Abstract
Background
Oral HPV infection elevates risk of oropharyngeal cancer, but its natural history is unknown. Natural history studies necessitate validation of an automated, high-throughput method for HPV genomic DNA detection in oral rinse samples (ORS).
Objectives
To compare agreement of oral HPV detection in ORS processed by a magnetic-bead based automated platform to a previous gold-standard, manual protein-precipitation method. Agreement was compared to that of repeat sampling and repeat HPV testing.
Study Design
HIV-infected individuals (n=100) provided two ORS collected 15 minutes apart. DNA was isolated from equal aliquots by either a protein-precipitation based (Puregene, Qiagen) or magnetic-bead based (QIAsymphonyTM SP, Qiagen) method. HPV DNA was detected and type-specified by consensus primer PCR and reverse line blot hybridization. The kappa statistic was used to assess overall agreement (OA) and agreement on a positive test (Ps+).
Results
The DNA purification methods had very high agreement for categorizing an individual as HPV infected (OA = 0.95; Ps+ = 0.94) as well as for detection of HPV type-specific infection (OA = 0.99; Ps+ = 0.88) in ORS. Agreement for detection of HPV type-specific infection was greater than that observed with repeat oral rinse sampling (OA = 0.99, Ps+ = 0.76) but comparable to inter-assay agreement (OA = 1.00, Ps+ = 0.90).
Conclusions
HPV detection in ORS processed with a magnetic-bead based automated platform will facilitate large natural history studies of oral HPV infection necessary to evaluate the potential use of oral HPV detection in oral cancer screening.
Keywords: Human Papillomavirus, Oral Rinse Samples, HPV detection, Qiasymphony, Puregene DNA isolation
Background
Case-control studies estimate oral human papillomavirus (HPV) 16 infection to confer an approximate 14-fold increase in oropharyngeal cancer risk, and ∼1% of healthy adults have a prevalent infection.1-3 While HPV detection has been incorporated into cervical cancer screening, its utility in oral cancer screening is unknown,4 despite the availability of a commercial assay for oral HPV detection, which to our knowledge remains to be validated. Natural history studies of oral HPV infection must be performed to guide the interpretation of an oral HPV detection test.
We have demonstrated that detection of oral HPV infection is affected by methods of sample collection and subsequent DNA purification.5-7 A protein-precipitation based DNA purification method (Puregene, Gentra Systems, Minneapolis, MN) yields high DNA purity and quantity and detected the greatest number of oral HPV infections in oral rinse samples (ORS) in comparison to several other methods.7 This method is labor intensive, not easily amenable to automation and therefore unfit for large-scale natural history studies. Magnetic bead-based DNA purification from body fluids has demonstrated high nucleic acid yields and utility for isolation of viral nucleic acid from a multitude of sample types.8-10
Objectives
We compared agreement for oral HPV detection in ORS processed by use of an automated, magnetic-bead-based DNA purification method to a previously established standard protein-precipitation method. To guide in data interpretation, this agreement was compared to that of paired ORS collected from the same individual 15 minutes apart and for repeat HPV testing of the same sample on two separate days (inter-assay agreement) using the current “gold standard” protein precipitation method.
Study Design
Study population
One hundred HIV-infected men and women enrolled in a prospective cohort study (Human Oral Papillomavirus Etiology [HOPE] Study) of oral HPV infection at the Johns Hopkins Hospital participated in this sub-study in 2008. The HOPE protocol was approved by the Johns Hopkins Hospital Institutional Review Board and written consent was obtained. Subjects were compensated $20.
Study schema
The study schema is shown in Figure 1. Two ORS (first and second) were collected 15 minutes apart from 100 study subjects by means of a 30-second oral rinse and gargle with Scope mouthwash. Samples were centrifuged, washed and re-suspended with phosphate-buffered saline (PBS), split into two equal aliquots (1.5mL each), and stored at -80°C until further processing.
FIG 1.
