Abstract
A microfluidic device capable of two-dimensional reversed-phase liquid chromatography-capillary electrophoresis with integrated electrospray ionization (LC-CE-ESI) for mass spectrometry (MS)-based proteomic applications is described. Traditional instrumentation was used for the LC sample injection and delivery of the LC mobile phase. The glass microfabricated device incorporated a sample-trapping region and an LC channel packed with reversed-phase particles. Rapid electrokinetic injections of the LC effluent into the CE dimension were performed at a cross channel intersection. The CE separation channel terminated at a corner of the square device, which functioned as an integrated electrospray tip. In addition to LC-CE-ESI, this device was used for LC-ESI without any instrumental modifications. To evaluate the system, LC-MS and LC-CE-MS analysis of protein digests were performed and compared.
INTRODUCTION
For comprehensive analysis of complex biological samples, separation methods should have high resolving power and are often coupled with mass spectrometry (MS) detection for analyte identification.1 One-dimensional separation methods lack the peak capacity to adequately separate large numbers of proteins or peptides. As a result, multidimensional separations are desired for their ability to dramatically increase peak capacity by combining two or more modes of separation.2 Provided that the coupled separations are orthogonal (i.e., the separation methods are not correlated) and the resolution obtained in any dimension is retained throughout the analysis, the total peak capacity for the multidimensional method is the product of the peak capacities for the individual separation modes. 3–4
One common multidimensional separation method for protein or peptide analysis is two-dimensional poly-acrylamide gel electrophoresis (2D-PAGE).5 Although 2D-PAGE can generate peak capacities of several thousand, there are several key limitations.6–7 The method is time consuming, labor intensive, and difficult to automate. In addition, several classes of proteins are not amenable to this method. Very hydrophobic proteins may not efficiently load into the gel and very acidic or basic proteins are often poorly separated. Most importantly, 2D-PAGE cannot be coupled online with MS detection, making comprehensive identification of complex samples largely impractical.
Several column-based methods for performing comprehensive online 2D separations that can be directly interfaced with electrospray ionization (ESI)-MS have been developed.8–13 The majority of these methods employ two different types of liquid chromatography (LC), such as size exclusion chromatography followed by reversed phase chromatography. Alternatively, LC can be coupled with capillary electrophoresis (CE), which enables improved sampling of the first dimension because of the rapid separation capability of CE.8–9 Coupling LC with CE is challenging because injections between the dimensions have to be performed very rapidly without dilution. The Jorgenson group developed a “transverse flow gating” injector to interface these two techniques.14–15 Although there has been considerable development of 2D LC-CE instrumentation,8–9 there have only been a few reports that describe direct interfacing with MS detection.16–18
Lewis et al. described the first online system for performing comprehensive LC-CE with ESI-MS.16 This system utilized a transverse flow-gating interface machined from polyetherimide to couple a capillary LC column to a CE capillary. Injections into the CE dimension were accomplished by modulating a valve that controlled the pressure-driven transverse flow. More recently Bergström et al. described similar systems utilizing a polydimethylsiloxane interface, which simplified fabrication and reduced dead-volume between capillaries. 17–18
There are several inherent advantages of performing 2D LC-CE-ESI on microfluidic devices as opposed to capillary-based systems. All functional elements are easily integrated with near zero dead-volume connections on a single monolithically fabricated platform. 19 Extra-column band broadening is minimized or eliminated, enabling the use of small diameter channels and low flow rates. Small diameter LC channels improve sensitivity for mass limited samples20 and low flow rates increase the ionization efficiency of the electrospray.21–24 Finally, microchip CE separations may be performed at higher speed due to extremely narrow injection plugs and high electric field strengths.25 Previously we have demonstrated highly efficient 2D microfluidic separations using micellar electrokinetic chromatography and capillary electrochromatography coupled with CE .26–28
In this work we describe the fabrication and operation of a glass microfluidic device capable of LC-CE-ESI. This work builds on a previously described, fully integrated microchip ESI interface that was previously used for CE-MS.29–30 The new device incorporates a sample-trapping region and LC channel packed with commercial porous particles. Effluent from the LC channel is transferred to the CE dimension using electrokinetically gated injections.28 In addition to performing LC-CE-ESI, no instrumental modifications are necessary to perform LC-ESI experiments with the device.
EXPERIMENTAL
Reagents and Materials
Deionized water was obtained from a Nanopure water purifier fitted with a 0.2-µm filter (Barnstead International, Dubuque, IA). Acetone, acetonitrile, methanol, and ammonium acetate were all HPLC grade from Fisher Chemical (Fairlawn, NJ). The PolyE-323 polymer was synthesized as previously described from 1,2-bis(3-aminopropylamino)ethane and epichlorohydrin both obtained from Sigma Chemical Co. (St. Louis, MO).31 The PolyE-323 solution was adjusted to pH 7 with acetic acid (Fisher) and diluted with water to 15% (by mass) polymer. The trichloro(1H,1H,2H,2H-perfluorooctyl)silane and rhodamine B were also acquired from Sigma Chemical Co. Tryptic digests of bovine serum albumin (BSA), yeast enolase, and Escherichia coli (MassPREP, Waters Corp., Milford, MA) were reconstituted in 0.1% (v/v) formic acid (99%, Fisher) in water prior to use.
