Abstract
Androgen depletion for advanced prostate cancer (PCa) targets activity of the androgen receptor (AR), a steroid receptor transcription factor required for PCa growth. The emergence of lethal castration-resistant PCa (CRPCa) is marked by aberrant re-activation of the AR despite ongoing androgen depletion. Recently, alternative splicing has been described as a mechanism giving rise to COOH-terminally truncated, constitutively active AR isoforms that can support the CRPCa phenotype. However, the pathologic origin of these truncated AR isoforms is unknown. The goal of this study was to investigate alterations in AR expression arising in a cell-based model of PCa progression driven by truncated AR isoform activity. We show that stable, high-level expression of truncated AR isoforms in 22Rv1 CRPCa cells is associated with intragenic rearrangement of a ~35kb AR genomic segment harboring a cluster of previously-described alternative AR exons. Analysis of genomic data from clinical specimens indicated that related AR intragenic copy number alterations occured in CRPCa, in the context of AR amplification. Cloning of the break fusion junction in 22Rv1 cells revealed long interspersed nuclear elements (LINE-1) flanking the rearranged segment, and a DNA repair signature consistent with microhomology-mediated break-induced replication. This rearrangement served as a marker for the emergence of a rare sub-population of CRPCa cells expressing high levels of truncated AR isoforms during PCa progression in vitro. Together, these data provide the first report of AR intragenic rearrangements in CRPCa, and an association with pathologic expression of truncated AR isoforms in a cell-based model of PCa progression.
Keywords: prostate cancer, androgen receptor, castration-resistant, intragenic rearrangement, alternative splicing
Introduction
Prostate cancer (PCa) is the most frequently diagnosed male cancer in the United States, and the second leading cause of male cancer deaths (1). Normal prostate tissue requires androgens for healthy function and cellular homeostasis. Androgens exert their cellular action by binding to the androgen receptor (AR), a 110kDa transcription factor and member of the steroid nuclear receptor family (2). Initially, PCa depends on normal androgenic activation of the AR for ongoing growth and survival, and presents as an androgen- and AR-dependent disease. Therefore, androgen depletion is the standard systemic therapy for locally advanced or metastatic PCa (3). The limitation of androgen depletion is that PCa eventually recurs with a lethal, castration-resistant phenotype. Although this stage of the disease appears to be independent of normal androgenic signaling, it is well-established that castration-resistant PCa (CRPCa) remains AR-dependent through various mechanisms of aberrant AR activation and the AR remains an important therapeutic target for CRPCa (4).
AR mutations, which can broaden AR ligand specificity, and AR amplification, which can lead to AR protein overexpression, are two genomic mechanisms that can support the CRPCa phenotype (5-14). Ligand-independent AR activation has also been described, and can occur through enhanced dependence on mitogenic signaling cascades that converge on the AR and associated transcriptional coregulators (15). More recently, alternative splicing was described as a mechanism of aberrant AR activation in CRPCa (16-20). Wild-type AR is a modular protein, with an NH2-terminal (NTD) transcriptional activation function-1 (AF-1) domain, a central DNA binding domain (DBD), and a dual-function COOH-terminal ligand binding domain (LBD)/AF-2 domain. Splicing of cryptic exons or exon-skipping can yield truncated AR isoforms consisting of the NTD, DBD, and short, variable-length C-terminal extensions (16-20). These truncated AR isoforms are constitutively active and can support various features of the CRPCa phenotype, such as the androgen-independent activation of AR target genes and androgen-independent growth. Importantly, truncated AR isoforms have been observed in various PCa cell lines, xenografts, and clinical samples, which supports an important role in disease progression (16-20).
Alternative splicing is a widespread mechanism for increasing diversity from a single gene (21), and normal regulation of this process is disrupted in pathologic conditions such as cancer (22). The discovery of alternatively spliced AR isoforms has underscored the importance of understanding how AR splicing may be disrupted in CRPCa. This could provide clues to how truncated AR isoforms play a pathologic role at later stages of the disease. Therefore, the purpose of this study was to investigate the mechanisms underlying changes in AR isoform expression in a cell-based model of PCa progression.
Materials and Methods
Cell Culture
Benign prostate BPH-1 cells were generously provided by Dr. Haojie Huang (University of Minnesota) and cultured in RPMI 1640 (Invitrogen) with 10% FBS (Invitrogen). The CRPCa 22Rv1 cell line was obtained from ATCC and cultured in RPMI 1640 medium with 10% FBS. Androgen-dependent PCa CWR22Pc cells were generously provided by Dr. Marja Nevalainen (Thomas Jefferson University (23)) and cultured in RPMI 1640 supplemented with 10% FBS, 2.5 mM L-glutamine, and 0.8nM dihydrotestosterone (Sigma). Cell growth in RMPI 1640 medium containing 10% charcoal-stripped serum (CSS) +/- 1nM DHT was monitored by crystal violet staining. For androgen response experiments, cells were cultured in RPMI 1640 + 10% CSS for 48h, treated at t=0 with 1nM DHT (Sigma) or vehicle (EtOH), and then harvested at indicated time points. For long-term androgen deprivation, 22Pc cells were cultured in RPMI 1640 + 10% CSS for 7 days, and then split to fresh plates in RPMI 1640 + 10% CSS. Cells were trypsinized and re-seeded in RPMI 1640 + 10% CSS after an additional 10 days to disperse emerging foci of growth. Samples were harvested following 7, 12, 17, 22, 27, and 32 days of culture in RPMI 1640 + 10% CSS.