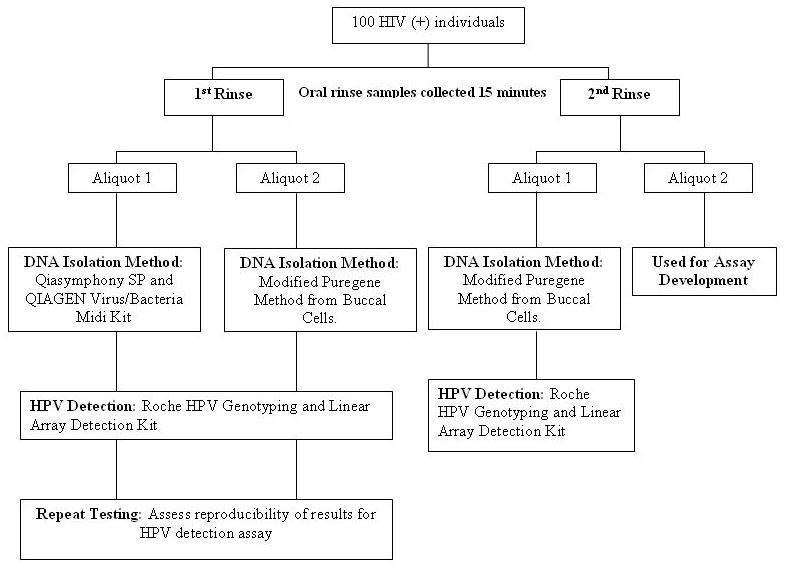
Flow chart of study. HPV detection is stratified by DNA isolation method (1st rinse, Puregene versus Qiasymphony SP) and by oral rinse number (1st or 2nd, both Puregene purified). Repeat HPV detection testing was also performed as a measure of inter-assay agreement (1st rinse, Qiasymphony SP or Puregene purified, repeat testing).
DNA purification
A single aliquot from the first and the second ORS was processed by use of a modified protocol for DNA purification from buccal cells in mouth-wash (Puregene DNA purification kit, Gentra Systems, Minneapolis, MN) as previously described.7 The second aliquot from the first rinse was processed by use of the Qiagen Virus/Bacteria Midi Kit (Qiagen Inc.; Hilden, Germany) on the Qiasymphony SP instrument as recommended by the manufacturer with the following modifications. Briefly, a 1.5-mL aliquot of the ORS (in PBS) was pelleted by centrifugation at 4,000 × g for 10 min. The supernatant was decanted and the pellet was resuspended in 1.4mL of Qiagen cell lysis solution and incubated at room temperature (RT) for 15 min. The sample was then digested with DNase-free RNase A (5ug/mL) for 30 minutes at 37°C followed by Proteinase K digestion (final concentration of 0.5mg/ml) overnight at 55°C. Following heat inactivation at 95°C for 10 minutes, the sample was placed in the Qiasymphony SP to undergo DNA isolation using the Qiagen Virus/Bacteria Midi Kit and Pathogen Complex 800 program. Isolated nucleic acid was eluted in 60ul of AVE buffer.
DNA quantity and quality for all samples were measured by spectrophotometry (Nanodrop Technologies Inc.; Wilmington, DE).
HPV detection
To account for difference in processing between the two DNA isolation methods, 10 μl of Puregene-purified and 12μl for Qiasymphony-purified DNA (representing equivalent proportions of the starting material) were used in PCR reactions. The presence of any of 37 HPV DNA types and ß-globin was detected in purified DNA from ORS by PGMY primer PCR followed by reverse line blot hybridization (Roche Linear Array HPV Genotyping Test, Roche Molecular Systems, Inc.). ß-globin positive samples were considered evaluable and classified as HPV-positive if any of the 37 HPV DNA types were detected.
ERV-3 (human cell number) quantification
Nonhuman DNA (e.g., bacteria) precludes use of total DNA concentration as a direct measure of total human DNA in ORS. Therefore, human diploid-cell genome equivalents were quantified by use of TaqMan real-time PCR targeting a single copy human gene on chromosome 7, ERV-3.11 The number of ERV-3 diploid genome equivalents was used as an estimate of the human cell number per PCR reaction and to adjust for the potential confounding effect of sampling variability on detection of HPV infections.