Microchip Fabrication
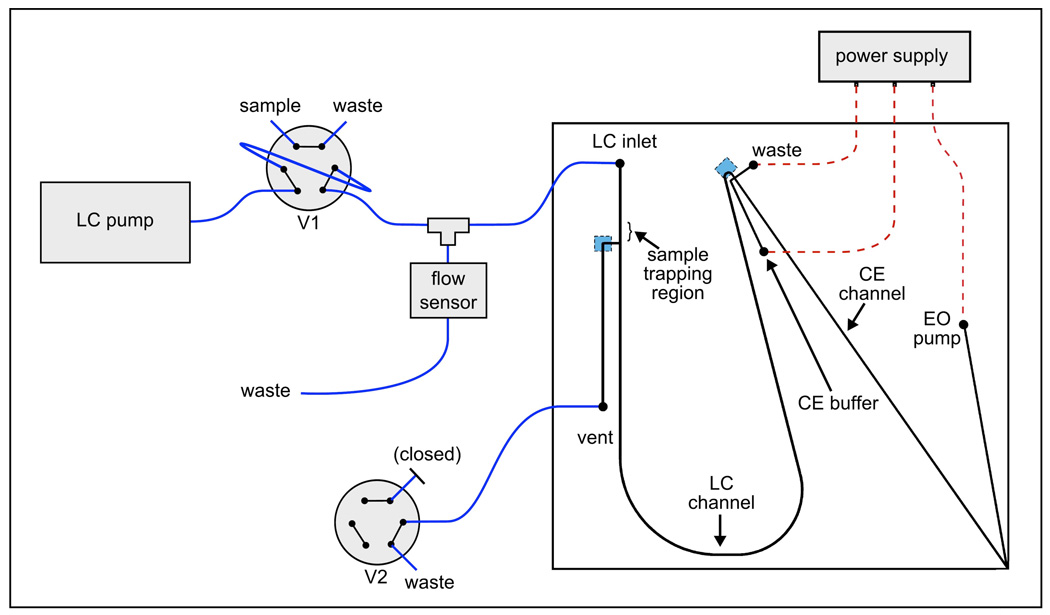
Microchips were fabricated using standard photolithography and wet-chemical etching methods as previously described.29 A schematic for the microchip is shown in Figure 1. The channel design was created using computer-aided drafting software (TurboCAD, ISMI Design, Novato, CA) and plotted on a mylar photomask (Infinite Graphics Inc., Minneapolis, MN). The device was fabricated from 150-µm-thick glass substrates (Corning 0211 borosilicate, Erie Scientific Co., Portsmouth, NH) that were coated with chrome and photoresist by a commercial vender (Telic Co., Valencia, CA). A simple multi-step etching process was used to create channels with different depths. First, all channels were etched to the desired depth for the shallowest feature. After the substrate was rinsed and dried, photoresist (Microposit S1813, Microchem Corp., Newton, MA) was manually applied and cured at the location of the shallowest feature to prevent further etching. The remaining uncoated channels were then etched to the second depth and the process was repeated until all three selective depths were achieved. Access ports with a 400-µm internal diameter were created in the etched substrates by powder blasting (Microblaster, Comco, Inc., Burbank, CA). Channel depths and widths were measured with a profilometer (P15, KLA-Tencor Corp., San Jose, CA). A blank substrate was fusion bonded to the etched substrate by standard procedures.29 The electrospray tip was machined by cutting the bonded microchip with a precision dicing saw (Dicing Technology, San Jose, CA) so that the CE channel terminated at a 90° corner of the microchip.29 The microchip was attached to 0.9-mm-thick glass with transparent UV epoxy (68, Norland Products Inc., Cranbury, NJ) to protect the device from mechanical damage. Glass cylinders with a volume of 200 µL were attached to the buffer, waste and electroosmotic (EO) pump channel access ports using chemically resistant epoxy (Hysol E-120HP; Henkle Corp., Morrisville, NC) and used as fluid reservoirs. The channel lengths were as follows: LC inlet, 10 mm; vent, 22 mm; LC channel, 100 mm; CE buffer, 11 mm; waste, 3 mm; CE channel, 50 mm; and EO pump, 30 mm. The LC inlet, vent and LC channels were 25-µm deep and 120 µm at full width. The CE buffer, waste, CE and EO pump channels were 8-µm deep and 50 µm at full width. Both weirs were etched to a depth of 6 µm. The same microfabricated device was used over a 5 week period to collect all data presented in this report.
Figure 1.
Schematic for the microchip-based LC-CE-MS system. The blue squares on the microchip denote the location of the weirs (channel segments etched 6 µm deep) that were used to retain the packed particles. Valve 1 (V1) was used to perform LC injections and valve 2 (V2) was used to open and close the vent line. Valves are shown in the “sample loading” configuration. Electrospray was performed from the lower right corner of the microchip.
Capillary-to-Microchip Fittings
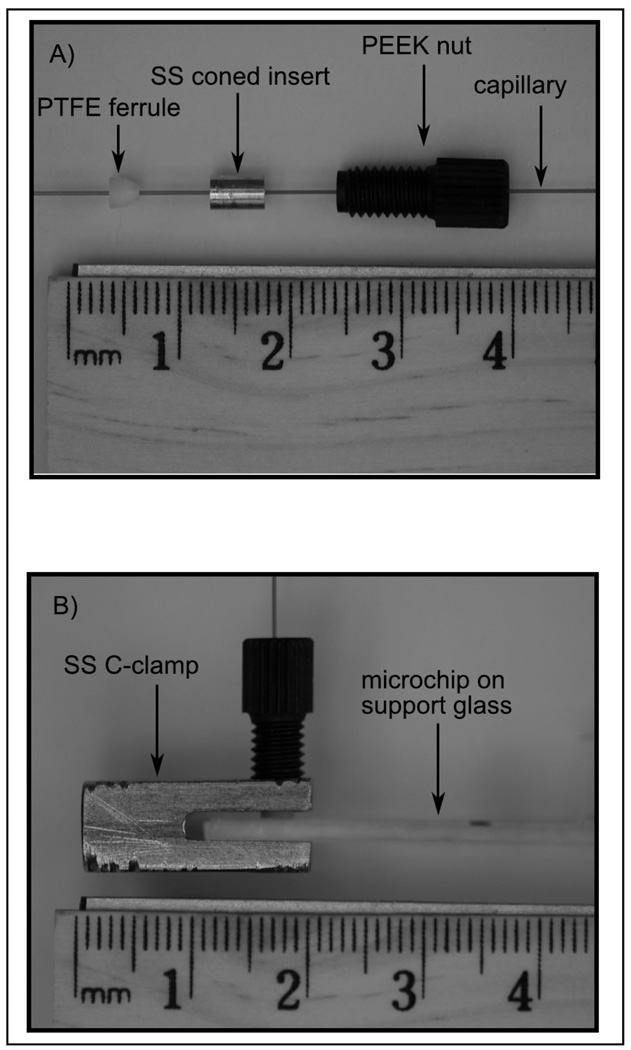
Figure 2 shows photographs of fittings that were developed to connect capillary tubing to microchips. A C-clamp was used to compress a modified LC fitting against a microchip. The C-clamp was machined from an 8-mm square stainless steel rod cut to a length of 20 mm. A 3-mm-wide slot was cut in one end of the rod to accept the microchip. A hole was drilled perpendicularly to the slot and tapped to match the thread of the fitting assembly. The fitting assembly consisted of a polyetheretherketone nut (F121-H, Upchurch Scientific, Oak Habor, WA) modified to hold a stainless steel coned insert. The cone of the insert was used to press a polytetrafluoroethylene (PTFE) ferrule against the surface of the microchip. The PTFE ferrule deformed easily to seal against both the capillary and the glass microchip surface. To install the fitting, a capillary was inserted through the loose fitting/C-clamp assembly and into the access port on the microchip. As the nut was finger tightened, the capillary would remain aligned with the access port on the microchip. The alignment could be checked by inspecting the capillary’s position through the hole drilled in the bottom of the C-clamp. By finger tightening the fittings, these connectors routinely held 100 bar without leakage. The fittings could be reused many times (> 25) before the PTFE ferrule required replacement.
Figure 2.
Photographs of the capillary-to-microchip fittings: A) disassembled fitting, B) side view of the assembled fitting in use.