Western Blot
Western blotting of CWR22Pc and 22Rv1 lysates with AR (Santa Cruz N-20), ERK-2 (Santa Cruz D-2), and ARV-7 (Precision Antibody # AG10008) antibodies was performed exactly as described (16)..
Quantitative Real-Time RT-PCR
Total cellular RNA was isolated from CWR22Pc and 22Rv1 cells as described (24). RNA was reverse transcribed using a RT kit and an oligo(dT) primer (Roche). Absolute quantitation of AR mRNA species was performed using forward and reverse primers listed in Supplementary Table 1. Concurrently, quantitative PCR with serial dilutions of plasmids harboring wild-type AR, AR 1/2/2b, AR 1/2/3/2b, AR 1/2/3/CE1, AR 1/2/3/CE2, and AR 1/2/3/CE3 cDNAs was performed using a SYBRGreen fastmix (PerfeCTa, VWR Life Sciences) and an iCycler instrument (BioRad) exactly as described (16). Threshold cycle of amplification (Ct) values obtained from cDNA standards were used to construct Ct vs. cDNA standard copy number standard curves. Ct values obtained from real-time RT-PCR were plotted on these standard curves to derive copy number values for individual AR mRNA isoforms. For relative quantitation, fold expression change relative to GAPDH was determined by the comparative Ct method (2-ΔΔCt).
Genomic PCR
Genomic DNA was isolated from BPH-1, CWR22Pc, and 22Rv1 cells using a Nucleospin Kit (Clontech). Genomic DNA from clinical CRPCa tissues was isolated as described previously (25). PCR primers designed using the Primer3 program of the MacVector software package and are listed in Supplementary Table 1. For copy number determination, quantitative PCR with serial dilutions of BPH-1 genomic DNA was performed for each primer pair using SYBRGreen fastmix and an iCycler instrument. Ct values obtained from BPH-1 genomic DNA dilutions were used to construct Ct vs. genomic copy number standard curves, with the inference that one BPH-1 genome contains one copy of the X chromosome and therefore one copy of the target region. Ct values obtained from test genomic DNA in real-time PCR reactions were plotted on these standard curves to derive genomic copy numbers for each of the PCR target regions. For conventional PCR, genomic DNA was amplified using a Taq Polymerase PCR kit (Qiagen) according to the manufacturer's protocol. For long range PCR, genomic DNA was amplified using outward facing primers (Supplemental Table 1) and a LongRange PCR kit (Qiagen). Cloned PCR products originating from the AR locus were completely sequenced to identify the 22Rv1 AR locus break fusion junction.
Affymetrix Genome-Wide Human SNP Array 6.0 Analysis
Affymetrix SNP6.0 profiling of primary PCa (26) and metastatic CRPCa (25) was performed in previous studies. Raw data in .CEL format was obtained from the Gene Expression Omnnibus website (accession numbers GSE18333 and GSE14996). Copy numbers were calculated for each probeset using Partek Genomics Suite 6.4 analysis software with default settings. Briefly, for each probeset, raw intensity was corrected for fragment length and sequence, and the geometric means of allele intensity values were scaled to 1 (0 in Log2 space). Copy number was calculated from these summarized intensities by normalizing intensity of each individual tumor samples to the mean intensity of the pooled noncancerous samples. Probe level copy number data was used as input in an algorithm designed to determine the collection of breakpoints that satisfy the maximum likelihood between the input data and the noise-free version. The detailed algorithm is described in the Supplemental Methods section and is available in MATLAB (The MathWorks) upon request.
