Abstract
Genome analysis revealed a mosquito orthologue of adenosine kinase in Anopheles gambiae (AgAK; the most important vector for the transmission of Plasmodium falciparum in Africa). P. falciparum are purine auxotrophs and do not express an adenosine kinase but rely on their hosts for purines. AgAK was kinetically characterized and found to have the highest affinity for adenosine (Km 8.1 nM) of any known adenosine kinase. AgAK is specific for adenosine at the nucleoside site but several nucleotide triphosphate phosphoryl donors are tolerated. The AgAK crystal structure with a bound bisubstrate analogue Ap4A (2.0 Å resolution) reveals interactions for adenosine, ATP and the geometry for phosphoryl transfer. The polyphosphate charge is partly neutralized by a bound Mg2+ ion and an ion pair to a catalytic site Arg. The AgAK structure consists of a large catalytic core in a three-layered α/β/α sandwich, and a small cap domain in contact with adenosine. The specificity and tight-binding for adenosine arises from hydrogen bond interactions of Asn14, Leu16, Leu40, Leu133, Leu168, Phe168 and Thr171 and the backbone of Ile39 and Phe168 with the adenine ring as well as through hydrogen bond interactions between Asp18, Gly64 and Asn68 and the ribosyl 2′- and 3′-hydroxyl groups. The structure is more similar to human adenosine kinase (48% identity) than to AK from Toxoplasma gondii (31% identity). With this extraordinary affinity for AgAK, adenosine is efficiently captured and converted to AMP at near the diffusion limit, suggesting an important role of this enzyme to maintain the adenine nucleotide pool. mRNA analysis verifies that AgAK transcripts are produced in the adult insects.
Keywords: adenosine kinase, malaria, Anopheles gambiae, kinetics, structure
Anopheles gambiae is the most important malaria vector in Africa, responsible for the transmission of Plasmodium falciparum. Malaria is a leading cause of illness and death in the tropics, with 300 to 500 million clinical cases and over a million deaths per year (1). P. falciparum is a purine auxotroph, salvaging host cell purines for synthesis of cofactors and nucleic acids (2, 3). Parasite cell growth and division demands robust purine salvage, in particular adenosine, since the parasite contains the most (A+T)-rich genome sequenced to date (approximately 80%). Adenosine metabolism has been well characterized during P. falciparum growth in the erythrocytic stages (4) but not in the mosquito vector. Human AK uses adenosine efficiently (Km = 41 nM) but is inhibited when the concentration is high, thus preventing excess accumulation of intracellular purine nucleotide levels (5). Adenosine kinase (EC 2.7.1.20) catalyzes the phosphorylation of adenosine using MgATP2− as the phosphoryl donor to release AMP and adenosine 5′-diphosphate (ADP). Most studies on this enzyme have been directed towards inhibitor design, especially in parasitic protozoa, bacteria and humans (6–11). Adenosine kinases have been purified from human, yeast and protozoa sources and extensively characterized at kinetic and structural levels. Sequence analysis has shown that human adenosine kinase has similarities to microbial ribokinases and fructokinases from the phosphofructokinase B type (PFK-B) family (5, 12). Despite the common feature of ATP-binding sites, these kinases have different functions and share a low overall sequence identity (around 20%), preventing precise determination of the structural and functional characteristics of new members.
Purine metabolism in Anopheles spp. remains poorly explored. Characterization and understanding of mosquito purine metabolism might identify new targets for specific insecticides to control this malaria vector and to facilitate study of purine metabolism in the mosquito and in the malaria parasite in the sexual stages. Purine metabolism in A. gambiae mosquitoes can be predicted through the genome sequence (13), allowing in silico comparison of parasite, vector and human metabolism. Anopheline genome analysis revealed a mosquito orthologue of adenosine kinase (AgAK). The anopheline AK was expressed, kinetically characterized and found to exhibit the lowest Km known for any adenosine kinase, and found to be genetically expressed in the insect by direct mRNA analysis. Crystallographic analysis revealed a monomeric structure similar to Homo sapiens adenosine kinase (HsAK), reflecting the 48% amino acid sequence identity.
EXPERIMENTAL PROCEDURES
In silico A. gambiae Adenosine Kinase identification
The protein sequence of Homo sapiens adenosine kinase (HsAK) was used as a query in a tBLASTn search of the A. gambiae genome sequence (available in 2003), through the NCBI web site (default settings). The A. gambie genome was analyzed for the presence of introns, and translations of the exon regions were used for further analyses.
RNA isolation, cDNA Synthesis, and PCR Analysis
Genomic DNA from A. gambiae G3 was obtained from MR4. Total RNA was prepared directly from 7–10 frozen adults of A. gambiae G3 generation F5 (MRA-132B, MR4) in 2 mL of Trizol (Invitrogen), and RNA was extracted according to the manufacturer’s instructions. Subsequent cDNA synthesis was performed with 1 μg of total RNA. The extracted RNA was treated with DNaseI (RNase-free, Invitrogen) before cDNA synthesis, according to the manufacturer’s instructions. First strand cDNA was synthesized by use of Superscript III reverse transcriptase (Invitrogen) and a gene-specific primer mix with 2 pmol of each antisense oligonucleotide or (dT)20 as described by the manufacturer. Gene-specific oligonucleotide primers were used for the AgAK gene and for AgPNP (used as a positive control; Table 1, (14)). PCR reactions were performed with Platinum PCR super master mix High Fidelity (Invitrogen) as described by the manufacturer. PCR products from cDNA and gDNA of each gene were sequenced after gel purification.
Table 1.
Gene-specific oligonucleotides used for the analysis of AgAK and its transcript
| Template | Primers | Expected size (bp) with cDNA | Expected size (bp) with gDNA |
|---|---|---|---|
| AgPNP (control) | F: 5′-CACCATGAGCAAATTTAGCTACCTTCAAA -3′ R: 5′-CTACTTCTTCGCCTCGTAGTGG -3′ |
1062 | 2278 |
| AgAK | F: 5′-GCGCTTCCTTTTTATATAATC -3′ R: 5′-TTAATTGTCAGCACAGAAGGACG -3′ |
1074 | 1547 |
bp, base pairs
Anopheles gambiae adenosine kinase gene construction, protein expression and purification
A chemically synthesized gene encoding the adenosine kinase from A. gambiae was synthesized and cloned into a pDONR221 vector (DNA 2.0). The gene sequence was synthesized using optimized codons for Escherichia coli protein expression with an N-terminal thrombin-cleavable hexa-histidine tag. The synthetic gene was transferred to the pDEST-14 vector (Invitrogen) with LR-Clonase II (Invitrogen). The pDEST14-AgAK construct was transformed into TOP10 competent cells (Invitrogen) and cultured on LB-carbenicillin agar plates. The plasmid was isolated using a Miniprep Kit (Qiagen) and sequenced using a T7 promotor primer and a T7 terminator primer. The pDEST14-AgAK construct was then introduced into E. coli BL21-AI cells (Invitrogen) for expression. The cells were grown in LB-carbenicillin at 37 °C to an OD600 of 0.6, induced with L-arabinose at 0.2% (w/v) final concentration, and grown at 20 °C overnight. Cells were harvested by centrifugation (4000 × g for 30 min) and were ruptured by passage through a French press. Cell debris was pelleted by centrifugation (16000 × g for 30 min) and the supernatant was purified over a 3 mL Ni-NTA affinity column (Qiagen) with elution by a step gradient of 25, 50, 100, 200, 250, 300 and 500 mM imidazole in 50 mM HEPES (pH 8), 300 mM NaCl and 1 mM DL-dithiothreitol (DTT). The purified recombinant protein was dialyzed overnight against 50 mM Tris-base (pH 7.5) and 1 mM DTT. After dialysis, glycerol was added at 10% final concentration and 50 μL aliquots were frozen in liquid nitrogen and stored at −80 °C. Under these conditions, AgAK retained full activity for several months. Protein concentration was determined by UV absorbance by use of the extinction coefficient 27765 M−1 cm−1 at 280 nm. All enzymatic parameters and crystal structures were obtained with the recombinant AgAK containing the N-terminal hexa-histidine tag.