Particle Packing
The sample-trapping region and LC channel were packed with C18-bonded, 3.5-µm porous particles (X-Bridge, Waters Corp.). The particles were suspended in acetone at 5 mg/mL by vortexing and brief sonication. This slurry was then placed in a vial within a stainless steel vessel and helium gas pressure (70 bar) was used to force the slurry through a 50-µm i.d. capillary (Polymicro Technologies, Phoenix, AZ) into the LC inlet port of the microchip. The vent was initially closed during the packing procedure (see Figure 1) to direct the particles towards the LC channel. Particles were retained by the weir near the end of the LC channel. The weirs in this device were simply channels etched to a shallower depth and are denoted in Figure 1 with blue squares. Even though the weirs in the channels were 6-µm deep, they successfully retained the 3.5 µm particles. It is suspected that the particles were partially trapped by the “keystone effect,” where particle aggregation at a taper is sufficient for retention of a packed bed.32–33 Packing of the device was monitored using an optical microscope. When the packing reached the vent channel junction, the gas cylinder was shut off and the system was slowly depressurized to avoid decompression of the bed. As long as the device was not rapidly depressurized to atmospheric pressure there was no need to frit the inlet of the packed channel. The vent was then opened and low pressure was used to pack the particles in the vent channel against the weir. Packing was continued until the bed extended 3 mm into the LC inlet, beyond the intersection of the vent channel. This 3 mm segment of packed particles functioned as the sample-trapping region. The microchip was then connected to the LC pump (nanoAcquity Binary Solvent Manager, Waters Corp.) and the packed LC channel was flushed overnight using a linear gradient from acetonitrile to water. Using a light microscope, no voids or other defects were visible in the packed LC channel. Figure 3 shows a scanning electron microscope image of a cross-section of a channel with similar dimensions packed with the same 3.5-µm particles.
Figure 3.
A scanning electron microscope image of the cross section of a microchip channel, 33-µm deep and 131-µm at full width, packed with 3.5-µm particles.
Surface Modifications
The surface of the CE buffer, waste, and CE channels was coated with PolyE-323.31,34 PolyE-323 is a polyamine that adheres to glass surfaces through electrostatic and hydrogen bonding forces to provide stable anodic (reversed) electroosmotic flow (EOF) when using a neutral to acidic CE background electrolyte. This surface coating reduces the adsorption of positively charged peptides to improve CE separation efficiency and does not increase the background in the MS signal. The coating procedure has been previously described.29 The exterior of the integrated electrospray tip was coated with trichloro(1H,1H,2H,2H-perfluorooctyl)silane to make the surface hydrophobic as previously reported.30 The use of this coating on the electrospray tip prevents wetting and droplet formation improving electrospray stability over a wide range of conditions without raising the background in the MS signal.
Microchip Operation
The experimental setup is shown in Figure 1. All of the fluidic connections outside of the microchip were made with 20-µm i.d. fused silica capillary tubing using the in-house connectors mentioned above. The LC pump (nanoAcquity Binary Solvent Manager, Waters Corp.) delivered the LC mobile phase at 700 nL/min. Mobile phase A was 0.1% formic acid in water and mobile phase B was 0.1% formic acid in acetonitrile. The LC injections were performed with a 6-port valve (Valco Instruments Co. Inc., Houston, TX) with a 1 µL sample loop (labeled as V1 in Figure 1). After the injector, a low dead volume tee (MT.5XCS6, Valco) was used as a splitter to reduce the flow rate delivered to the inlet of the microchip device. A flow sensor (µ-Flow, Bronkhurst, Bethlehem, PA) was connected to the waste line to measure the split flow rate and to monitor flow stability. The capillary connected to the vent channel on the microchip was connected to a second 6-port valve (labeled as V2 in Figure 1) to open or close the vent. The vent was open during the LC injection providing a low flow resistance path through the sample-trapping region on the microchip. This resulted in 80% of the flow (570 nL/min) being delivered to the microchip. After the sample was loaded the vent was closed, reducing the flow rate through the microchip to 65 nL/min. This flow rate corresponds to approximately twice the theoretical optimum linear velocity for moderately sized peptides. The pressure required to deliver this flow rate was less than 50 bar and was well within the pressure limitation of the capillary-to-microchip connectors. A linear mobile phase gradient from 5 to 50% B in 30 min was used for all separations.
The LC channel terminated at a cross channel intersection used to perform the CE injections. The CE injections were accomplished using the gated injection scheme to route the fluids.35 The only modification was that pressure-driven flow was used to introduce the sample to the injection cross. During CE separations, the EOF from the CE buffer to the waste channel prevented the LC effluent from entering the CE channel. This use of EOF to gate a sample introduced to the injection cross by pressure-driven flow has been described in detail.36–37 During CE injections, the EOF counterbalanced the pressure-driven flow in the CE buffer and waste channels to allow the LC effluent to flow into the CE channel. The electric potentials were applied by a custom power supply built using three (two negative and one positive) 10-kV power supply modules (UltraVolt Inc., Ronkonkoma, NY.) The power supply was computer controlled using an analog output board (PCI-6713; National Instruments, Austin, TX) and an in-house written LabVIEW program (National Instruments). Electrical connections were made to the buffer, waste and EO pump channel reservoirs with platinum electrodes. The background electrolyte (BGE) for the CE separations was an aqueous solution of 0.1% formic acid with 25% acetonitrile, pH 2.5. Typical voltages applied to the CE buffer, waste and EO pump reservoirs were −0.8 kV, −0.8 kV, and +8.0 kV for injections and −4.0 kV, 0.0 kV, and +8 kV for separations, respectively. Due to the electrical resistance of the channels in the device, both sets of applied voltages resulted in a potential of −0.6 kV at the injection cross and +5.0 kV at the electrospray tip. The relatively low potential at the injection cross minimized the electric field dropped across the LC channel to the valves and flow sensor, which were all held at ground. Furthermore, because the potentials at the beginning and the end of the CE channel did not change during injections, the separation field strength of 1.1 kV/cm was held constant. This provided stable electrospray and allowed the use of “overlapping injections.”38–39 For the tryptic digests studied, the CE analysis time was 18 s from sample injection to detection of the last eluting peptide. The earliest eluting peptides were detected 9 s after injection yielding a migration time window of only 9 s. Therefore, the CE injections were spaced 10 s apart to increase the sampling of peaks eluting from the first dimension while ensuring that peaks from adjacent CE runs would not overlap before arrival at the ESI emitter. If the separation field strength had not been held constant during injections, detection of analytes would have been impaired as their linear velocity (and therefore the temporal peak width) would have been variable. When LC-MS analyses were performed, the CE injection interface was left in the “injection” mode for the duration of the LC run.
The operation of the integrated electrospray tip relies on an electrokinetic pumping strategy as previously reported.29 The EO pump channel intersected the CE channel less than 700 µm from the electrospray tip. The PolyE-323 coated CE channel had anodic (reversed) EOF while the uncoated EO pump channel had cathodic (normal) EOF. When the potentials described above were applied, the direction of the EOF in both the CE and EO pump channels was toward the electrospray tip. This created a positive pressure at the intersection, which pumped fluid out of the microchip through the short channel segment leading to the electrospray orifice. The voltage at the intersection of the CE and EO pump channels provided the ESI potential. One important feature is that the ESI potential can be independently optimized by offsetting the voltages applied to the device. For example, a 500 V increase in all voltages applied to the device will result in a 500 V increase at the ESI tip without changing the electric field strength in the CE channel. The microchip was positioned with the electrospray tip approximately 5 mm from the inlet of the mass spectrometer (Micromass QTof Micro, Waters Corp.) For optimal ESI, a voltage of approximately +5 kV was required at the electrospray tip with the inlet of the MS held at ground. The electrospray plume was visualized using a 3-mW green diode laser and a CCD camera. For all experiments, the mass spectrometer recorded a mass-to-charge (m/z) range of 300–1000 in “Tof-only” mode using 0.24 s per summed scan and 0.1 s interscan delay. These settings were selected to maximize the data acquisition rate of the instrument.