Statistical analyses
The HPV DNA detection results for each study subject were stratified by oral rinse number (1st or 2nd, both Puregene purified) and DNA isolation method (1st rinse, Puregene versus Qiasymphony purified). Repeat HPV detection testing was also performed as a measure of inter-assay agreement (1st rinse, Puregene or Qiasymphony SP purified, repeat testing). The HPV status of each sample was categorized as HPV positive or negative for any of the 37 HPV types and for each type-specific HPV detected. Median values for DNA purity (A260:A280 ratio), quantity (in ng per PCR reaction), and human cell number (ERV-3 diploid genome copies per PCR) for paired samples were compared by use of the Wilcoxon sign-rank test. HPV detection results were compared using McNemar's test. The kappa statistic was used to assess the agreement of HPV infection status (infected or uninfected), and the agreement for HPV type-specific infection. To account for the high number of concordant negative-negative tests, agreement on a positive test (Ps+) was also determined.12 The 95% confidence intervals (CI) for the overall and positive test agreements were computed using the bias corrected and accelerated (BCa) method of non-parametric bootstrapping. Generalized linear mixed effects models were used to analyze type specific infections and determine the possible effects that DNA isolation may have on HPV detection after adjusting for ERV-3 and the within-sample correlation. All P-values reported are two-sided, and P-values less than 0.05 were considered statistically significant. The statistical packages Stata 10 and R 2.9.2 were used for the data analyses.
Results
A total of 100 HIV-positive individuals enrolled in this study provided two ORS collected fifteen minutes apart. An HIV-positive cohort was chosen as this population is at increased risk for both tonsillar cancer and oral HPV infection.13,14 The median age was 48 years (IQR, 45 to 51 year) with the majority of participants being male (60 men, 61.9% of total). The median CD4-cell count was 414 cells per microliter (IQR, 231-605) and the median HIV viral load was 128 copies per microliter (IQR, 25-1783).
A summary of results for measures of DNA purity, quantity and HPV detection are shown in Table 1. Aliquots from the first oral rinse that were Qiasymphony or Puregene purified had similar DNA purity (A260:A280 = 1.84). Although total DNA yield was significantly lower with the Qiasymphony SP, total human DNA yield was significantly higher. A comparison of the 1st and 2nd ORS collected from the same individual 15 minutes apart and purified by the Puregene method indicated that total DNA yield was significantly lower with the second rinse, however, human DNA yield was not (Table 1).
TABLE 1.
Assessment of DNA quality, quantity and HPV status on oral rinse samples stratified by rinse number and DNA purification method.
| Rinse No.- DNA Purification Method | No. of samples tested | Spectrophotometry | ERV-3 TaqMan PCR d | HPV detection by line blotc | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| DNA purity (A260: A280) | Total nucleic acid (ng) per PCRc | No. of human genome copies per PCRc | No. β-globin positivee | No. HPV positivee | Total no. of HPV infections detected | |||||
| Mediana | IQRb | Mediana | IQRb | Mediana (×103) | IQRb (×103) | |||||
| 1st rinse -Qiasymphony SP | 100 | 1.84 f | 1.77 -1.89 | 436 g | 205 - 840 | 576 g | 213-1,256 | 100 of 100 f | 45 f | 95 |
| 1st rinse- Puregene | 100 | 1.84 | 1.74 -1.89 | 732 | 326 - 1,395 | 104 | 44-290 | 100 of 100 | 42 | 85 |
| 1st rinse- Puregene (repeat HPV detection) | 100 | 100 of 100 f | 41 f | 73 | ||||||
| 2nd rinse- Puregene | 100 | 1.81 f | 1.74 -1.88 | 470 g | 251- 792 | 118 f | 59-216 | 100 of 100 f | 36 f | 80 |
P-values were determined by the Wilcoxon sign-rank and are for comparison to 1st rinse-Puregene data.