Adenosine kinase enzymatic assays and inhibition studies
Adenosine kinase activity was measured using a modified radiometric assay (15). Reactions were conducted in 1 mL at 27 °C in 50 mM Tris buffer (pH 7.4) containing 5 mM MgCl2, 50 mM KCl, 0.2 mM DTT and varying concentrations of ATP and [2-3H]adenosine (23 Ci/mmol, Amersham Biosciences) as appropriate. Reactions were initiated by the addition of enzyme, and terminated by spotting 40 μL onto DE81 filters (Whatman). Aliquots were taken at 1 min intervals over the course of a 5 min assay. Dried filters were washed three times with 5 mL water, once with 5 mL 95% ethanol, and dried at room temperature. Filters were subjected to liquid scintillation counting in 10 mL of Betaplate Scint (PerkinElmer). An enzyme-free control for each concentration was used to subtract background. The apparent Km value for adenosine was determined at 1 mM ATP and 5 mM MgCl2 using [2-3H]adenosine concentrations ranging from 1 to 75 nM and 0.1 nM AgAK. The apparent Km value for ATP was obtained at 0.5 μM [2-3H]adenosine with ATP concentrations ranging from 0.1 μM to 50 μM, 5 mM MgCl2 and 5 nM AgAK. Kinetic parameters were calculated from Michaelis-Menten analysis of initial rate data.
Enzyme inhibition assays to determine Ki values were performed using different concentrations of iodotubercidin (ITU, 7-Deaza-7-iodoadenosine, Berry and Associates), P1,P3-di(adenosine-5′) triphosphate ammonium salt (Ap3A, Sigma) or P1,P4-di(adenosine-5′) tetraphosphate ammonium salt (Ap4A, Sigma), 50 nM [2-3H]adenosine, 1 mM ATP and 5 mM MgCl2. The inhibition constants were determined as described previously (16) with inhibitor concentrations ranging from 1 nM to 100 nM. Reactions using different ITU concentrations were filtered through a YM10 Centricon spin column (MW retention = 10000; Amicon) and analyzed by High Performance Liquid Chromatography (HPLC) to test for substrate activity with AgAK.
Analysis of enzymatic reactions by high-performance liquid chromatography (HPLC)
Samples were analyzed by reverse-phase (Luna C18(2), 150 × 4.6 mm, 5 μm, Phenomenex) ion pair HPLC system. The mobile phases were 8 mM tetrabutylammonium bisulfate (Fluka) and 100 mM KH2PO4 with the pH adjusted to 6.0 with KOH (solution A) and 30% acetonitrile containing 8 mM tetrabutylammonium bisulfate and 100 mM KH2PO4 (pH 6) as solution B. The HPLC gradient was from 0% to 100% solution B in 20 min. The eluant was monitored at 254 nm and the flow rate was 1 mL/min.
Adenosine kinase substrate specificity assays
AgAK nucleoside specificity was measured using the method described above. Reactions were conducted in 100 μL at 27 °C in 50 mM Tris buffer (pH 7.4) containing 5 mM MgCl2, 50 mM KCl, 0.2 mM DTT, 1 mM ATP and 0.5 μM [2-3H]adenosine, [8-3H]guanosine (15 Ci/mmol, American Radiolabeled Chemicals Inc.), [5-3H]cytidine (25 Ci/mmol, Sigma), [5-3H]uridine (27.2 Ci/mmol, Perkin Elmer), [8-3H]inosine (27 Ci/mmol, American Radiolabeled Chemicals Inc.), [8-14C]2′-deoxyadenosine (48.8 mCi/mmol, Sigma), [8-14C]2′-deoxyguanosine (55 mCi/mmol, Sigma), [5-3H]2′-deoxycytidine (20 Ci/mmol, American Radiolabeled Chemicals Inc.), [5-3H]thymidine (12.9 Ci/mmol, Amersham Biosciences) or [1-3H]ribose (15 Ci/mmol, Sigma). For inhibitor or subversive substrate screening, reactions were performed with 0.5 μM [2-3H]adenosine, 1 mM ATP and 75 μM 9-β-D-arabinofuranosyladenine (Ara-A, Sigma) or ITU. In all cases, reactions were initiated by the addition of enzyme and terminated after 10 min incubation by spotting 40 μL onto DE81 filters (Whatman). A blank (no enzyme) for each radiolabeled substrate was used to subtract unmetabolized radiolabeled substrate bound to DE81 filters. The specific activity is expressed relative to activity with adenosine.
Adenosine kinase activity as a function of GTP, UTP, CTP and TTP concentrations was measured using a luminescence assay (17). In this assay, AMP generated from adenosine and NTPs by reaction with AgAK is converted to ATP by pyruvate orthophosphate dikinase (PPDK). ATP and luciferin are converted to light and AMP by firefly luciferase. The AMP formed in the luciferase reaction is cycled via PPDK to give a sustained luminescence signal. Km values for adenosine were determined with fixed concentrations of 100 μM of the NTPs, adenosine concentrations from 0.2 to 2 μM, and 4 nM AgAK. In a similar way, the Km values for the NTPs were measured with a fixed concentration of 10 μM adenosine with NTP concentrations varied from 30 to 200 μM and 4 nM AgAK. Kinetic parameters (kcat, Km and catalytic efficiencies) were determined from fits to the Michaelis-Menten equation.
Crystallization, Data Collection, and Structural Determination
AgAK was concentrated to 11.7 mg/mL and incubated on ice with 1 mM Ap4A and 5 mM MgCl2 in 50 mM HEPES buffer (pH 7.5), containing 500 mM NaCl. The enzyme was crystallized in 25% PEG3350, 0.2 M MgCl2, 5% 2-propanol, 25% glycerol and 0.1 M BisTris buffer pH 5.5 using the sitting drop vapor diffusion method at 18 °C. Crystals were freshly frozen in liquid N2 before data collection.