Data Processing
Peak capacities for the LC-MS runs were calculated using open source software (Peak Finder, available at http://omics.pnl.gov/software). LC-CE-MS data were processed into 2D image plots by initially segmenting the linear string of 2D data according to the individual CE separation windows using an in-house written spreadsheet program. The segmented data were then loaded into an open source imaging program (Image J, US National Institutes of Health, available at http://rsb.info.nih.gov/ij) using an available plug-in (XYZ2DEM importer). The resulting images were then imported into analysis and imaging software (Igor Pro, WaveMetrics, Lake Oswego, OR) for additional graphing options.
RESULTS AND DISCUSSION
ESI-MS
The integrated electrospray interface was previously characterized and found to provide sensitivity and stability comparable to a commercial fused-silica nanospray emitter.29 Initial stability tests were performed over 5 min intervals, which was adequate for the rapid CE-MS separations demonstrated on the device. In the current work the electrospray stability was measured for longer periods of time to ensure stable operation throughout the LC-CE-MS analysis. For this experiment rhodamine B (10 µM) was added to the BGE in the buffer reservoir and it was continuously infused using the “separation” voltage configuration of the CE injection interface. The relative standard deviation (RSD) for the total ion count and the rhodamine B [M+H]+ ion signal over 60 min was 4.7% and 4.3%, respectively.
LC-MS
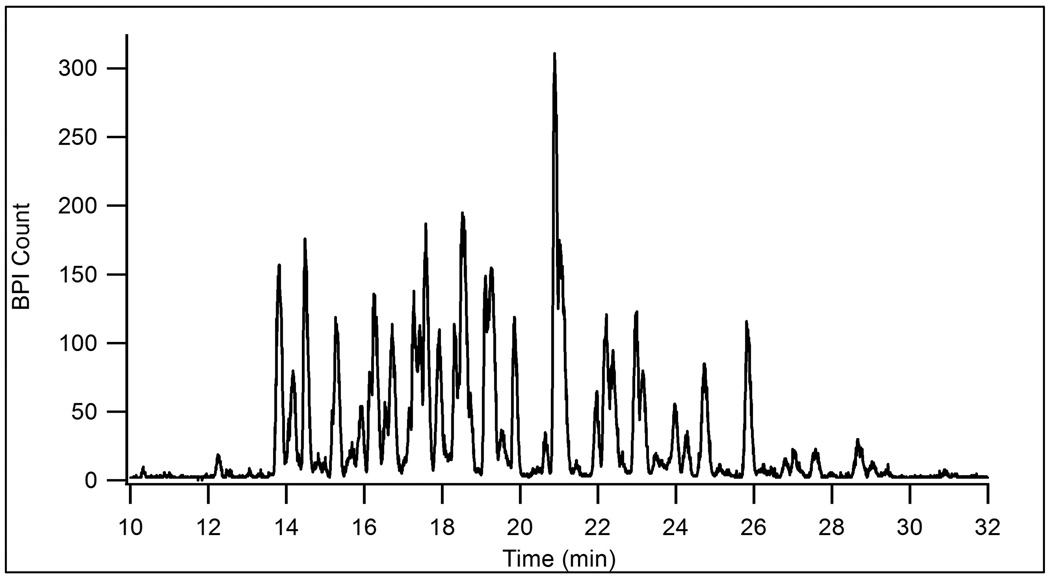
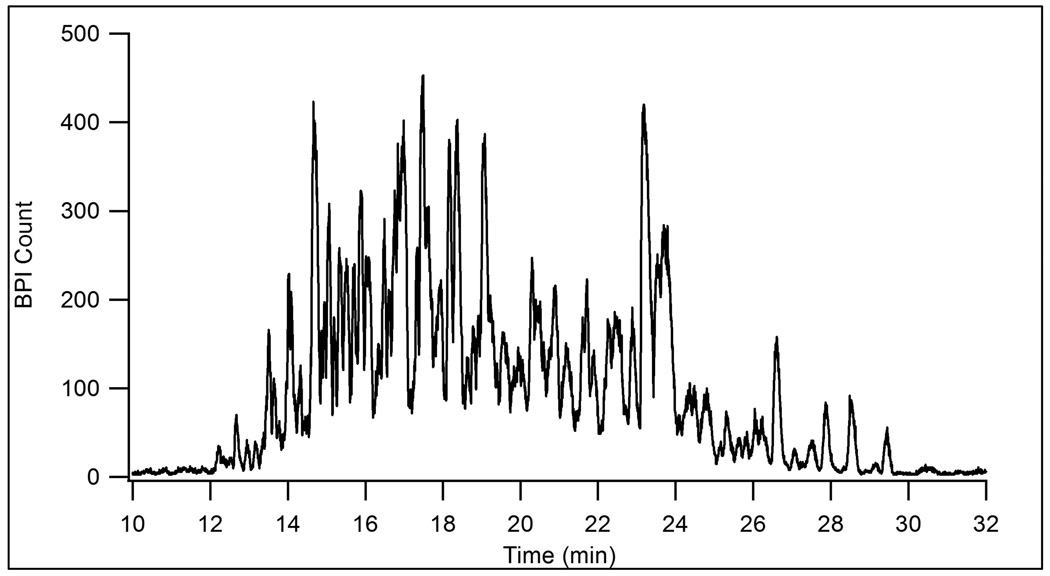
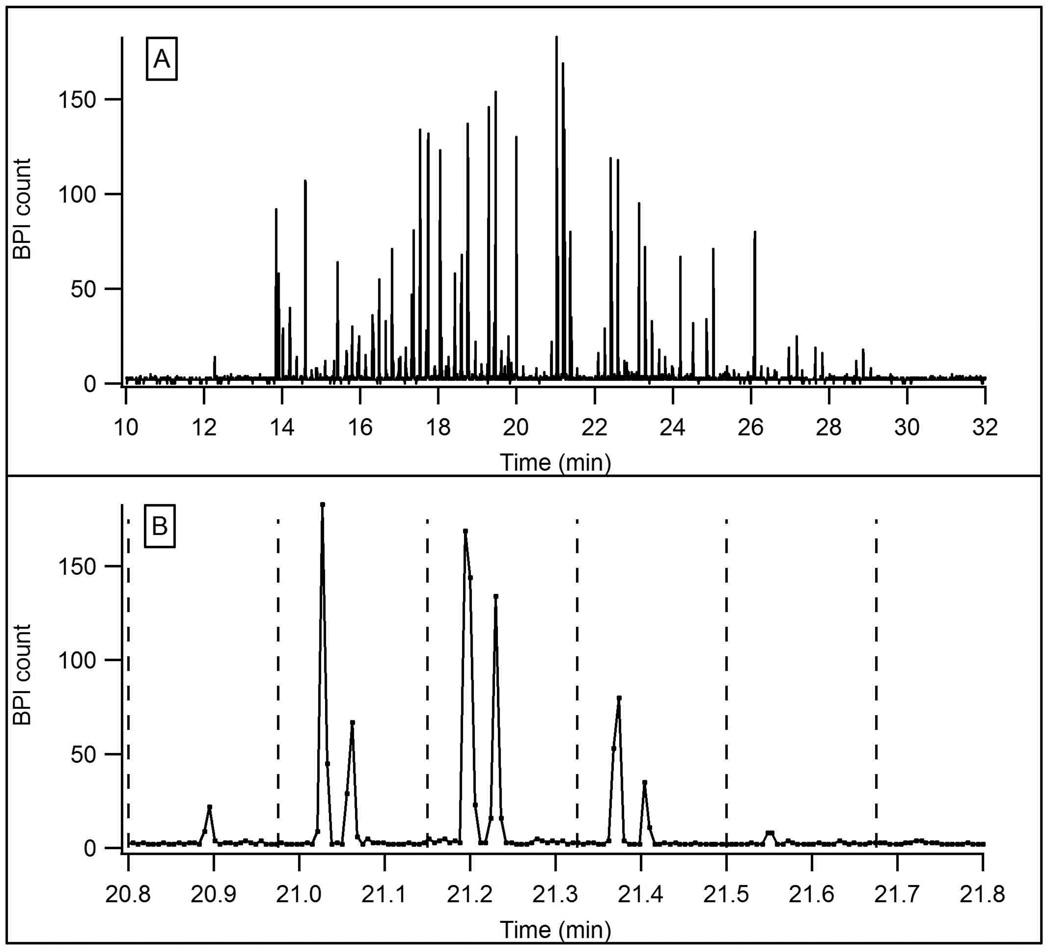
To test the chromatographic performance of the packed microchip channel, the device was utilized for reversed phase LC-MS. Figure 4 shows a base peak index (BPI) chromatogram for an LC-MS analysis of tryptic peptides from BSA and yeast enolase. The low flow resistance of the integrated sample-trapping region allowed dilute samples to be rapidly loaded onto the device. For this LC-MS analysis, 800 nL of a 0.25 µM sample was loaded in less than 2 min. In addition to preconcentration, the sample-trapping region may also be used to perform sample clean-up, such as desalting peptides, prior to the LC run. The peak capacity for this one-dimensional separation was 64. To illustrate the potential for analyzing more complicated samples, Figure 5 shows a separation of 800 ng of a tryptic digest of E. coli cell lysate. The peak capacity for this separation was 58. Both separations show that the device is capable of efficient LC separations in a relatively short analysis time. Tandem MS could be used to identify analytes in these microchip LC-MS experiments, but was not performed in this work.
Figure 4.
BPI chromatogram for the LC-MS analysis of 200 fmol of a tryptic digest of a BSA and enolase mixture. LC flow rate: 65 nL/min, MPA: 0.1% formic acid in water, MPB: 0.1% formic acid in acetonitrile, gradient: 5 to 50% B in 30 min, flow rate: 65 nL/min.
Figure 5.
BPI chromatogram for the LC-MS analysis of 800 ng of an E. coli cell lysate tryptic digest. Conditions as in Figure 4.
LC-CE-MS