Inter-quartile range (IQR) reports the range between the 25th and 75th percentiles and is a measure of data variability.
For Puregene method, 10ul of sample was added to each PCR mixture; for Qiasymphony SP, 12ul of sample was added to each PCR mixture.
ERV-3 is a single-copy human gene used to measure the number of human diploid genome equivalents.
P-values were determined by exact McNemar's test and are used for comparison to 1st rinse-Puregene data.
P > 0.05.
P ≤ 0.0001.
All samples were positive for ß-globin and therefore evaluable for HPV status (Table 1). The oral HPV infection prevalence estimates for the study population were similar when 1st rinse samples were purified either with the Puregene or Qiasymphony method (42 vs. 45%, p-value = 0.38). Prevalence estimates were slightly, although non-significantly, higher in the first ORS when compared to the second sample collected 15 minutes later (42 vs. 36%, p = 0.07). Repeat HPV testing of the first Puregene purified ORS indicates reproducible results for HPV infection prevalence with the Roche HPV genotyping and linear array detection system (42 vs. 41%, p-value = 1.00).
The HPV type distribution detected in the study population is shown in Table 2 stratified by oral rinse number and DNA isolation method. High-risk HPV types accounted for ∼44-45% of HPV infections, and HPV16 was one of the most frequently detected types. The Qiasymphony DNA isolation method detected more type-specific HPV infections (95 vs. 85) in 1st rinse samples when compared to the Puregene method. However, the difference was not significant after adjusting for ERV3 and within-sample correlation between DNA isolation methods, consistent with the interpretation that the higher human DNA yield (as measured by ERV-3) from the Qiasymphony SP accounted for the increase in HPV detection.
TABLE 2.
HPV type distribution in oral rinse samples stratified by DNA isolation method, repeat sampling and repeat HPV testing.
| HPV type | No. of Samples | |||
|---|---|---|---|---|
| Qiagen Qiasymphony 1st rinse | Puregene 1st rinse | Puregene 1st rinse repeated | Puregene 2nd rinse | |
| High-risk HPV | ||||
| 16 | 6 | 5 | 4 | 5 |
| 18 | 5 | 4 | 4 | 6 |
| 26 | 1 | 1 | 1 | 0 |
| 31 | 1 | 0 | 0 | 1 |
| 33 | 3 | 3 | 3 | 2 |
| 35 | 4 | 4 | 3 | 3 |
| 39 | 2 | 2 | 2 | 2 |
| 45 | 2 | 2 | 2 | 2 |
| 51 | 2 | 2 | 2 | 2 |
| 52/33/35/58 | 2 | 1 | 0 | 1 |
| 53 | 4 | 4 | 3 | 1 |
| 56 | 0 | 0 | 0 | 0 |
| 58 | 0 | 0 | 0 | 0 |
| 59 | 3 | 1 | 1 | 3 |
| 66 | 2 | 3 | 2 | 3 |
| 68 | 1 | 1 | 1 | 1 |
| 73 | 2 | 2 | 2 | 1 |
| 82 | 3 | 2 | 2 | 2 |
| No. high-risk HPVs (% of total, 95% CI) | 43 (45, 35 - 56) | 37 (44, 33 - 55) | 32 (44, 32 - 56) | 35 (44, 33 - 55) |
| Low-risk HPV | ||||
| 6 | 3 | 3 | 2 | 4 |
| 11 | 0 | 0 | 0 | 1 |
| 40 | 0 | 0 | 0 | 0 |
| 42 | 2 | 5 | 2 | 2 |
| 54 | 4 | 1 | 1 | 0 |
| 55 | 8 | 8 | 8 | 6 |
| 61 | 4 | 4 | 3 | 3 |
| 62 | 7 | 6 | 6 | 8 |
| 64 | 0 | 0 | 0 | 0 |
| 67 | 0 | 0 | 0 | 0 |
| 69 | 2 | 1 | 0 | 2 |
| 70 | 1 | 1 | 1 | 0 |
| 71 | 1 | 1 | 1 | 1 |
| 72 | 5 | 5 | 5 | 6 |
| 81 | 4 | 4 | 4 | 4 |
| 83 | 8 | 5 | 4 | 4 |
| 84 | 3 | 4 | 4 | 4 |
| 89 | 0 | 0 | 0 | 0 |