X-ray diffraction data of Ap4A bound AgAK was collected at the beamline X29A of Brookhaven National Laboratory on an ADSC Q315 detector at 100 K. Data was processed with the HKL2000 program suite and data processing statistics are provided in Table 3 (18).
Table 3.
Data collection and refinement statistics
| PDB codes | Ap4A bound AgAK 3LOO |
|---|---|
| Data collection | |
| Space group | P21 |
| Cell dimension | |
| a, b, c (Å) | 50.4, 76.8, 140.5 |
| α, β, γ (°) | 90.0, 92.1, 90.0 |
| Resolutions (Å) | 20.00–2.00 (2.07–2.00) |
| Rsym (%) | 6.8 (54.7) |
| I/σI | 19.5 (2.1) |
| Completeness (%) | 100.0 (99.8) |
| Redundancy | 3.7 (3.6) |
| Refinement | |
| Resolution (Å) | 20.00–2.0 |
| No. reflections | 72380 |
| Rwork/Rfree (%) | 19.8/24.7 |
| B-factors (Å2) | |
| Protein | |
| (main chain) | 38.8 |
| (side chain) | 41.0 |
| Water | 39.9 |
| Ligand | 37.4 |
| No. of Atoms | |
| Protein | 7753 |
| Water | 242 |
| Ligand | 165 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.014 |
| Bond angles (°) | 1.50 |
| Ramanchran analysis | |
| allowed region | 99.3% |
| disallowed region | 0.7% |
| Coordinate Error by Luzzati plot (Å) | 0.25 |
Numbers in parentheses are for the highest-resolution shell. One crystal was used for each data set.
The crystal structure of AgAK was determined by molecular replacement in Molrep using the published structure of human adenosine kinase (PDB ID: 1BX4) as the search model (19). The initial atomic model was rebuilt by PHENIX (20). The model without ligand was first rebuilt in COOT and refined in Refmac5 (21, 22). The ligands were later added in COOT using the Fo − Fc map when the Rfree is below 30% and refined in Refmac5. A chloride ion near the adenine binding site was added based on the crystallization condition and the presence of a chloride ion at similar position in the human AK structure (PDB ID: 1BX4). Two of three catalytic Arg131 side chain residues where the electron density is weak were built by pointing the side chain away from bound Ap4A. The final model was validated by Procheck (23). Refinement statistics are summarized in Table 3. The coordinates and structure factors for Ap4A bound AgAK were deposited in the Protein Data Bank under accession code 3LOO.
RESULTS
Identification and expression of recombinant A. gambiae adenosine kinase
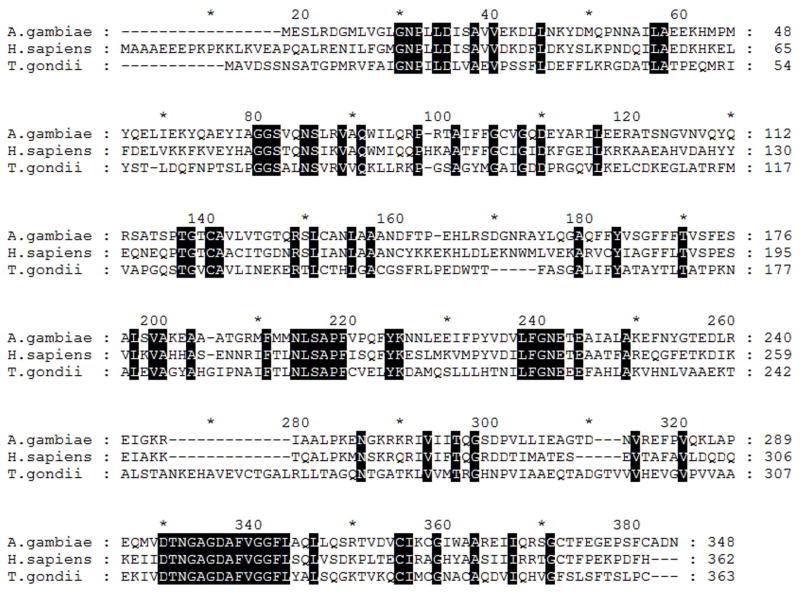
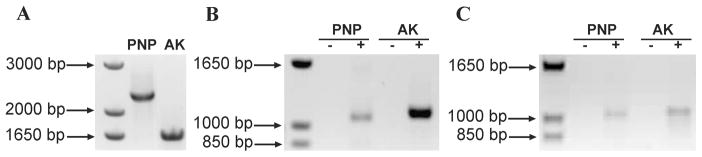
The presence of adenosine kinase in the A. gambiae genome was suggested previously but not established (14). The A. gambiae AK was located from similarity to the amino acid sequence of the Homo sapiens AK (Gen Bank accession number: CAI39671.1) by BLAST analysis. A genomic region whose proposed translation showed high level of similarity to the H. sapiens enzyme was retrieved (Figure 1). Conceptual translation of this region resulted in a 348-amino acid protein, representing a putative A. gambiae AK. The presence of this gene and its transcript in A. gambiae were confirmed by PCR amplification of A. gambiae gDNA and cDNA (Figure 2). RT-PCR analysis of the mosquito total RNA and mRNA revealed that AgAK is transcribed in adult mosquitoes.
Figure 1.
Multiple alignment of the predicted amino acid sequence of A. gambiae with the known adenosine kinases from human (Homo sapiens) and Toxoplasma gondii. The amino acid sequence of the HsAK was used to search the A. gambiae genome by BLAST analysis. The AgAK sequence is 48% identical and 19% similar to the HsAK while only shares 31% identity and 20% similarity to the TgAK.
Figure 2.
The presence of adenosine kinase gene and its transcript in A. gambiae were confirmed by PCR of A. gambiae gDNA and cDNA in adult mosquitoes. A: gDNA analysis revealed the expected size for the adenosine kinase predicted gene. B: PCR performed using cDNA generated by RT-PCR using gene-specific oligonucleotide primers (total RNA). C: PCR performed using cDNA generated by RT-PCR using oligo(dT)20 primers (mRNA).