Results
AR intragenic rearrangement and aberrant AR mRNA splicing
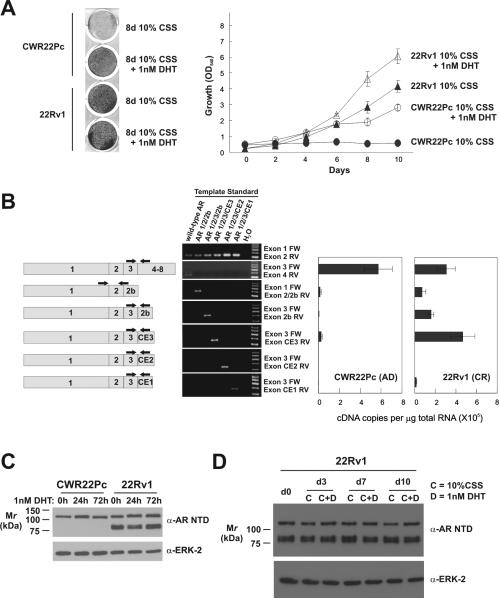
The CWR22Pc cell line was recently established from the CWR22 human PCa xenograft (23). This cell line is androgen-dependent for growth, which is in contrast to the CWR22-derived CRPCa 22Rv1 cell line (Fig. 1A and (27)). Recent reports demonstrated that alternatively spliced, truncated AR isoforms support constitutive AR-mediated transcription and androgen-independent proliferation of 22Rv1 cells (16-18). We therefore examined whether these isoforms were also synthesized in CWR22Pc cells. Using different PCR primer sets with different amplification efficiencies to identify the various AR mRNA isoforms precludes the use of the differential threshold cycle (Ct) of amplification (2-ΔΔCt) method for determining relative expression by real-time PCR. We therefore pursued RT-PCR based absolute quantification (Fig. 1B). As previously reported, full-length AR expression as well as high-level expression of the AR 1/2/2b, AR 1/2/3/2b, and AR 1/2/3/CE3 isoforms was observed at the mRNA and protein level in 22Rv1 cells (Figs. 1B and S1). In androgen-dependent CWR22Pc cells, full-length AR expression was predominant, but expression of AR 1/2/3/CE3 mRNA and protein was also detectable (Figs 1B and S2). No substantial change in these AR expression patterns was observed following 24 or 72 hours of treatment with androgens (Fig. 1C and S2). Similarly, AR isoform expression was stable during 10 days of 22Rv1 cell culture in the presence or absence of androgens (Fig. 1D). Together, these data demonstrate that both androgen-dependent CWR22Pc and CRPCa 22Rv1 cells can synthesize truncated AR isoforms, but 22Rv1 cells are able to sustain stable, high-level expression.
Figure 1.
Efficient and stable synthesis of alternatively spliced AR mRNA isoforms in CRPCa cells. A, Growth of CWR22Pc and 22Rv1 cells in the presence or absence of androgens. B, Plasmid templates harboring depicted cDNAs were subjected to PCR with indicated primer pairs. Right panels, mRNA from CWR22Pc and 22Rv1 cells was subjected to quantitative RT-PCR using indicated primer sets. Ct values obtained from qRT-PCR reactions were converted to copy number by plotting sample Ct values on Ct vs. copy number standard curves constructed from concurrent qPCR analysis of serial dilutions of plasmid templates. Data represent Mean +/- Standard Error from two independent experiments, each performed in triplicate (n = 6). C, AR Western blots of CWR22Pc and 22Rv1 cells following 3 day treatment with 1nM DHT. ERK-2, loading control. D, AR Western blot of 22Rv1 cells following 10 day culture in steroid depleted medium containing 1nM DHT or vehicle control. ERK-2, loading control.
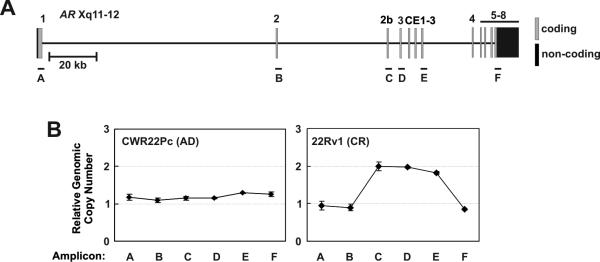
Because the full-length AR mRNA in 22Rv1 cells has two copies of Exon 3, resulting in an AR DBD with 3 zinc fingers (28), and because 22Rv1 cells can efficiently synthesize mRNAs with contiguously-spliced Exons 1, 2, 3, and 2b (16), we hypothesized that a genomic aberration in the 180kb AR locus at Xp11-12 may underlie the stable splicing alterations observed in these cells. We therefore interrogated copy number at distributed loci along the length of the AR gene. Strikingly, in castration-resistant 22Rv1 cells, we observed increased copy number of AR Exons 2b, 3, and CE3, suggesting a rearrangement involving this genomic segment (Fig 2). This aberration was not observed in the androgen-dependent CWR22Pc cell line (Fig. 2). These data therefore suggest that alternative AR isoforms, which support the castration-resistant phenotype of 22Rv1 cells, may arise via enhanced splicing of alternative exons harbored on a rearranged genomic segment in the AR locus.
Figure 2.
Alternatively spliced AR exons are contained on a rearranged genomic segment in 22Rv1 cells. A, Schematic of the ~180kb AR locus at Xp11-12. PCR amplicons used for copy number determination are labeled A-F. B, Genomic DNA from CWR22Pc and 22Rv1 cells was subjected to quantitative PCR using amplicon primer pairs indicated in A. Ct values were converted to copy number by plotting sample Ct values on Ct vs. copy number standard curves constructed from serial dilutions of BPH-1 genomic DNA. Data represent Mean +/- Standard Error from two independent experiments, each performed in triplicate (n = 6).