| No. low-risk HPVs (% of total, 95% CI) | 52 (55, 44 - 65) | 48 (56, 45 - 67) | 41 (56, 44 - 68) | 45 (56, 45 - 67) |
| Total no. of HPV infections | 95 | 85 | 73 | 80 |
Agreement was evaluated with regard to categorization of an individual as HPV infected or uninfected and for detection of type-specific infection. To account for the high number of concordant negative tests, agreement on a positive test (Ps+), in addition to overall agreement, was determined (Table 3). Qiasymphony and Puregene purification on aliquots from the first ORS had very high agreement for categorizing an individual as HPV infected (OA = 0.95; Ps+ = 0.94) as well as for type specific infection (OA = 0.99; Ps+ = 0.88). Similarly, repeat testing of the Puregene purified 1st ORS revealed high inter-assay agreement for oral HPV infection status by individual (OA = 0.99, Ps+ = 0.99) and for type-specific infection (OA = 1.00, Ps+ = 0.90). Repeat HPV testing of the Qiasymphony SP purified 1st ORS also revealed high inter-assay agreement for oral HPV infection status by individual (OA = 0.95, Ps+ = 0.94) and for type-specific infection (OA = 1.00, Ps+ = 0.91). Type-specific agreement for oral HPV infection was not as strong for repeat sampling (OA = 0.99, Ps+ = 0.76).
TABLE 3.
Measures of agreement in HPV detection stratified by individual and type-specific infection.
| Summary by Individuala | Summary by Type Specific Infectionb | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Measures of Agreementc | Measures of Agreementc | ||||||||||
| Puregene 1st rinse | Ps+ (95% CI d) |
Overall Agreement (95% CI d) |
Puregene 1st rinse | Ps+ (95% CI d) |
Overall Agreement (95% CI d) |
||||||
| No | Yes | No | Yes | ||||||||
| Qiasymphony 1st rinse | No | 54 | 1 | 0.94 (0.87 – 0.98) |
0.95 (0.87 – 0.97) |
Qiasymphony 1st rinse | No | 3,599 | 6 | 0.88 (0.82 – 0.93) |
0.99 (0.99 – 1.00) |
| Yes | 4 | 41 | Yes | 16 | 79 | ||||||
| Puregene 1st rinse repeated | No | 58 | 1 | 0.99 (0.93 – 1.00) |
0.99 (0.94 – 1.00) |
Puregene 1st rinse repeated | No | 3,613 | 14 | 0.90 (0.81 – 0.95) |
1.00 (0.99 – 1.00) |
| Yes | 0 | 41 | Yes | 2 | 71 | ||||||
| Puregene 2nd rinse | No | 57 | 7 | 0.90 (0.80 – 0.95) |
0.92 (0.84 – 0.96) |
Puregene 2nd rinse | No | 3,598 | 22 | 0.76 (0.67 – 0.83) |
0.99 (0.98 – 0.99) |
| Yes | 1 | 35 | Yes | 17 | 63 | ||||||
Positive HPV status is assigned to an individual with at least one oral HPV infection.
The Roche linear array can detects a maximum of 37 different types of HPV infections. Thus, the maximum number of type specific infections possible for 100 patients is 3,700 infections.
Measures of agreement are stratified by overall agreement and agreement among the positive samples (Ps +) only.
The 95% confidence interval (CI) was computed by using the bias corrected and accelerated (BCa) method of non-parametric bootstrapping.
As samples from the above cohort were exhausted, duplicate aliquots of ORS collected from an additional 100 HIV-infected individuals (data not shown) were used to evaluate intra-assay agreement for Qiasymphony purification. High intra-assay agreement was observed for oral HPV infection status by individual (OA = 0.93, Ps+ = 0.91) and for type specific infection (OA = 0.99, Ps+ = 0.86).