AgAK kinetic parameters, substrate specificity and inhibition
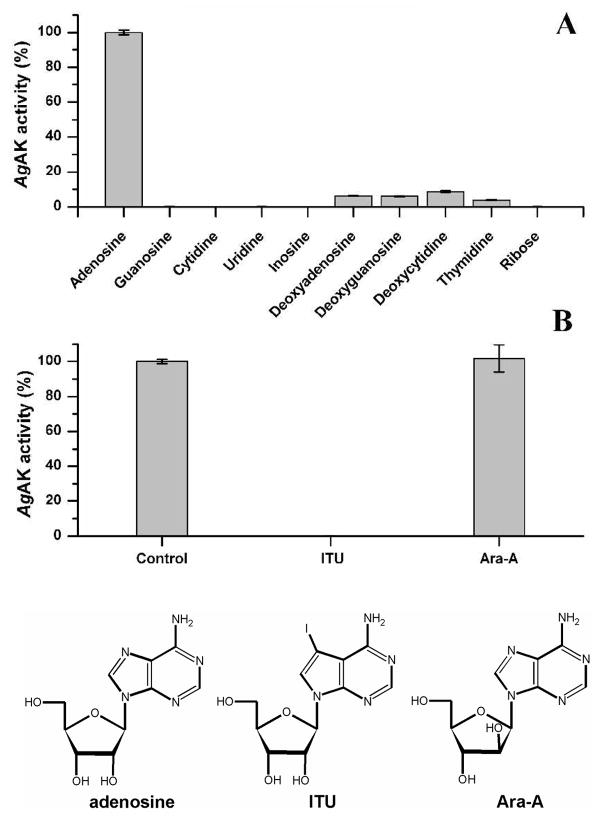
AgAK activity was measured by the production of [3H]AMP from [3H]adenosine. Steady-state kinetics for AgAK indicated a Km value of 8.1 ± 0.6 nM for adenosine and a Km of 1.4 ± 0.4 μM for ATP (Table 2). The turnover number of the enzyme was calculated to be 0.16 s−1 at saturating ATP concentration and 0.3 s−1 at saturating adenosine concentration. The kcat/Km values with respect to ATP and adenosine are 2.4 × 105 and 1.97 × 107 M−1 sec−1 and are near the diffusion limit for enzymatic catalysis. With these Km values, the enzyme in vivo is saturated with respect to ATP and is designed to capture adenosine and phosphorylate with high efficiency. The low kcat is a kinetic requirement because of the unusually tight binding by adenosine. Similar to other adenosine kinases, the AgAK enzyme is inhibited at high concentrations of adenosine, explaining the increased kcat value when higher concentration of adenosine are used. To determine the Km for ATP, a higher concentration of enzyme was used. The AgAK enzyme exhibits narrow nucleoside specificity with weak deoxyribonucleoside kinase activity (Figure 3A). The 9-β-D-arabinofuranosyladenine (Ara-A) analogue of adenosine at concentrations in 150-fold excess over adenosine did not affect the AgAK activity (Figure 3B) while iodoturbecidin (ITU) inhibited with an apparent Ki of 1 nM (Table 2). AgAK recognition of ITU as substrate was also assayed by HPLC analysis since the radiochemical assay does not distinguish conventional inhibition from competition as substrate. No phosphorylation was detected indicating that ITU acts as inhibitor of the enzymatic activity and not as a subversive substrate. Inhibition assays to determine the Ki values for ITU, Ap3A and Ap4A used 50 nM [2-3H]adenosine and 1 mM ATP (Table 2). Ap4A and Ap3A are well-known bisubstrate analogues of adenosine kinases and surprisingly, these bind with relative weak affinity of 0.86 and 61 μM, respectively (Table 2).
Table 2.
Kinetic parameters and inhibition constants of AgAK
| Km (nM) | kcat (s−1) | kcat/Km (M−1 s−1) | Ki (nM) | |
|---|---|---|---|---|
| Adenosinea | ||||
| ATP | 8.1 ± 0.6 | 0.160 ± 0.003 | 1.97 ± 0.04 × 107 | - |
| GTP | 230 ± 10 | 0.63 ± 0.01 | 2.7 ± 0.1 × 106 | - |
| UTP | 740 ± 50 | 0.035 ± 0.002 | 4.7 ± 0.4 × 106 | - |
| CTP | 630 ± 50 | 0.029 ± 0.001 | 4.6 ± 0.4 × 106 | - |
| TTP | 790 ± 80 | 0.056 ± 0.004 | 7.1 ± 0.9 × 106 | - |
| NTPb | Km (μM) | |||
| ATP | 1.4 ± 0.4 | 0.33 ± 0.02 | 2.4 ± 0.5 × 105 | - |
| GTP | 54 ± 5 | 1.39 ± 0.07 | 2.6 ± 0.3 × 104 | - |
| UTP | 56 ± 2 | 0.061 ± 0.001 | 1.09 ± 0.04 × 103 | - |
| CTP | 49 ± 3 | 0.062 ± 0.002 | 1.27 ± 0.09 × 103 | - |
| TTP | 245 ± 74 | 0.30 ± 0.07 | 1.2 ± 0.5 × 103 | - |
| ITU | - | - | - | 1.0 ± 0.2 |
| Ap4A | - | - | - | 860 ± 130 |
| Ap3A | - | - | - | 61,000 ± 13,000 |
Km values for adenosine were determined at fixed concentrations of 100 μM of each NTP with other conditions as described in the methods.
Km values for the NTPs were determined at fixed concentrations of 10 μM adenosine with other conditions as described in the methods.
Figure 3.
Substrate specificity for AgAK. A: The nucleoside specificity activities are relative to the activity with adenosine (shown to give a kcat of 0.16 s−1 and a kcat/Km of 2.0 × 107) that was set to 100% for each fraction. AgAK showed partial deoxyribonucleoside kinase activity. B: Adenosine analog Ara-A and ITU tested as potential inhibitor/subversive substrate of AgAK at concentrations in 150-fold excess over adenosine. Activity was relative to the activity of control were only adenosine and ATP were present in the reaction.
AgAK is highly specific for adenosine as the nucleoside substrate but the substrate specificity for the phosphoryl donor is broader. The enzyme utilizes the γ-phosphoryl from ATP, GTP, UTP, CTP and TTP (Table 2) and with all phosphoryl donors AgAK is relatively efficient in converting adenosine to AMP (kcat/Km values with respect to adenosine are 2.7 × 106 to 1.97 × 107 M−1 sec−1). The kcat/Km values with respect to NTPs are less, 1.09 × 103 to 2.4 × 105 M−1 sec−1 because of weaker binding of the phosphoryl donors. Purine triphosphates ATP and GTP, have better efficiencies (2.4 × 105 and 2.6 × 104 M−1 sec−1) while pyrimidine triphosphates UTP, CTP and TTP have lower efficiencies (1.09 × 103 to 1.27 × 103 M−1 sec−1).