AR intragenic copy number alterations in metastatic CRPCa tissues
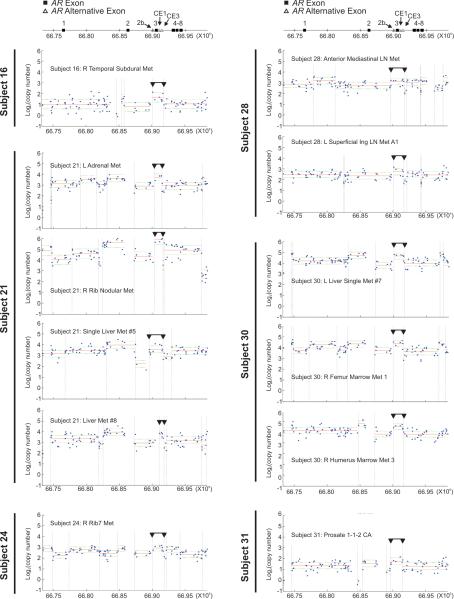
To determine whether AR intragenic copy number alterations occurred in human CRPCa, we analyzed high-resolution Affymetrix Genome-Wide Human SNP Array 6.0 (SNP6.0) data derived from clinical primary PCa (n=44 tissues from 44 patients) and metastatic CRPCa (n = 58 tissues from 14 patients) (25, 26). To localize the boundaries of putative breakpoints, we used a dynamic program that estimates the number and locations of segments adaptively based on probe-level data. This analysis revealed a high incidence of rearrangement in conjunction with AR amplification, only in in CRPCa, which to our knowledge is a novel phenomenon that has not been described (Fig. S3). Because the outcome of the 22Rv1 AR rearrangement appeared to be a focal copy number increase of a segment between AR Exons 2 and 4, resulting in higher dosage of Exon 3 and alternative exons relative to AR Exon 4 (Fig. 2), we asked whether these phenomena occurred in clinical PCa. Indeed, focal copy number increases were observed between AR Exons 2/3 and 3/4 in 12/58 (20.7%) metastases from 6/14 (42.9%) subjects, which presented as rearrangement of a segment harboring some or all alternative AR Exons 2b or CE1-3 (Fig. 3). For most of these CRPCa samples, the outcome was higher gene content of a segment containing AR Exon 3 and alternative exons compared with AR Exon 4 (Fig. S4). These alterations were not observed in genomic DNA samples from these subjects’ normal tissue (Fig. S5). Focal copy number increase of this segment in one CRPCa subject was confirmed using targeted quantitative PCR (Fig. S6). SNP6.0 analysis revealed no changes in overall AR copy number or focal alterations in this region in 44 primary PCa samples (Fig. S4), indicating that CRPCa patients are more likely to harbor this rearrangement in at least one of their tumors than patients with localized, androgen dependent PCa (6/14 vs. 0/44, P = .000074, Fisher's exact test). Overall, these data suggest that the region encompassing AR Exon 3 may represent a “hotspot” for intragenic rearrangement in CRPCa.
Figure 3.
AR intragenic rearrangements in CRPCa detected by Affymetrix Genome Wide SNP 6.0 Array analysis of metastatic tissues. Top, Exon organization of the AR locus on Xq11-12 and chromosome position (human genome build 19, hg19) is indicated at the top of each panel. All panels shown are individual tissue samples from CRPCa metastases. Blue dots represent probe-level copy number, horizontal red lines represent mean segment copy number, horizontal green dashed lines represent standard deviation, and dashed vertical lines represent segment boundaries defined by the segmentation algorithm. Black horizontal lines with downward-facing arrowheads denote a region of focal copy number alteration similar to 22Rv1 cells.
AR breakpoint junction boundaries lie within LINE-1 elements in 22Rv1 cells
To establish with more precision the breakpoint junctions between AR Exons 2 and 2b as well as AR Exons CE3 and 4 in 22Rv1 cells, we carried out higher-resolution copy number interrogation (Fig. 4A). Using this approach, we mapped the 5’ breakpoint between AR Exons 2 and 2b to a resolution of 4kb (Fig. 4B). Concurrently, we mapped the 3’ breakpoint between AR Exons CE3 and 4 to a resolution of 8kb (Fig. 4B). Attempts to map the 5’ or 3’ breakpoints with higher resolution yielded real-time PCR products associated with very low Ct values in both reference and test DNA samples, indicating repetitive sequence. Indeed, analysis of public reference genome sequence revealed long interspersed nuclear elements (LINE-1) and low complexity (TA)n repeats and AT rich sequence in both of these regions (Fig. 4C).
Figure 4.
