and beta-catenin signaling. Mol Cancer. 2010;9:145. doi: 10.1186/1476-4598-9-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu YY, Yu JY, Yin D, Patwardhan GA, Gupta V, Hirabayashi Y, et al. A role for ceramide in driving cancer cell resistance to doxorubicin. FASEB J. 2008;22:2541–51. doi: 10.1096/fj.07-092981. [DOI] [PubMed] [Google Scholar]
- 36.Ogretmen B, Safa AR. Expression of the mutated p53 tumor suppressor protein and its molecular and biochemical characterization in multidrug resistant MCF-7/Adr human breast cancer cells. Oncogene. 1997;14:499–506. doi: 10.1038/sj.onc.1200855. [DOI] [PubMed] [Google Scholar]
- 37.De Feudis P, Debernardis D, Beccaglia P, Valenti M, Graniela Sire E, Arzani D, et al. DDP-induced cytotoxicity is not influenced by p53 in nine human ovarian cancer cell lines with different p53 status. Br J Cancer. 1997;76:474–9. doi: 10.1038/bjc.1997.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J, et al. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell. 2003;4:321–8. doi: 10.1016/s1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- 39.Villunger A, Michalak EM, Coultas L, Mullauer F, Bock G, Ausserlechner MJ, et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science. 2003;302:1036–8. doi: 10.1126/science.1090072. [DOI] [PubMed] [Google Scholar]
- 40.Vega FJ, Iniesta P, Caldes T, Sanchez A, Lopez JA, de Juan C, et al. p53 exon 5 mutations as a prognostic indicator of shortened survival in non-small-cell lung cancer. Br J Cancer. 1997;76:44–51. doi: 10.1038/bjc.1997.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Obeid LM, Linardic CM, Karolak LA, Hannun YA. Programmed cell death induced by ceramide. Science. 1993;259:1769–71. doi: 10.1126/science.8456305. [DOI] [PubMed] [Google Scholar]
- 42.Burger H, Nooter K, Boersma AW, van Wingerden KE, Looijenga LH, Jochemsen AG, et al. Distinct p53-independent apoptotic cell death signalling pathways in testicular germ cell tumour cell lines. Int J Cancer. 1999;81:620–8. doi: 10.1002/(sici)1097-0215(19990517)81:4<620::aid-ijc19>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 43.Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–60. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 44.Selivanova G, Iotsova V, Okan I, Fritsche M, Strom M, Groner B, et al. Restoration of the growth suppression function of mutant p53 by a synthetic peptide derived from the p53 C-terminal domain. Nat Med. 1997;3:632–8. doi: 10.1038/nm0697-632. [DOI] [PubMed] [Google Scholar]
- 45.Foster BA, Coffey HA, Morin MJ, Rastinejad F. Pharmacological rescue of mutant p53 conformation and function. Science. 1999;286:2507–10. doi: 10.1126/science.286.5449.2507. [DOI] [PubMed] [Google Scholar]
- 46.Tang X, Zhu Y, Han L, Kim AL, Kopelovich L, Bickers DR, et al. CP-31398 restores mutant p53 tumor suppressor function and inhibits UVB-induced skin carcinogenesis in mice. J Clin Invest. 2007;117:3753–64. doi: 10.1172/JCI32481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hupp TR, Sparks A, Lane DP. Small peptides activate the latent sequence-specific DNA binding function of p53. Cell. 1995;83:237–45. doi: 10.1016/0092-8674(95)90165-5. [DOI] [PubMed] [Google Scholar]
- 48.Chalfant CE, Rathman K, Pinkerman RL, Wood RE, Obeid LM, Ogretmen B, et al. De novo ceramide regulates the alternative splicing of caspase 9 and Bcl-x in A549 lung adenocarcinoma cells. Dependence on protein phosphatase-1. J Biol Chem. 2002;277:12587–95. doi: 10.1074/jbc.M112010200. [DOI] [PubMed] [Google Scholar]
- 49.Hait NC, Allegood J, Maceyka M, Strub GM, Harikumar KB, Singh SK, et al. Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science. 2009;325:1254–7. doi: 10.1126/science.1176709. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.