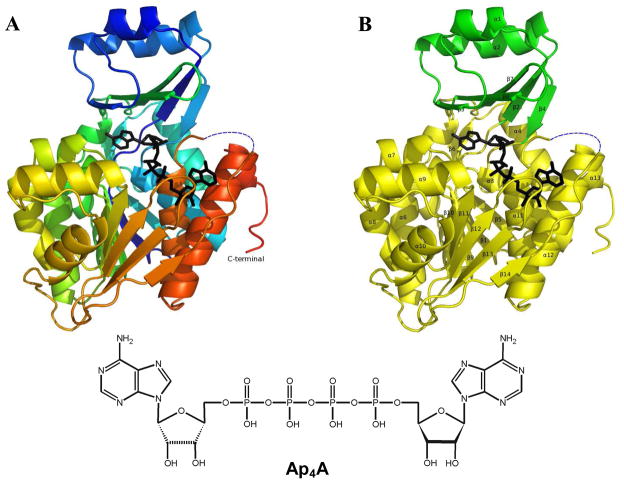
Overall structure and inhibitor binding
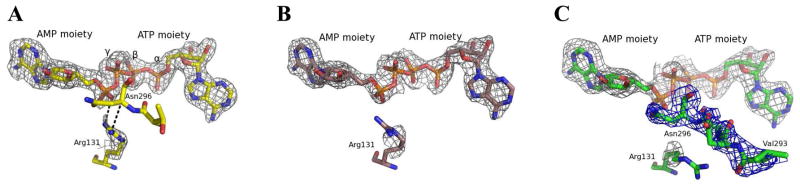
The crystal structure of AgAK was determined to 2.0 Å resolution. The enzyme is a monomer with three AgAK molecules in the asymmetric unit. All contained Ap4A and Mg2+ in the active sites. AgAK consists of two domains (Figure 4). The small domain located at the N-terminus is responsible for adenosine interactions and consists of the secondary structure elements helix α1, α2 and beta strands β2, β3, β4, β7 and β8. The large ATP-binding domain has a typical α/β structure as well as conserved kinase anion hole motifs where the phosphates of ATP bind (24). The overall structure of the three AgAK molecules in the asymmetric unit is almost identical (the root-mean-square deviations of Cα backbone from residues 10 to 333 are 0.2–0.3 Å). However, subtle differences are seen in the region near the catalytic site Arg131. In two AgAK molecules the electron density of the side chain of Arg131 is well defined while the region from residue 288 to 295 is disordered (Figure 5A and 5B). In the third AgAK molecule, the electron density of the side chain of Arg131, is weak but the residues in the region 288 to 295 are ordered with well-defined electron density (Figure 5C). We interpret these differences to represent catalytic site motions linked to formation of a catalytically competent complex (see discussion).
Figure 4.
The Ap4A bound AgAK structure depicted as a ribbon diagram. A: The Ap4A bound AgAK is ramped from blue to red (N terminus to C terminus) and Ap4A is depicted as sticks in black. The disordered region (residue 289 to 294) was indicated as a blue dash line. B: The small domain (residue 17 to 62 and residue 121 to 136) of AgAK and the large domain (the rest residues) of AgAK are drawn in green and yellow, respectively. The disordered region (residue 289 to 294) was indicated as a blue dash line.
Figure 5.
Ap4A interactions with the side chain of Arg131 and corresponding Fo−Fc omit electron density (gray) contoured at 2.5σ. Strong electron density is observed for all three Ap4A molecules bound to AgAK. A: First AgAK molecule where electron density is clearly shown at the side chain of Arg131. The side chain of Arg131 hydrogen bonds to the phosphate from AMP moiety and is in proximity to the backbone of Asn296. Distances shorter than 3.2 Å are indicated as black dash lines. B: Second AgAK molecule, the electron density of the guanidine group of Arg131 is missing. C: Third AgAK molecule, the electron density of the side chain of Arg131 is almost absent but strong density of the backbone of the residues 288 to 295 is observed. The 2Fo−Fc electron density map (at 1σ) from Val293 to Asn296 is colored in blue.
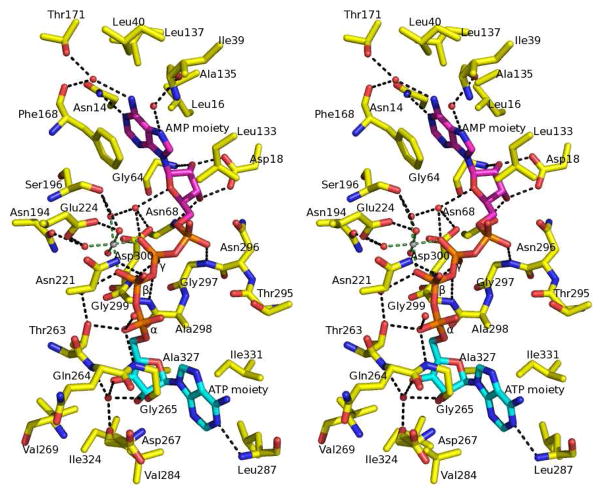
AgAK accommodates the tri-polyphosphate from ATP in the catalytic site prior to phosphoryl transfer. However, the Ap4A bound to AgAK has a tetraphosphate and is a better mimic ATP and AMP than the normal reactants (ATP + adenosine or ADP + AMP). Polyphosphate chains are flexible and instead of pushing aside the adenosine moieties, the four phosphoryl groups fit tightly into the tri-polyphosphate binding site in a geometry to simulate phosphoryl transfer. The oxygen equivalent to the 5′-hydroxyl of adenosine (the nucleophilic oxygen) is 2.9 Å from the γ-phosphorus of the ATP mimic. By comparison, the nucleophilic adenosine oxygen is located 4 Å from the β-phosphorus of the ATP moiety (Figure 6). A magnesium ion is positioned between the β- and γ-phosphoryl groups to form an octahedral (hexa-coordinate) complex with two oxygen atoms from the β- and γ-phosphoryl of the ATP moiety and four coordinated water molecules (Figure 7). Three of the coordinated water molecules form hydrogen bonds with neighboring residues, including side chains of Asn194, Ser196, Glu224 and Asp300. The γ-phosphoryl of the ATP moiety accepts hydrogen bonds from the amides of Gly299 and Asp300. The α-phosphoryl of the ATP moiety hydrogen bonds the side chains of Thr263 and the amide of Gly265. The phosphoryl from AMP makes a hydrogen bond to the amide of Gly297 (Figure 7). The adenine of ATP is partially buried in a hydrophobic pocket formed by Val269, Leu287, Ile324, Ala327, Ile331. The N1 of ATP forms a hydrogen bond to the amide of Leu287, and the O3′ interacts with a hydrogen bond to the amide of Gln264 via a water molecule. The adenosine from the AMP moiety forms an anticlinal conformation and the ribose of AMP has an O4′-endo sugar pucker conformation. The adenine of the AMP moiety is surrounded by hydrophobic residues, including Leu16, Leu40, Leu133, Ala135, Leu137 and Phe168. In the AgAK molecule where residues 288 to 294 are ordered, Met292 is near the adenine. The N1 of AMP accepts a hydrogen bond from the side chain of Asn14 and a water molecule mediates the hydrogen bonding between N9 of AMP and the amide of Ile39. Water molecules also mediate the hydrogen bonding between the N6 amino of AMP, the side chain of Thr171 and the carbonyl of Phe168. The O2′ and O3′ of AMP both make hydrogen bonds to the side chain of Asp18. In addition, the O2′ and O3′ from AMP moiety make hydrogen bonds to the amide of Gly64 and the side chain of Asn68, respectively. A chloride molecule, which is also present in the crystal structures of the liganded human and T. gondii AKs, is found 3.5 Å from C2 of the AMP moiety. The function of this chloride is not apparent from the structure and may be charge neutralization. The root mean square deviation (r.m.s.d.) of Cα backbone in reference to liganded HsAK and TgAK is between 1 to 1.5 Å.
Figure 6.