Fine mapping of AR intragenic rearrangement segment boundaries in 22Rv1 cells. A, Schematic of the AR locus at Xq11-12. PCR amplicons used for copy number determination are labeled B, C, E, F, and G-S. B, Genomic DNA from 22Rv1 cells was subjected to quantitative PCR using amplicon primer pairs indicated in A. Ct values were converted to copy number by plotting sample Ct values on Ct vs. copy number standard curves constructed from serial dilutions of BPH-1 genomic DNA. C, Schematic of repetitive element organization at the 5’ and 3’ boundaries of the 22Rv1 duplicated AR segment in the reference human genome. Elements were defined by RepeatMasker 3.0 (41). Black arrows indicate the directional orientation of L1 elements. L1 elements are named based on their evolutionary origin and sequence divergence, with relative ages (oldest to youngest) L1M1>L1MA3>L1PB2>L1PREC2>L1PA7>L1PA5 (41, 42).
It is common for the endpoints of genomic deletions or insertions to map to repetitive elements such as LINE-1, although the underlying mechanisms are not fully-established (29, 30). One possibility is that extensive homology between LINE-1 elements at breakpoint junctions could lead to deletion on one sister chromatid and duplication on the other via non-allelic homologous recombination (NAHR) (31). Pairwise alignments between the 5’ LINE-1 fragments and the full-length 3’ LINE-1 element identified a >1kb stretch of 87% sequence identity with one particular 5’ LINE-1 fragment, implicating NAHR as the basis for this rearrangement (Fig. S7). Therefore, we performed long-range PCR using two pairs of outward facing primers to isolate the breakpoint junction in 22Rv1 cells (Fig. 5A and S8). This resulted in long PCR products of 6723 and 4762 (Fig. S8). Sequencing of cloned PCR products revealed that they were identical over the common 4762bp, and localized the 22Rv1-specific 5’ and 3’ breakpoints to genomic positions 66,889,976 and 66,924,525, respectively (Fig. 5B and C). Analysis of the break fusion junction revealed 27bp of inserted sequence (Fig. 5C). The origin of this sequence was not apparent by BLASTN and BLAT searches; however, the first 8bp of this sequence perfectly matched an 8bp motif at the 5’ breakpoint. Sequence alignments of the cloned break fusion junction and the 5’ and 3’ breakpoints demonstrated virtually no extended homology through this region (Fig. 5D). However, regions of 3bp microhomology were found at the breakpoints (Fig. 5D). Microhomology at the breakpoints, as well as inserted sequence at the fusion site argues against NAHR and supports a microhomology-mediated break-induced replication (MMBIR) (32) mechanism of segmental duplication in 22Rv1 cells.
Figure 5.
Outward-facing PCR to isolate the 22Rv1 AR tandem duplication. A, Schematic of the AR locus at Xq11-12 with locations of primers used for outward-facing long-range PCR. B, Schematic of the AR locus in 22Rv1 cells as revealed by sequencing of cloned long-range PCR products. C, Electropherogram sequence of the AR break fusion junction in 22Rv1 cells, including a novel 27 bp insert. D, Sequence alignments the 3’ breakpoint, the 22Rv1 break fusion junction, and the 5’ breakpoint. Sequence contained in the break fusion junction is shaded in gray. Regions of microhomology are boxed.
Emergence of CRPCa cells during long-term CWR22Pc castration
Using conventional and nested PCR strategies, we confirmed that the AR breakpoint observed in 22Rv1 cells was indeed restricted to this cell line (Figs. S8C and 6B). Previous studies have demonstrated that androgen-dependent CWR22Pc xenograft tumors initially regress during castration, but eventually recur with a CRPCa phenotype (23). To probe the link between AR intragenic rearrangement and CRPCa, we cultured CWR22Pc cells over a 1 month period in androgen-depleted medium. During the first 12 days of culture, no changes in AR protein expression patterns were observed (Fig. 6C). Interestingly, the 22Rv1 breakpoint was detected in CWR22Pc cells by nested PCR after 7 days of castration (Fig. 6D). The sensitivity of this nested PCR approach was determined to be as low as 1-2 genomes in limiting dilution assays (Fig. S8D), indicating that the sub-population of cells harboring this rearrangement was very rare. By day 17, discrete proliferative foci were apparent, which coincided with faint expression of truncated AR isoforms (Fig. 6C) and detection of the 22Rv1 breakpoint via conventional PCR (Fig. 6D). On day 17, cells were trypsinized and re-seeded to disperse these proliferative foci. By day 22 and onward, androgen-independent cell growth was apparent, as was the expression of truncated AR isoforms. Together, these findings demonstrate that AR intragenic rearrangement is linked to high-level truncated AR isoform expression and CRPCa growth in a cell-based model of PCa progression.
Figure 6.