Discussion
Our data indicate that the magnetic bead-based automated platform for DNA purification chosen for this study provided excellent agreement for HPV DNA detection in ORS when compared to our previously established “gold-standard” method. Our study design demonstrated that Qiasymphony performance as measured by agreement with the gold standard was similar to inter-assay agreement for repeat testing with that gold standard.
We have shown that PCR inhibitors in ORS samples and total human DNA yield affect detection of HPV DNA.7 In our prior work we demonstrated that thorough digestion of both RNA and protein in samples successfully removed PCR inhibitors, thus precluding need for spiking samples with an internal control. In this study, both DNA purification methods yielded similar levels of DNA purity but the magnetic-bead based method provided higher human DNA yield as measured by ERV3. Given HPV replicates inside infected cells without cell lysis, the DNA isolation method that resulted in the highest human DNA yield also resulted in detection of the highest number of HPV infections.
To guide in data interpretation, we compared agreement between the two DNA isolation methods to that observed with repeat HPV testing (inter-assay agreement) and repeat sampling. With repeat sampling, a lower HPV prevalence and type-specific agreement were observed. The affect that such a short sampling interval may have on HPV detection has never been fully assessed. However, the higher number of HPV infections detected with the initial oral rinse demonstrates the utility of the first rinse in collecting HPV infected buccal cells.
Previous research from our laboratory demonstrated higher detection of HPV infections in volume-standardized samples as compared to cell number-standardized samples per PCR.7 Therefore, no normalization in DNA concentration was implemented for this study. The Roche HPV detection kit is a multiplex PCR reaction for which a competition platform exists for the PGMY L1 consensus primers. The lower limit of assay sensitivity is variable by HPV type and may differ in the presence of multiple type-infections. Indeed, low viral load infections (as indicated by the intensity of the line-blot signal) in individuals with multiple-type infections accounted for the majority of the discrepancies observed in HPV detection among sample pairs. Nevertheless, this detection system proves to be highly reliable given the high level of agreement observed in repeat HPV testing of samples.
This study was performed using an HIV-positive cohort because of their high prevalence of oral HPV infection which facilitated comparison of DNA isolation methods, sampling interval and repeat detection. We acknowledge that this study did not include immunocompetent subjects for whom the effects of DNA isolation method and sampling interval are equally important. This research has relevance for the study design and interpretation of future natural history studies. The Qiasymphony SP allows for a greater number of samples to be run and also requires less technician time making it a more efficient technique for high-throughput DNA isolation. Rinse with SCOPE is considered the gold standard for oral specimen collection and given the Qiasymphony's high throughput and automated properties, this technique will be used by our group henceforth to conduct future natural history studies necessary for appropriate interpretation of a single positive test result.
Acknowledgments
Authors thank R. Pickard, W. Xiao, and Y. Chen for aiding in data analysis.
Funding: This study was funded by a grant (DE016631) from the National Institute of Dental and Craniofacial Research (NIDCR).