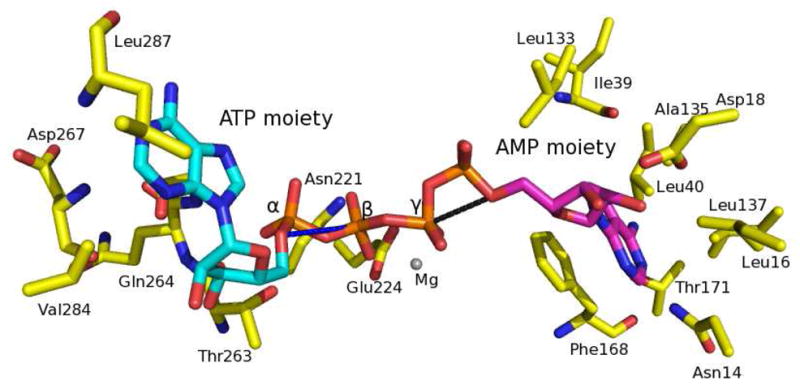
Conformation of Ap4A in the actives sites. The adenine from ATP moiety and the adenine from AMP moiety are colored in cyan and purple, respectively. The α-, β- and γ-phosphate molecules from ATP moiety are labeled. The distance of 2.9 Å between 5′-hydroxyl of AMP and the γ-phosphate of ATP is indicated as a black line. The distance of 4.0 Å between 5′-hydroxyl of ATP and the β-phosphate of ATP is indicated as a blue line. Only part of the active site residues are depicted as sticks.
Figure 7.
The stereo view of Ap4A bound to AgAK and its surrounding residues. The adenine from ATP moiety and the adenine from AMP moiety are colored in cyan and purple, respectively. The magnesium and water molecules are depicted as gray and red dots, respectively. The α-, β- and γ-phosphate molecules from ATP moiety are labeled. The magnesium-oxygen and hydrogen bonds are indicated as green and black dash lines, respectively.
DISCUSSION
Identification and characterization of adenosine kinase from A. gambiae
Mosquitoes grown in semisynthetic media revealed dietary requirements for AMP (or IMP), TMP, and UMP (or CMP) or the corresponding nucleosides (25). Requirement for these metabolites suggested that purine and pyrimidine salvage pathways may be important in Anopheles nucleotide metabolism. Adenosine kinase has a key role in adenosine salvage to maintain the ATP pool, particularly in cells with no de novo purine synthesis, including human erythrocytes. Ribonucleoside and deoxyribonucleoside kinases serve to trap nucleosides within cells by phosphorylation and may be involved both in the maintenance of the ATP pool and in buildup of other nucleic acid precursors. Here, the full-length ORF of AgAK (Figure 1) was over-expressed as a fusion protein with a His-tag. The presence of an AK gene and its transcript in A. gambiae adult mosquitoes was confirmed by PCR of A. gambiae genomic DNA and its cDNA (Figure 2). The kinetic parameters for the recombinant protein revealed the lowest Km reported for any adenosine kinase (Table 2). The low Km, high kcat/Km and the substrate inhibition observed for the recombinant AgAK suggest that this enzyme provides an efficient salvage mechanism in the adenosine metabolism of A. gambiae. In humans, AK is the most important enzyme for maintenance of the ATP pool in erythrocytes. Although its kinetic properties suggest an important role in adenosine salvage, further metabolic studies are needed to reveal the biological function of this enzyme in mosquitoes.
AgAK has narrow nucleoside specificity supporting a specific role in adenosine salvage (Figure 3A). Previous work suggested three proteins with deoxyribonucleoside kinase activity in cell extracts from A. gambiae cell-culture on DEAE-ion exchange chromatography. The major peak was a multisubstrate deoxyribonucleoside kinase (AgdNK) and the other peaks had adenosine kinase activity with partial deoxyribonucleoside kinase activity (26). One of these showed substrate specificity consistent with the AgAK described here, supporting expression of this activity in vivo (26). The nucleoside specificity of adenosine kinases characterized from other organisms is usually broader than their nucleotide triphosphate specificity (7, 10, 15, 27, 28). For example the AK from T. gondii uses ITU and other 6-substituted analogues as nucleoside substrates (8, 24) whereas AgAK cannot. The AgAK enzyme is unusual in its high specificity for adenosine with lower activity on deoxyribonucleosides (Figure 3A and B). The enzyme has a broad NTP specificity using ATP, GTP, UTP, TTP and CTP as phosphoryl donors (Table 2).
AgAK inhibition by ITU showed a Ki value of 1 nM, a surprisingly powerful inhibitor considering the narrow nucleoside specificity of this enzyme (Table 2). However, the unusually tight interaction with adenosine (Km = 8.1 nM) gives a modest Km/Ki of 8.1. Ap4A and Ap3A are well-known bisubstrate analogues of adenosine kinases and surprisingly, these bind with relative weak affinity of 0.86 and 61 μM, respectively (Table 2).
AgAK crystal structure
The overall topology of AgAK showed high similarity to the human (HsAK) and T. gondii AK (TgAK) structures. Like the human and T. gondii AK structures, the AgAK structure contains a mixed fold and consists of small and large domains (Figure 4). The fold belongs to the ribokinase-like family with a small and large Rossmann fold domain. The large domain, termed the catalytic core, is a three-layered α/β/α sandwich, and the small domain, termed the cap, is in contact with the substrate, adenosine. In TgAK, the cap domain rotates 30° upon adenosine or ITU binding, inducing a fully closed conformation (24), but this rotation is not observed when ATP analogues are bound (29). Furthermore, a semi-closed or semi-opened conformation where the small lid region undergoes a rigid-body rotation of about 12° was observed in TgAK bound to adenosine analogues N6,N6-dimethyladenosine (DMA) and 6-methylmercaptopurine riboside (MMPR) (30). Despite our efforts, the AgAK failed to crystallize in the apo form or with ITU. And the co-crystallization with Ap4A succeeded only after an extensive screening to give a crystal structure similar to the closed conformation.
The catalytic site of AgAK normally accommodates three phosphoryl groups together with the 5′-hydroxyl group as the incipient nucleophile. However, the Ap4A bound to AgAK consists of four phosphoryl groups that fit tightly into the phosphate binding site keeping the adenosine and ATP binding groups in place. Interestingly, Ap4A (Ki = 860 nM) showed tighter binding than Ap3A (Ki = 61,000 nM) (Table 2). Ap3A is one atom short of the productive catalytic site complex and a similar binding pattern has been found for Ap5A with adenylate kinase, where the extra phosphoryl group is required to allow alignment of groups in the catalytic site to mimic the geometry similar to a trigonal bipyramid needed for terminal phosphoryl transfer (31).