One challenging aspect of performing online 2D separations with MS detection is ensuring adequate sampling between dimensions and between the second dimension and the mass spectrometer. With a gated injection scheme, a fraction of the LC effluent is injected into the CE dimension at regular intervals. For this method to be comprehensive, each component eluting from the LC channel must be injected into the CE dimension. To retain the resolution achieved in the first dimension, a frequency of at least 3–4 injections per peak eluting from the first dimension to the second is required.27 A limiting factor for obtaining this frequency is the time required to complete the second dimension separation and detection. A peak capacity of 38 in less than 1 s has been reported for microchip CE using laser-induced fluorescence.28 Commercially available mass spectrometers, however, provide sampling rates that are much slower than obtainable using fluorescence detection. As a result the CE peaks must be significantly wider to ensure adequate peak detection. In this work, we have purposely lengthened the CE injection time to produce peaks sufficiently wide for detection with our mass spectrometer (in effect reducing our separation efficiency to make it more compatible with the MS scan rate).
A BPI chromato-electropherogram for the 2D analysis of tryptic peptides from BSA and enolase is shown in Figure 6A. This figure shows all of the consecutive CE separations as they were recorded in a single data file. The overall pattern of this LC-CE-MS data appears similar to the LC-MS data in Figure 4 because the sample and LC conditions were the same. The median base peak width for components eluting from the LC dimension was 15.7 s (as determined from the LC-MS analysis, Figure 4) and the calculated peak volume was 17 nL. Since the time between successive CE injections was 10 s the components eluting from the LC channel were only sampled 1.6 times on average. Each 0.5 s CE injection corresponded to a volume of 0.8 nL. In Figure 6B an expanded time interval of the 2D analysis shows a few of the CE runs in greater detail, with dashed lines indicating individual CE separation windows. The peaks shown in the first four CE separation windows are not the same two peptides but rather multiple components. Extracted ion chromato-electropherograms (available as Supporting Information, Figure S-2) show that each component was injected 1–2 times into the CE dimension. This data agrees with the calculation above that states that each component eluting from the LC dimension was analyzed by the CE dimension 1.6 times on average. Due to the relatively slow data acquisition rate of the mass spectrometer (~3 Hz), the CE peaks were undersampled as only 1–2 MS data points were obtained per component. It was estimated that the CE base peak widths for individual components were slightly greater than 0.5 s.
Figure 6.
A) BPI chromato-electropherogram for the LC-CE-MS analysis of the same tryptic digest of BSA and enolase shown in Figure 4. LC conditions were the same as in Figure 4. Injections were made into the CE dimension every 10 s. CE BGE: 1% formic acid, 25% acetonitrile, field strength: 1.1 kV/cm. B) An expanded view of a 1 min interval in the separation above. Dashed lines indicate CE separation windows.