Abbreviations
- ORS
oral rinse samples
- OA
overall agreement
- Ps+
positive test
- HPV
Human Papillomavirus
- HOPE
Human Oral Papillomavirus Etiology
- PBS
phosphate-buffered saline
- RT
room temperature
- CI
confidence interval
- IQR
inter-quartile range
- BCa
bias corrected and accelerated
- NIDCR
National Institute of Dental and Craniofacial Research
Footnotes
Competing Interests: None declared
Ethical approval: Not required
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mork J, Lei AK, Glattre E, Hallmans G, Jellum E, Koskela P, et al. Human papillomavirus infection as a risk factor for squamous-cell carcinoma of the head and neck. N Engl J Med. 2001;344:1125–31. doi: 10.1056/NEJM200104123441503. [DOI] [PubMed] [Google Scholar]
- 2.D'Souza G, Kreimer AR, Viscidi R, Pawlita M, Fakhry C, Koch WM, et al. Case-control study of human papillomavirus and oropharyngeal cancer. N Engl J Med. 2007;356:1944–56. doi: 10.1056/NEJMoa065497. [DOI] [PubMed] [Google Scholar]
- 3.Kreimer AR, Bhatia RK, Messeguer AL, González P, Herrero R, Giuliano AR. Oral human papillomavirus in healthy individuals: a systematic review of the literature. Sex Transm Dis. 2010;37(6):386–91. doi: 10.1097/OLQ.0b013e3181c94a3b. [DOI] [PubMed] [Google Scholar]
- 4.Solomon D, Papillo JL, Davey DD, for the Members of the Cytopathology Education and Technology Consortium Statement on Human Papillomavirus DNA Test Utilization. Arch Pathol Lab Med. 2009;133(8):1276–7. doi: 10.5858/133.8.1276. [DOI] [PubMed] [Google Scholar]
- 5.Lawton G, Thomas S, Schonrock J, Monsour F, Frazer I. Human papillomaviruses in normal oral mucosa: a comparison of methods for sample collection. J Oral Pathol Med. 1992;21:265–9. doi: 10.1111/j.1600-0714.1992.tb01008.x. [DOI] [PubMed] [Google Scholar]
- 6.Heath EM, Morken NW, Campbell KA, Tkach D, Boyd EA, Strom DA. Use of buccal cells collected in mouthwash as a source of DNA for clinical testing. Arch Pathol Lab Med. 2001;125:127–33. doi: 10.5858/2001-125-0127-UOBCCI. [DOI] [PubMed] [Google Scholar]
- 7.D'Souza G, Sugar E, Ruby W, Gravitt P, Gillison ML. Analysis of the Effect of DNA Purification on Detection of Human Papillomavirus in Oral Rinse Samples by PCR. J Clin Microbiol. 2005;43:5526–35. doi: 10.1128/JCM.43.11.5526-5535.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knepp JH, Geahr MA, Forman MS, Valsamakis A. Comparison of automated and manual nucleic acid extraction methods for detection of enterovirus RNA. J Clin Microbiol. 2003;41:3532–6. doi: 10.1128/JCM.41.8.3532-3536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riemann K, Adamzik M, Frauenrath S, Egensperger R, Schmid KW, Brockmeyer NH, et al. Comparison of manual and automated nucleic acid extraction from whole-blood samples. J Clin Lab Anal. 2007;21:244–8. doi: 10.1002/jcla.20174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller S, Seet H, Khan Y, Wright C, Nadarajah R. Comparison of QIAGEN automated nucleic acid extraction methods for CMV quantitative PCR testing. Am J Clin Pathol. 2010 Apr;133(4):558–63. doi: 10.1309/AJCPE5VZL1ONZHFJ. [DOI] [PubMed] [Google Scholar]
- 11.Yuan CC, Miley W, Waters D. A quantification of human cells using an ERV-3 real time PCR assay. J Virol Methods. 2001;91:109–17. doi: 10.1016/s0166-0934(00)00244-5. [DOI] [PubMed] [Google Scholar]
- 12.Fitzmaurice G. Statistical methods for assessing agreement. Nutrition. 2002;18(7-8):694–6. doi: 10.1016/s0899-9007(02)00802-x. [DOI] [PubMed] [Google Scholar]
- 13.Frisch M, Biggar RJ, Goedert JJ. Human papillomavirus-associated cancers in patients with human immunodeficiency virus infection and acquired immunodeficiency syndrome. J Natl Cancer Inst. 2000;92:1500–10. doi: 10.1093/jnci/92.18.1500. [DOI] [PubMed] [Google Scholar]
- 14.Kreimer AR, Alberg AJ, Daniel R, Gravitt PE, Viscidi R, Garrett ES, et al. Oral human papillomavirus infection in adults is associated with sexual behavior and HIV serostatus. J Infect Dis. 2004;189:686–98. doi: 10.1086/381504. [DOI] [PubMed] [Google Scholar]