The AgAK active site residues that involve hydrogen bonding and hydrophobic interaction to adenosine are either conserved or replaced with similar amino acids when compared to HsAK and TgAK (Figure 1). From the structural point of view, there is no clear evidence to illustrate why AgAK can capture adenosine more efficiently than HsAK and TgAK. However, the relatively high Km value of TgAK for adenosine may due to the replacement of Phe168 to Tyr that involves hydrophobic stacking with adenosine. Substrate on-off rates from catalytic sites dictate the affinity, thus dynamic components not readily apparent from the crystal structure are likely to be involved. The sequence identity of HsAK and TgAK relative to AgAK are 48% and 31%, respectively. Thus, catalytic site efficiency of AgAK may also be contributed by residues remote from the adenosine binding site. As an example, the 30° rotation of the cap domain required for adenosine binding in TgAK (24) may differ in the AgAK since amino acid residues in the hinge region differ and may change the dynamics of the rotation motion and therefore the on-off frequency and the binding affinity. The structural comparison showed Tyr60, Tyr94 and Asn136 in AgAK are replaced by Ser, Arg and His, respectively, in the TgAK structure. Furthermore, the Tyr49 in AgAK, that forms a hydrogen bond to carbonyl Cys134, is replaced by a Phe in HsAK.
Tight binding of ITU
Factors contributing to the binding (1.6 μM) of ITU to TgAK include the displacement of an ordered water molecule near N7 of adenosine being replaced by the relatively large and hydrophobic iodine atom (24). As this region in TgAK is hydrophobic, surrounded by Ala44, Leu46 (adenosine domain), Cys127, Leu138, Thr140, Tyr169 and Phe201 (ATP domain), more favorable interactions are made. These residues are completely conserved in AgAK except for the conservative Thr140Ala and Tyr169Phe replacements found in AgAK. Although AgAK did not crystallize with ITU, the sequence similarity and high affinity of ITU (1 nM) supports an even more favorable interaction with the catalytic site residues in AgAK.
The catalytic arginine
It has been proposed that the closure of the small domain of AK brings the conserved catalytic arginine to the active site when adenine is bound (24). The catalytic arginine forms a hydrogen bond to the γ-phosphate of ATP and orientates the γ-phosphate into the catalytic position for the SN2 reaction (24). In the crystal structure of Ap4A bound to AgAK, the small domains of all three AgAK molecules in the asymmetric unit are in the closed conformation. However, the side chains of the catalytic Arg131 either interact with the phosphate from the AMP moiety or move freely (Figure 5). The side chain of the catalytic Arg131 is closer to the phosphate from AMP than to the γ-phosphate of ATP. The arginine is near the backbone of Asn296 when the catalytic Arg131 interacts with the phosphate from AMP moiety (Figure 5A). Thus, residues near Asn296 are disordered when the side chain of Arg131 forms a hydrogen bond to Ap4A. This motion may be linked to the phosphoryl transfer in the normal reaction.
Conclusions
Properties of adenosine kinase from the mosquito suggest an important role in adenylate nucleotide pool regulation. The ability to salvage adenosine by a kinase separates the insect host from the parasite and provides a rationale for metabolic and inhibitor design studies to investigate targets in host-parasite interactions. Since malaria infections involve only a few parasites transmitted from human host to human recipient, the insect vector represents a biological bottleneck where interruption of metabolism of only a few parasites can break the infective cycle. Characterization of AgAK adenosine kinase provides biochemical access to this unusual step in host-parasite metabolism.
Acknowledgments
We thank MR4 for providing us with Anopheles gambiae mosquitoes contributed by Mark Q. Benedict and William E. Collins and genomic DNA from A. gambiae contributed by William E. Collins. Data of X-ray diffraction for this study were measured at Beamline X29A of the National Synchrotron Light Source. Financial support of Beamline X29A of the National Synchrotron Light Source comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy, and from the National Center for Research Resources of the National Institutes of Health. This research was supported by NIH Research Grant AI049512.
Abbreviations
- AMP
adenosine 5′-monophosphate
- ADP
adenosine 5′-diphosphate
- ATP
adenosine 5′-triphosphate
- GTP
guanosine 5′-triphosphate
- UTP
uridine 5′-triphosphate
- TTP
thymidine 5′-triphosphate
- CTP
cytidine 5′-triphosphate
- IMP
inosine 5′-monophosphate
- TMP
thymidine 5′-monophosphate
- UMP
uridine 5′-monophosphate
- CMP
cytidine 5′-monophosphate
- T
thymidine
- dC
2′-deoxycytidine
- dA
2′-deoxyadenosine
- dG
2′-deoxyguanosine
- ITU
7-Deaza-7-iodoadenosine
- Ap3A
P1,P3-di(adenosine-5′) triphosphate
- Ap4A
P1,P4-di(adenosine-5′) tetraphosphate
- Ara-A
9-β-D-arabinofuranosyladenine
- NTPs
nucleotides triphosphates
- NDPs
nucleotides diphosphates
- Arg
arginine, Asp, aspartic acid
- Glu
glutamic acid
- His
Histidine
- Ala
alanine
- Gly
glycine
- Thr
threonine
- Phe
phenylalanine
- Met
methionine
- Ile
isoleucine
- Asn
asparagine
- Ser
serine
- Val
valine
- Leu
leucine
- Gln
glutamine
- Cys
cysteine
- ORF
open reading frame
References
- 1.Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Koning HP, Bridges DJ, Burchmore RJS. Purine and pyrimidine transport in pathogenic protozoa: From biology to therapy. FEMS Microbiol Rev. 2005;29:987–1020. doi: 10.1016/j.femsre.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Hyde JE. Targeting purine and pyrimidine metabolism in human apicomplexan parasites. Curr Drug Targets. 2007;8:31–47. doi: 10.2174/138945007779315524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cassera MB, Hazleton KZ, Riegelhaupt PM, Merino EF, Luo M, Akabas MH, Schramm VL. Erythrocytic adenosine monophosphate as an alternative purine source in Plasmodium falciparum. J Biol Chem. 2008;283:32889–32899. doi: 10.1074/jbc.M804497200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spychala J, Datta NS, Takabayashi K, Datta M, Fox IH, Gribbin T, Mitchell BS. Cloning of human adenosine kinase cDNA: sequence similarity to microbial ribokinases and fructokinases. Proc Natl Acad Sci USA. 1996;93:1232–1237. doi: 10.1073/pnas.93.3.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boison D. Adenosine kinase, epilepsy and stroke: mechanisms and therapies. Trends Pharmacol Sci. 2006;27:652–658. doi: 10.1016/j.tips.