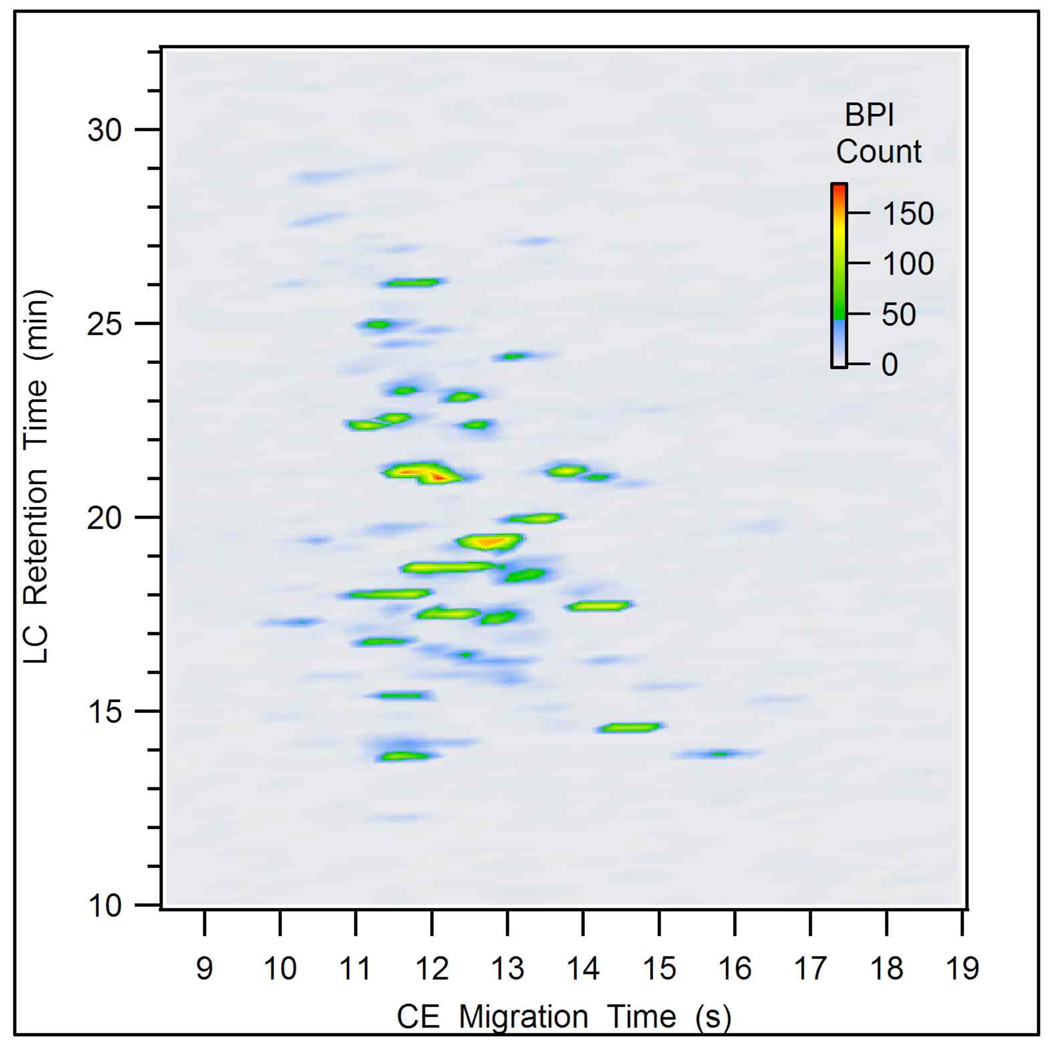
The sequential CE separation windows were stacked according to the corresponding LC retention to simulate a 2D plot as shown in Figure 7. The components from Figure 6B are located in the 2D plot at a retention time of 21 min. The 2D separation resolves these components into two groups and some separation is observed between individual components in each group. Overall, analytes were fairly well distributed across both dimensions and there were a relatively large number of baseline resolved peaks (spots). The estimated peak capacities for the LC and CE dimensions were 64 and 10, respectively. If higher sampling rates could be achieved (4 CE injections per LC peak and 6–10 MS data points per CE peak), the theoretical peak capacity for this separation would be 640 for the 30 min analysis.
Figure 7.
A two-dimensional plot for an LC-CE-MS analysis of 200 fmol of a tryptic digest of a BSA and enolase mixture. LC flow rate: 65 nL/min, MPA: 0.1% formic acid in water, MPB: 0.1% formic acid in acetonitrile, gradient: 5 to 50% B in 30 min, CE BGE: 1% formic acid, 25% acetonitrile, field strength: 1.1 kV/cm.
Three replicate 2D separations were performed using the same tryptic digest of BSA and enolase to evaluate the reproducibility of the system (data available as Supporting Information, Figure S-1). The positions of 15 components (spots) in the 2D plots were used to compare the LC retention times and CE migration times. The average RSD for the LC retention times and CE migration times was 0.35% and 2.4%, respectively. The slow data acquisition rate (~3 Hz) likely contributed significantly to the observed variation in CE migration times.
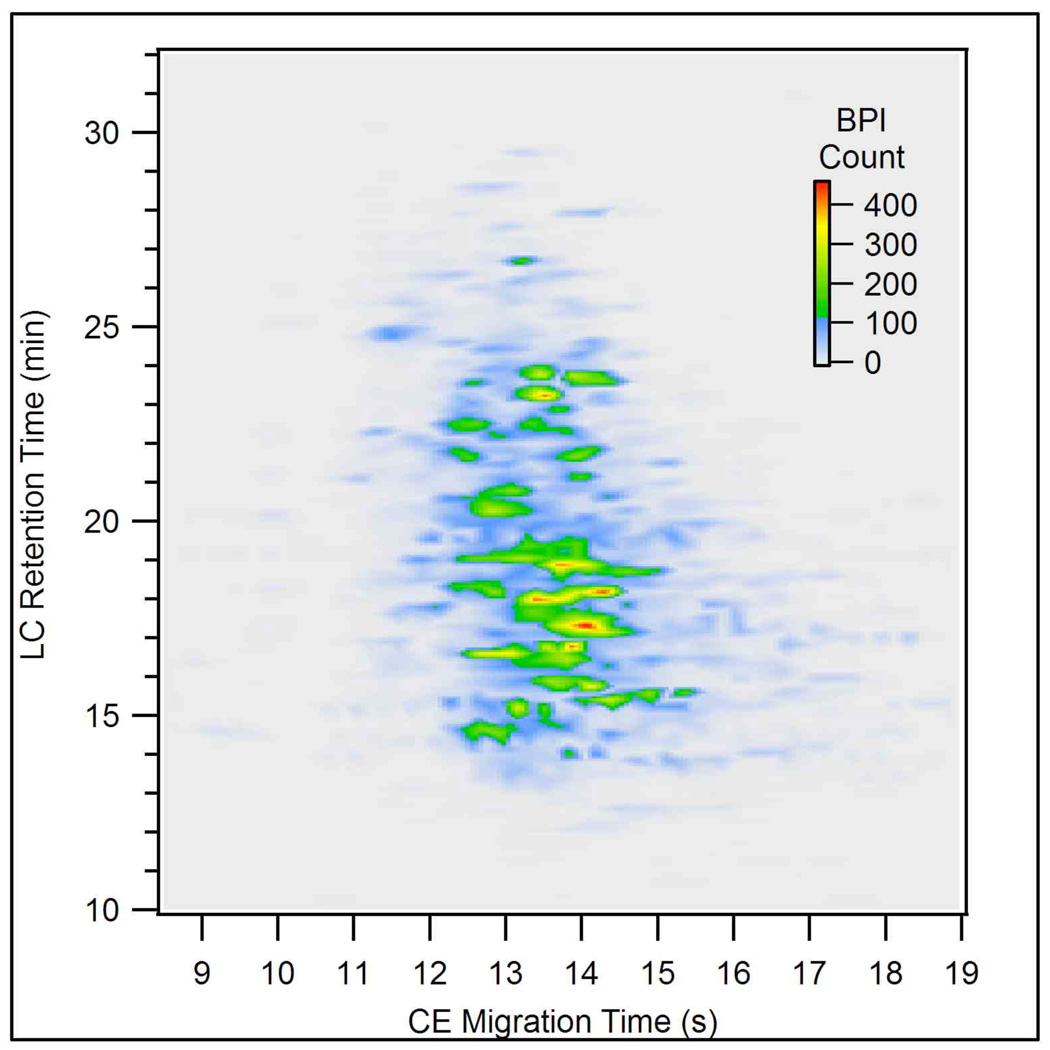
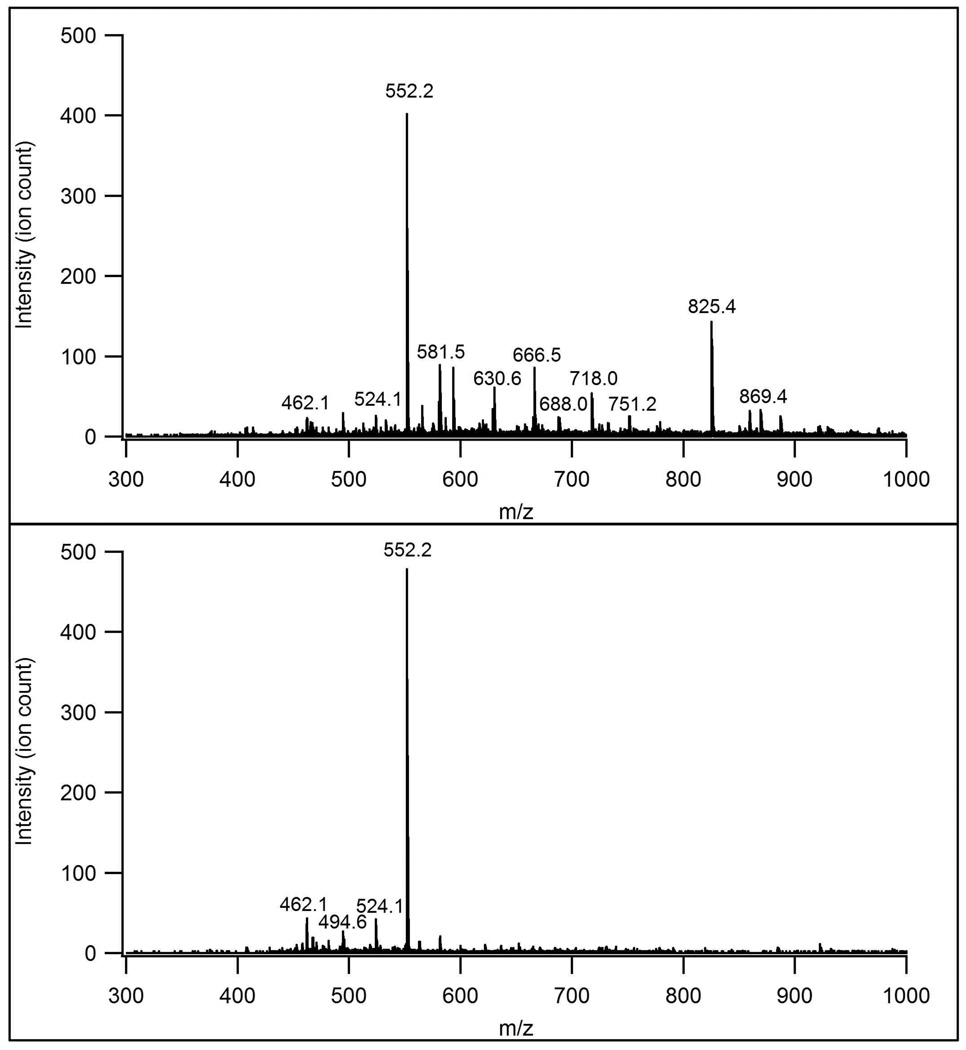
To demonstrate the ability to separate more complex samples, a tryptic digest of E. coli cell lysate was analyzed by LC-CE-MS. The 2D plot for this separation is shown in Figure 8. Although most peaks were not baseline resolved due to the complexity of the sample, the 2D analysis clearly offered higher resolution as compared to the LC-MS analysis shown in Figure 5. The increase in separation power is illustrated by the reduction of the complexity of the mass spectra. Figure 9 shows single scan spectra containing the highest intensity for an abundant ion (552.2 m/ z) from the LC-MS and LC-CE-MS analyses. Fewer ions are present in the spectrum obtained from the 2D run. The intensity for the 552.2 m/z ion is also 23% higher in the 2D analysis. Although there are numerous variables that affect MS intensity, coeluting components may induce ionization suppression in the ESI process, which may account for the reduced intensity of the 552.2 m/z ion in the 1D analysis.
Figure 8.
A two-dimensional plot of the LC-CE-MS analysis of 800 ng of an E. coli cell lysate tryptic digest. Conditions as in Figure 7.