Concurrent emergence of AR intragenic rearrangement, androgen-independent growth, and high-level truncated AR isoform expression during CWR22Pc castration. A, Schematic of the 22Rv1 AR locus and locations of primers used for nested PCR. B, Conventional PCR was performed using Tfwd/Trev primers and 40ng of input DNA from the indicated cell lines. An aliquot of this reaction was used in a second nested PCR reaction using Ufwd/Trev primers. C, AR Western blot of CWR22Pc castration time-course. CWR22Pc cells were cultured in androgen-depleted medium for the indicated time-points. ERK-2, loading control. D, Nested PCR of CWR22Pc castration time-course. Reactions were performed exactly as described in B.
Discussion
Recent reports describing the synthesis and function of truncated, constitutively active AR isoforms have provided a novel and conceptually simple mechanism for the resistance of CRPCa cells to androgen depletion (16-19). However, the mere presence of truncated AR isoforms does not correlate perfectly with androgen responsiveness, which highlights the importance of quantitative understanding in this area. This is especially apparent from a recent study demonstrating that AR 1/2/3/CE3 (also termed AR-V7 (18) or AR-3 (17)) increases during progression to CRPCa, but is also expressed in benign prostate tissue and hormone naive PCa, (17). Because truncated AR isoforms were originally identified in CRPCa cells derived from the CWR22 model, the recent establishment of an androgen-dependent cell line from CWR22 xenografts has permitted an evaluation of the changes in AR mRNA splicing regulation that may occur during PCa progression in a lineage-related context. One striking difference between androgen-dependent CWR22Pc cells and 22Rv1 CRPCa cells was the expression profile of full-length and alternatively-spliced AR mRNAs. Although alternatively spliced AR mRNAs and protein were detectable in both cell lines, we found that 22Rv1 cells had an enhanced capacity to efficiently synthesize AR 1/2/2b, AR 1/2/3/2b, and AR 1/2/3/CE3 mRNAs. We further demonstrated tandem duplication of a ~35kb segment harboring these alternative exons as a likely basis for the de-regulation of AR mRNA splicing observed in 22Rv1 cells. Interestingly, a recent study identified two additional alternative exons expressed in VCaP cells that are clustered on this segment between AR Exons CE1 and CE3 (20). Mechanistically, such a rearrangement could impair normal splicing by lengthening of the already vast distance between the AR transcription start site and AR Exon 4, increasing the likelihood of incorporating one of the 2 sets of alternative exons preceding Exon 4, disrupting the normal genomic organization of cis-acting intronic and exonic splicing elements, or any combination of these possibilities. It will be important to elucidate a clear cause/effect mechanism, but technical limitations such as the size of the AR locus (~180kb) and even larger aberrant locus in 22Rv1 cells (~215kb) will have to be addressed.
AR overexpression is common in CRPCa, and AR gene amplification is thought to be a main driver of increased AR protein expression (33, 34). Most prior assessments of AR amplification in PCa tissues employed FISH, which lacks resolution and does not permit accurate copy number assessment along the length of the AR gene (7, 13, 34-36). Our findings indicate that a subset of amplified AR loci in CRPCa harbor intragenic rearrangements similar to 22Rv1, in addition to other alterations, which would clearly lead to a reconfigured AR exon organization for many of these alleles. It will therefore be important to perform a comprehensive study of the relationship between AR intragenic rearrangements and levels of alternatively spliced AR isoforms in CRPCa to determine whether there is selection for intragenic rearrangement or whether intragenic rearrangement is simply arising as a byproduct AR gene amplification. Long-term castration of CWR22Pc cells suggests that AR intragenic rearrangement and the CRPCa growth phenotype are linked, and that enrichment for cells with this genomic alteration occurs because of a selective advantage under castrate conditions. It is also possible that larger-scale genomic rearrangements may play a role in disrupted AR splicing, as evidenced by the recent identification of the truncated mAR-V4 isoform in the Myc-CaP mouse model, which results from alternative splicing of a cryptic exon nearly 1Mb upstream of the mouse AR locus (20). Together, these findings indicate that tissues displaying AR intragenic rearrangements should be prioritized for further studies of AR splice variants and their importance to PCa prognosis and therapeutic response.
It will also be important to map and sequence the break fusion junctions within the AR locus in individual CRPCa metastases to obtain a more complete understanding of the mechanisms and significance of AR intragenic rearrangements. Our work revealed LINE-1 elements at the 5’ and 3’ ends of the 22Rv1 AR rearrangement. Transposable elements (TEs) are implicated in the genesis of rearrangements underlying TE-related genetic diseases, including cancer (29), and often arise through non-allelic homologous recombination (NAHR). However, sequencing the 22Rv1 AR break fusion junction revealed a 27bp insertion of unknown origin, which opposes a NAHR-based model. Indeed, stressed cancer cells are deficient in NAHR (37) and cancer-specific rearrangements frequently contain insertions ranging from 1-154bp of so-called non-template sequence at the break fusion junction.(38-40). Therefore, a new model, termed microhomology-mediated break-induced replication (MMBIR) has recently been proposed to account for this class of break fusion junctions in cancer cells (32).