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 7.Galazka J, Striepen B, Ullman B. Adenosine kinase from Cryptosporidium parvum. Mol Biochem Parasitol. 2006;149:223–230. doi: 10.1016/j.molbiopara.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 8.Kim YA, Rawal RK, Yoo J, Sharon A, Jha AK, Chu CK, Rais RH, Al Safarjalani ON, Naguib FN, El Kouni MH. Structure-activity relationships of carbocyclic 6-benzylthioinosine analogues as subversive substrates of Toxoplasma gondii adenosine kinase. Bioorg Med Chem. 2010;18:3403–3412. doi: 10.1016/j.bmc.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Ugarkar BG, DaRe JM, Kopcho JJ, Browne CE, 3rd, Schanzer JM, Wiesner JB, Erion MD. Adenosine kinase inhibitors 1 Synthesis, enzyme inhibition, and antiseizure activity of 5-iodotubercidin analogues. J Med Chem. 2000;43:2883–2893. doi: 10.1021/jm000024g. [DOI] [PubMed] [Google Scholar]
- 10.Vodnala M, Fijolek A, Rofougaran R, Mosimann M, Maser P, Hofer A. Adenosine kinase mediates high affinity adenosine salvage in Trypanosoma brucei. J Biol Chem. 2008;283:5380–5388. doi: 10.1074/jbc.M705603200. [DOI] [PubMed] [Google Scholar]
- 11.Parker WB, Barrow EW, Allan PW, Shaddix SC, Long MC, Barrow WW, Bansal N, Maddry JA. Metabolism of 2-methyladenosine in Mycobacterium tuberculosis. Tuberculosis (Edinb) 2004;84:327–336. doi: 10.1016/j.tube.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Wu LF, Reizer A, Reizer J, Cai B, Tomich JM, Saier MH., Jr Nucleotide sequence of the Rhodobacter capsulatus fruK gene, which encodes fructose-1-phosphate kinase: evidence for a kinase superfamily including both phosphofructokinases of Escherichia coli. J Bacteriol. 1991;173:3117–3127. doi: 10.1128/jb.173.10.3117-3127.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro JM, Wides R, Salzberg SL, Loftus B, Yandell M, Majoros WH, Rusch DB, Lai Z, Kraft CL, Abril JF, Anthouard V, Arensburger P, Atkinson PW, Baden H, de Berardinis V, Baldwin D, Benes V, Biedler J, Blass C, Bolanos R, Boscus D, Barnstead M, Cai S, Center A, Chaturverdi K, Christophides GK, Chrystal MA, Clamp M, Cravchik A, Curwen V, Dana A, Delcher A, Dew I, Evans CA, Flanigan M, Grundschober-Freimoser A, Friedli L, Gu Z, Guan P, Guigo R, Hillenmeyer ME, Hladun SL, Hogan JR, Hong YS, Hoover J, Jaillon O, Ke Z, Kodira C, Kokoza E, Koutsos A, Letunic I, Levitsky A, Liang Y, Lin JJ, Lobo NF, Lopez JR, Malek JA, McIntosh TC, Meister S, Miller J, Mobarry C, Mongin E, Murphy SD, O’Brochta DA, Pfannkoch C, Qi R, Regier MA, Remington K, Shao H, Sharakhova MV, Sitter CD, Shetty J, Smith TJ, Strong R, Sun J, Thomasova D, Ton LQ, Topalis P, Tu Z, Unger MF, Walenz B, Wang A, Wang J, Wang M, Wang X, Woodford KJ, Wortman JR, Wu M, Yao A, Zdobnov EM, Zhang H, Zhao Q, Zhao S, Zhu SC, Zhimulev I, Coluzzi M, della Torre A, Roth CW, Louis C, Kalush F, Mural RJ, Myers EW, Adams MD, Smith HO, Broder S, Gardner MJ, Fraser CM, Birney E, Bork P, Brey PT, Venter JC, Weissenbach J, Kafatos FC, Collins FH, Hoffman SL. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- 14.Taylor EA, Rinaldo-Matthis A, Li L, Ghanem M, Hazleton KZ, Cassera MB, Almo SC, Schramm VL. Anopheles gambiae purine nucleoside phosphorylase: catalysis, structure, and inhibition. Biochemistry. 2007;46:12405–12415. doi: 10.1021/bi7010256. [DOI] [PubMed] [Google Scholar]
- 15.Darling JA, Sullivan WJ, Jr, Carter D, Ullman B, Roos DS. Recombinant expression, purification, and characterization of Toxoplasma gondii adenosine kinase. Mol Biochem Parasitol. 1999;103:15–23. doi: 10.1016/s0166-6851(99)00109-7. [DOI] [PubMed] [Google Scholar]
- 16.Tyler PC, Taylor EA, Frohlich RF, Schramm VL. Synthesis of 5′-methylthio coformycins: specific inhibitors for malarial adenosine deaminase. J Am Chem Soc. 2007;129:6872–6879. doi: 10.1021/ja0708363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sturm MB, Schramm VL. Detecting ricin: sensitive luminescent assay for ricin A-chain ribosome depurination kinetics. Anal Chem. 2009;81:2847–2853. doi: 10.1021/ac8026433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otwinowski Z, Minor W. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 19.Vagin AA, Teplyakov A. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 20.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 22.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 23.Laskowski RA, McArthur MW, Moss DS, Thornton JM. J Appl Cryst. 1993;265:283–291. [Google Scholar]
- 24.Schumacher MA, Scott DM, Mathews II, Ealick SE, Roos DS, Ullman B, Brennan RG. Crystal structures of Toxoplasma gondii adenosine kinase reveal a novel catalytic mechanism and prodrug binding. J Mol Biol. 2000;296:549–567. doi: 10.1006/jmbi.1999.3474. [DOI] [PubMed] [Google Scholar]
- 25.Clements AN. The biology of mosquitoes. Vol. 1. Chapman and Hall; New York: 1992. [Google Scholar]
- 26.Knecht W, Petersen GE, Sandrini MP, Sondergaard L, Munch-Petersen B, Piskur J. Mosquito has a single multisubstrate deoxyribonucleoside kinase characterized by unique substrate specificity. Nucleic Acids Res. 2003;31:1665–1672. doi: 10.1093/nar/gkg257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller RL, Adamczyk DL, Miller WH, Koszalka GW, Rideout JL, Beacham LM, 3rd, Chao EY, Haggerty JJ, Krenitsky TA, Elion GB. Adenosine kinase from rabbit liver II Substrate and inhibitor specificity. J Biol Chem. 1979;254:2346–2352. [PubMed] [Google Scholar]
- 28.Long MC, Escuyer V, Parker WB. Identification and characterization of a unique adenosine kinase from Mycobacterium tuberculosis. J Bacteriol. 2003;185:6548–6555. doi: 10.1128/JB.185.22.6548-6555.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, El Kouni MH, Ealick SE. Structure of Toxoplasma gondii adenosine kinase in complex with an ATP analog at 1.1 angstroms resolution. Acta Crystallogr D Biol Crystallogr. 2006;62:140–145. doi: 10.1107/S090744490503430X. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, El Kouni MH, Ealick SE. Substrate analogs induce an intermediate conformational change in Toxoplasma gondii adenosine kinase. Acta Crystallogr D Biol Crystallogr. 2007;63:126–134. doi: 10.1107/S0907444906043654. [DOI] [PubMed] [Google Scholar]
- 31.Muller CW, Schulz GE. Structure of the complex between adenylate kinase from Escherichia coli and the inhibitor Ap5A refined at 1.9 Å resolution A model for a catalytic transition state. J Mol Biol. 1992;224:159–177. doi: 10.1016/0022-2836(92)90582-5. [DOI] [PubMed] [Google Scholar]