Figure 9.
Mass spectra containing the highest abundance of the 552.2 m/z ion from an E. coli cell lysate tryptic digest analyzed by (A) LC-MS and (B) LC-CE-MS.
In the 2D LC-CE-MS analyses presented, the greatest limitation was the relatively slow scan rate of the mass spectrometer employed. While the maximum data acquisition rate for our instrument was ~3 Hz, commercially available time-of-flight instruments are capable of speeds in excess of 100 Hz. This MS acquisition rate would enable a 100 ms wide CE peak to be sampled 10 times providing a peak capacity of 30 in the CE dimension in only 3 s (assuming 6 s CE runs and “overlapping injections”). Therefore LC peaks would only have to be 12 s wide to obtain optimal sampling between separation dimensions (4 CE injections per LC peak). This system could potentially provide a 30-fold increase in total peak capacity compared to 1D separation methods with currently available MS technology.
CONCLUSION
A microchip device for performing 2D LC-CE-ESI is described for the analysis of protein digests for MS-based proteomic applications. Integrating many functional elements on the microfluidic platform enabled the use of low flow rates without incurring substantial band-broadening. The electrokinetically-gated injections significantly reduced the complexity of the separation system and permitted rapid, sub-nanoliter CE injections of the LC effluent. Tryptic digests were separated to demonstrate multidimensional microfluidic separations directly coupled with MS detection. Faster data acquisition rates (without loss of resolution or mass accuracy) will be required to take full advantage of the high-speed separation capabilities of the device. In order to obtain tandem MS data, in particular, significant increases in data acquisition rates are needed. Future work includes modifying the LC-CE interface to incorporate sample preconcentration between the separation dimensions. In addition, enzymatic digestion and other processing steps may be integrated onto the device to further reduce the overall analysis time.
Supplementary Material
ACKNOWLEDGEMENT
Funding for this research was provided by the National Heart, Lung and Blood Institute proteomics initiative (N01-HV-28182) and Waters Corporation. Use of the Peak Finder software, written by Matthew Monroe, is acknowledged in the following statement. “Portions of this research were supported by the NIH National Center for Research Resources (Grant RR018522), and the W.R. Wiley Environmental Molecular Science Laboratory (a national scientific user facility sponsored by the U.S. Department of Energy's Office of Biological and Environmental Research and located at PNNL). PNNL is operated by Battelle Memorial Institute for the U.S. Department of Energy under contract DE-AC05-76RL0 1830.”