In summary, our work describes a novel AR intragenic rearrangement in the 22Rv1 model of PCa progression, which is linked to enhanced synthesis of truncated AR isoforms and androgen-independent growth. We further demonstrate that similar genomic rearrangements occur in metastatic CRPCa specimens. It will be important in future studies to define whether intragenic AR rearrangements directly cause disrupted AR splicing, because a scenario of AR intra-locus breaks leading to enhanced synthesis of truncated AR isoforms indicates there may be little plasticity in the repertoire of AR isoforms synthesized.. This would potentially limit the effectiveness of manipulating “alternative” splicing as a therapy for CRPCa. Nevertheless, a genomic basis for pathologic AR isoform expression may serve as a stable mechanism-based marker for resistance to androgen depletion therapies.
Supplementary Material
Acknowledgements
We are grateful to the Minnesota Supercomputing Institute and the Masonic Cancer Center Bioinformatics and Biostatistics Core for providing computing, bioinformatics, statistical, software, and data storage support for this project. We also thank Dr. Haojie Huang (Masonic Cancer Center, University of Minnesota) for critical reading of the manuscript. S.M.D. is a Masonic Scholar of the Masonic Cancer Center, University of Minnesota.
Grant Support: This work was supported by a Young Investigator Award from the Prostate Cancer Foundation (S.M.D.), DOD New Investigator Award PC094384 (S.M.D.), NCI Grants CA141011 (S.M.D.), and CA105217 (G.S.B.), and NCI Cancer Center Support Grant P30 077598
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Heemers HV, Tindall DJ. Androgen receptor (AR) coregulators: a diversity of functions converging on and regulating the AR transcriptional complex. Endocr Rev. 2007;28:778–808. doi: 10.1210/er.2007-0019. [DOI] [PubMed] [Google Scholar]
- 3.Taplin ME. Drug insight: role of the androgen receptor in the development and progression of prostate cancer. Nat Clin Pract Oncol. 2007;4:236–44. doi: 10.1038/ncponc0765. [DOI] [PubMed] [Google Scholar]
- 4.Chen Y, Clegg NJ, Scher HI. Anti-androgens and androgen-depleting therapies in prostate cancer: new agents for an established target. Lancet Oncol. 2009;10:981–91. doi: 10.1016/S1470-2045(09)70229-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tilley WD, Wilson CM, Marcelli M, McPhaul MJ. Androgen receptor gene expression in human prostate carcinoma cell lines. Cancer Res. 1990;50:5382–6. [PubMed] [Google Scholar]
- 6.Thompson J, Hyytinen ER, Haapala K, et al. Androgen receptor mutations in high-grade prostate cancer before hormonal therapy. Lab Invest. 2003;83:1709–13. doi: 10.1097/01.lab.0000107262.40402.44. [DOI] [PubMed] [Google Scholar]
- 7.Haapala K, Hyytinen ER, Roiha M, et al. Androgen receptor alterations in prostate cancer relapsed during a combined androgen blockade by orchiectomy and bicalutamide. Lab Invest. 2001;81:1647–51. doi: 10.1038/labinvest.3780378. [DOI] [PubMed] [Google Scholar]
- 8.Hyytinen ER, Haapala K, Thompson J, et al. Pattern of somatic androgen receptor gene mutations in patients with hormone-refractory prostate cancer. Lab Invest. 2002;82:1591–8. doi: 10.1097/01.lab.0000038924.67707.75. [DOI] [PubMed] [Google Scholar]
- 9.Taplin ME, Bubley GJ, Shuster TD, et al. Mutation of the androgen-receptor gene in metastatic androgen-independent prostate cancer. N Engl J Med. 1995;332:1393–8. doi: 10.1056/NEJM199505253322101. [DOI] [PubMed] [Google Scholar]
- 10.Taplin ME, Bubley GJ, Ko YJ, et al. Selection for androgen receptor mutations in prostate cancers treated with androgen antagonist. Cancer Res. 1999;59:2511–5. [PubMed] [Google Scholar]
- 11.Taplin ME, Rajeshkumar B, Halabi S, et al. Androgen receptor mutations in androgen-independent prostate cancer: Cancer and Leukemia Group B Study 9663. J Clin Oncol. 2003;21:2673–8. doi: 10.1200/JCO.2003.11.102. [DOI] [PubMed] [Google Scholar]
- 12.Dehm SM, Tindall DJ. Regulation of androgen receptor signaling in prostate cancer. Expert Rev Anticancer Ther. 2005;5:63–74. doi: 10.1586/14737140.5.1.63. [DOI] [PubMed] [Google Scholar]
- 13.Leversha MA, Han J, Asgari Z, et al. Fluorescence in situ hybridization analysis of circulating tumor cells in metastatic prostate cancer. Clin Cancer Res. 2009;15:2091–7. doi: 10.1158/1078-0432.CCR-08-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Attard G, Swennenhuis JF, Olmos D, et al. Characterization of ERG, AR and PTEN gene status in circulating tumor cells from patients with castration-resistant prostate cancer. Cancer Res. 2009;69:2912–8. doi: 10.1158/0008-5472.CAN-08-3667. [DOI] [PubMed] [Google Scholar]
- 15.Culig Z, Bartsch G. Androgen axis in prostate cancer. J Cell Biochem. 2006;99:373–81. doi: 10.1002/jcb.20898. [DOI] [PubMed] [Google Scholar]