Footnotes
SUPPORTING INFORMATION
Additional information as noted in text. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Aebersold R, Mann M. Nature. 2003;422:198. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 2.Cortes HJ. Multidimensional Chromatography: Techniques and Applications. New York: Marcel Dekker; 1990. [Google Scholar]
- 3.Giddings JC. Anal. Chem. 1984;56:1258A. doi: 10.1021/ac00276a003. [DOI] [PubMed] [Google Scholar]
- 4.Giddings JC. J. High Resolut. Chromatogr. 1987;10:319. [Google Scholar]
- 5.O’Farrell PH. J. Biol. Chem. 1975;250:4007. [PMC free article] [PubMed] [Google Scholar]
- 6.Gorg A, Weiss W, Dunn MJ. Proteomics. 2004;4:3665. doi: 10.1002/pmic.200401031. [DOI] [PubMed] [Google Scholar]
- 7.Lopez JL. J. Chromatogr. B. 2007;849:190. doi: 10.1016/j.jchromb.2006.11.049. [DOI] [PubMed] [Google Scholar]
- 8.Evans CR, Jorgenson JW. Anal. Bioanal. Chem. 2004;378:1952. doi: 10.1007/s00216-004-2516-2. [DOI] [PubMed] [Google Scholar]
- 9.Stroink T, Ortiz MC, Bult A, Lingeman H, de Jong GJ, Underberg WJM. J. Chromatogr. B. 2005;817:49. doi: 10.1016/j.jchromb.2004.11.057. [DOI] [PubMed] [Google Scholar]
- 10.Shellie RA, Haddad PR. Anal. Bioanal. Chem. 2006;386:405. doi: 10.1007/s00216-006-0516-0. [DOI] [PubMed] [Google Scholar]
- 11.Stoll DR, Li XP, Wang XO, Carr PW, Porter SEG, Rutan SC. J. Chromatogr. A. 2007;1168:3. doi: 10.1016/j.chroma.2007.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pol J, Hyotylainen T. Anal. Bioanal. Chem. 2008;391:21. doi: 10.1007/s00216-008-1879-1. [DOI] [PubMed] [Google Scholar]
- 13.Dugo P, Cacciola F, Kumm T, Dugo G, Mondello L. J. Chromatogr. A. 2008;1184:353. doi: 10.1016/j.chroma.2007.06.074. [DOI] [PubMed] [Google Scholar]
- 14.Lemmo AV, Jorgenson JW. Anal. Chem. 1993;65:1576. [Google Scholar]
- 15.Hooker TF, Jorgenson JW. Anal. Chem. 1997;69:4134. [Google Scholar]
- 16.Lewis KC, Opiteck GJ, Jorgenson JW, Sheeley DM. J. Am. Soc. Mass Spectrom. 1997;8:495. [Google Scholar]
- 17.Bergstrom SK, Samskog J, Markides KE. Anal. Chem. 2003;75:5461. doi: 10.1021/ac030117g. [DOI] [PubMed] [Google Scholar]
- 18.Bergstrom SK, Dahlin AP, Ramstrom M, Andersson M, Markides KE, Bergquist J. Analyst. 2006;131:791. doi: 10.1039/b601660j. [DOI] [PubMed] [Google Scholar]
- 19.Kutter JP, Fintschenko Y. Separation Methods in Microanalytical Systems. Boca Raton, FL: CRC Press Taylor and Francis Group; 2006. [Google Scholar]
- 20.Vissers JPC. J. Chromatogr. A. 1999;856:117. doi: 10.1016/s0021-9673(99)00692-5. [DOI] [PubMed] [Google Scholar]
- 21.Wilm MS, Mann M. Int. J. Mass Spectrom. Ion Processes. 1994;136:167. [Google Scholar]
- 22.Wilm M, Mann M. Anal. Chem. 1996;68:1. doi: 10.1021/ac9509519. [DOI] [PubMed] [Google Scholar]
- 23.Emmett MR, Caprioli RM. J. Am. Soc. Mass Spectrom. 1994;5:605. doi: 10.1016/1044-0305(94)85001-1. [DOI] [PubMed] [Google Scholar]
- 24.Valaskovic GA, Kelleher NL, Little DP, Aaserud DJ, McLafferty FW. Anal. Chem. 1995;67:3802. doi: 10.1021/ac00116a030. [DOI] [PubMed] [Google Scholar]
- 25.Jacobson SC, Hergenroder R, Koutny LB, Ramsey JM. Anal. Chem. 1994;66:1114. [Google Scholar]
- 26.Rocklin RD, Ramsey RS, Ramsey JM. Anal. Chem. 2000;72:5244. doi: 10.1021/ac000578r. [DOI] [PubMed] [Google Scholar]
- 27.Gottschlich N, Jacobson SC, Culbertson CT, Ramsey JM. Anal. Chem. 2001;73:2669. doi: 10.1021/ac001019n. [DOI] [PubMed] [Google Scholar]
- 28.Ramsey JD, Jacobson SC, Culbertson CT, Ramsey JM. Anal. Chem. 2003;75:3758. doi: 10.1021/ac0264574. [DOI] [PubMed] [Google Scholar]
- 29.Mellors JS, Gorbounov V, Ramsey RS, Ramsey JM. Anal. Chem. 2008;80:6881. doi: 10.1021/ac800428w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mellors JS, Jorabchi K, Smith LM, Ramsey JD. Anal. Chem. 2010;82:967. doi: 10.1021/ac902218y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hardenborg E, Zuberovic A, Ullsten S, Soderberg L, Heldin E, Markides KE. J. Chromatogr. A. 2003;1003:217. doi: 10.1016/s0021-9673(03)00739-8. [DOI] [PubMed] [Google Scholar]
- 32.Lord GA, Gordon DB, Myers P, King BW. J. Chromatogr. A. 1997;768:9. [Google Scholar]
- 33.Ceriotti L, de Rooij NF, Verpoorte E. Anal. Chem. 2002;74:639. doi: 10.1021/ac0109467. [DOI] [PubMed] [Google Scholar]
- 34.Ullsten S, Zuberovic A, Wetterhall M, Hardenborg E, Markides KE, Bergquist J. Electrophoresis. 2004;25:2090. doi: 10.1002/elps.200305787. [DOI] [PubMed] [Google Scholar]
- 35.Jacobson SC, Koutny LB, Hergenroder R, Moore AW, Ramsey JM. Anal. Chem. 1994;66:3472. [Google Scholar]
- 36.Lin YH, Lee GB, Li CW, Huang GR, Chen SH. J. Chromatogr. A. 2001;937:115. doi: 10.1016/s0021-9673(01)01326-7. [DOI] [PubMed] [Google Scholar]
- 37.Chen SH, Lin YH, Wang LY, Lin CC, Lee GB. Anal. Chem. 2002;74:5146. doi: 10.1021/ac0202886. [DOI] [PubMed] [Google Scholar]
- 38.Larmann JP, Lemmo AV, Moore AW, Jorgenson JW. Electrophoresis. 1993;14:439. doi: 10.1002/elps.1150140169. [DOI] [PubMed] [Google Scholar]
- 39.Moore AW, Jorgenson JW. Anal. Chem. 1995;67:3448. doi: 10.1021/ac00115a013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.