- 16.Dehm SM, Schmidt LJ, Heemers HV, Vessella RL, Tindall DJ. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 2008;68:5469–77. doi: 10.1158/0008-5472.CAN-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo Z, Yang X, Sun F, et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 2009;69:2305–13. doi: 10.1158/0008-5472.CAN-08-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu R, Dunn TA, Wei S, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 2009;69:16–22. doi: 10.1158/0008-5472.CAN-08-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun S, Sprenger CC, Vessella RL, et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J Clin Invest. 120:2715–30. doi: 10.1172/JCI41824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watson PA, Chen YF, Balbas MD, et al. Inaugural Article: Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor. Proc Natl Acad Sci U S A. doi: 10.1073/pnas.1012443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–98. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 22.Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8:749–61. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 23.Dagvadorj A, Tan SH, Liao Z, Cavalli LR, Haddad BR, Nevalainen MT. Androgen-regulated and highly tumorigenic human prostate cancer cell line established from a transplantable primary CWR22 tumor. Clin Cancer Res. 2008;14:6062–72. doi: 10.1158/1078-0432.CCR-08-0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dehm SM, Tindall DJ. Ligand-independent androgen receptor activity is activation function-2-independent and resistant to antiandrogens in androgen refractory prostate cancer cells. J Biol Chem. 2006;281:27882–93. doi: 10.1074/jbc.M605002200. [DOI] [PubMed] [Google Scholar]
- 25.Liu W, Laitinen S, Khan S, et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med. 2009;15:559–65. doi: 10.1038/nm.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mao X, Yu Y, Boyd LK, et al. Distinct genomic alterations in prostate cancers in Chinese and Western populations suggest alternative pathways of prostate carcinogenesis. Cancer Res. 70:5207–12. doi: 10.1158/0008-5472.CAN-09-4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sramkoski RM, Pretlow TG, 2nd, Giaconia JM, et al. A new human prostate carcinoma cell line, 22Rv1. In Vitro Cell Dev Biol Anim. 1999;35:403–9. doi: 10.1007/s11626-999-0115-4. [DOI] [PubMed] [Google Scholar]
- 28.Tepper CG, Boucher DL, Ryan PE, et al. Characterization of a novel androgen receptor mutation in a relapsed CWR22 prostate cancer xenograft and cell line. Cancer Res. 2002;62:6606–14. [PubMed] [Google Scholar]
- 29.Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res. 2008;18:343–58. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- 30.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10:551–64. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edwards J, Krishna NS, Grigor KM, Bartlett JM. Androgen receptor gene amplification and protein expression in hormone refractory prostate cancer. Br J Cancer. 2003;89:552–6. doi: 10.1038/sj.bjc.6601127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Linja MJ, Savinainen KJ, Saramaki OR, Tammela TL, Vessella RL, Visakorpi T. Amplification and overexpression of androgen receptor gene in hormone-refractory prostate cancer. Cancer Res. 2001;61:3550–5. [PubMed] [Google Scholar]
- 35.Bubendorf L, Kononen J, Koivisto P, et al. Survey of gene amplifications during prostate cancer progression by high-throughout fluorescence in situ hybridization on tissue microarrays. Cancer Res. 1999;59:803–6. [PubMed] [Google Scholar]
- 36.Visakorpi T, Hyytinen E, Koivisto P, et al. In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat Genet. 1995;9:401–6. doi: 10.1038/ng0495-401. [DOI] [PubMed] [Google Scholar]
- 37.Bindra RS, Crosby ME, Glazer PM. Regulation of DNA repair in hypoxic cancer cells. Cancer Metastasis Rev. 2007;26:249–60. doi: 10.1007/s10555-007-9061-3. [DOI] [PubMed] [Google Scholar]
- 38.Bignell GR, Santarius T, Pole JC, et al. Architectures of somatic genomic rearrangement in human cancer amplicons at sequence-level resolution. Genome Res. 2007;17:1296–303. doi: 10.1101/gr.6522707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campbell PJ, Stephens PJ, Pleasance ED, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–9. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stephens PJ, McBride DJ, Lin ML, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–10. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tarailo-Graovac M, Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics. 2009 doi: 10.1002/0471250953.bi0410s25. Chapter 4:Unit 4 10. [DOI] [PubMed] [Google Scholar]
- 42.Khan H, Smit A, Boissinot S. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 2006;16:78–87. doi: 10.1101/gr.4001406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.