Abstract
Folate-mediated one-carbon metabolism is required for the de novo synthesis of purines, thymidylate, and S-adenosylmethionine, the primary cellular methyl donor. Impairments in folate metabolism diminish cellular methylation potential and genome stability, which are risk factors for colorectal cancer (CRC). Cytoplasmic serine hydroxymethyltransferase (SHMT1) regulates the partitioning of folate-activated one-carbons between thymidylate and S-adenosylmethionine biosynthesis. Therefore, changes in SHMT1 expression enable the determination of the specific contributions made by thymidylate and S-adenosylmethionine biosynthesis to CRC risk. Shmt1 hemizygosity was associated with a decreased capacity for thymidylate synthesis, due to down regulation of enzymes in its biosynthetic pathway, namely thymidylate synthase and cytoplasmic thymidine kinase. Significant Shmt1-dependent changes to methylation capacity, gene expression and purine synthesis were not observed. Shmt1 hemizygosity was associated with increased risk for intestinal cancer in Apcmin/+ mice through a gene-by-diet interaction, indicating that the capacity for thymidylate synthesis modifies susceptibility to intestinal cancer in Apcmin/+ mice.
Keywords: Cytoplasmic serine hydroxymethyltransferase, thymidylate synthesis, folate, colon cancer, Apc
Introduction
The interactions among nutrients and genetic factors play an important role in the development of numerous cancers, including colorectal cancer (CRC). A strong, inverse association of folate status and CRC has been demonstrated; individuals with lowest dietary folate intake show a 40–60% increase in CRC risk when compared to individuals with highest folate intake (1–3). Genetic variation that alters folate metabolism and utilization also influences cancer risk (2). The mechanism by which folate metabolism alters CRC risk is not known, which has led to concerns regarding the potential impact of elevated dietary folate intake and folate fortification initiatives on CRC incidence (4, 5).
Folate-mediated one-carbon (1C) metabolism is a metabolic network of interdependent biosynthetic pathways required for the de novo biosynthesis of purines, dTMP, and the remethylation of homocysteine to methionine (Fig. 1)(6). Methionine can be converted to S-adenosylmethionine (AdoMet), the major 1C donor for cellular methylation reactions including the methylation of DNA, RNA, phospholipids, proteins and small molecules (6, 7). Impairments in 1C metabolism due to nutrient deficiencies and/or single nucleotide polymorphisms diminish dTMP synthesis, leading to elevated dUTP pools, increased rates of dUTP misincorporation into DNA, and consequently futile cycles of DNA excision repair and chromosomal strand breaks (8–10). Altered folate metabolism also influences chromatin methylation patterns, including genome-wide CpG hypomethylation and site-specific hypermethylation, and altered histone methylation, which modify gene expression patterns (11–13). Therefore, the loss of DNA integrity due to increased genome instability and/or changes in gene expression due to altered genome methylation are candidate causal pathways for folate-mediated CRC.
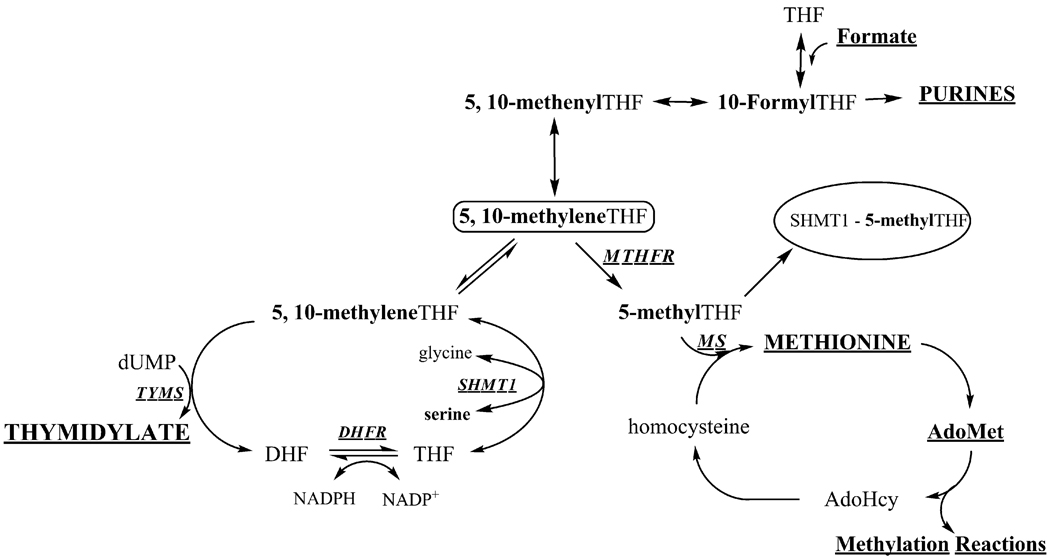
Figure 1. Folate-mediated one-carbon metabolism.
Tetrahydrofolate (THF)-mediated one-carbon metabolism is required for the synthesis of purines, thymidylate and the remethylation of homocysteine to methionine. The hydroxymethyl group of serine is the major source of one-carbon units which are generated in the mitochondria in the form of formate, or in the cytoplasm through the activity of SHMT1. Mitochondrial-derived formate can enter the cytoplasm and function as a one-carbon unit for folate metabolism. 5,10-methyleneTHF can be generated in the cytoplasm from formate or serine, and the sources of 5,10-methyleneTHF exist in equilibrium. The SHMT1 enzyme also inhibits homocysteine remethylation by sequestering 5-methylTHF in the cytoplasm. The de novo thymidylate synthesis pathway involves the three enzymes, SHMT1, TYMS and DHFR, while the salvage pathway involves TK1. Abbreviations: MTHFR, methylenetetrahydrofolate reductase; SHMT1, cytoplasmic serine hydroxymethyltransferase; DHFR, dihydrofolate reductase; TK1, cytoplasmic thymidine kinase; TYMS, thymidylate synthase; MS, methionine synthase, AdoMet, S-adenosylmethionine; AdoHcy, S-adenosylhomocysteine; THF, tetrahydrofolate. The one-carbon is labeled in bold.
Cytoplasmic serine hydroxymethyltransferase (SHMT1) acts as a metabolic switch that regulates the partitioning of 1C units between the dTMP and methionine biosynthetic pathways (Fig. 1)(14). Folate-activated 1Cs can be derived in the cytoplasm from formate through the Mthfd1 gene product or from serine through SHMT1 to support methionine and thymidylate biosynthesis (Figure 1). Thymidylate synthesis can occur in both the cytoplasm and nucleus (7, 15). When expressed, SHMT1 preferentially partitions methylenetetrahydrofolate (methyleneTHF) into the dTMP synthesis pathway through the small ubiquitin-like modifier (SUMO)-mediated compartmentalization of the dTMP biosynthetic pathway components in the nucleus during S-phase (14, 16). In the cytoplasm, SHMT1 tightly binds and sequesters 5-methylTHF making it unavailable for the methionine cycle, inhibiting AdoMet synthesis and reducing the cellular methylation potential (14).
Aberrant Wnt signaling resulting from altered Apc gene expression is observed in approximately 85% of cases of sporadic CRC (reviewed in (17)). In rats, folate deficiency has previously been associated with strand breaks in the mutation cluster region within the Apc gene, which were inversely correlated with Apc gene expression (18). Folate-mediated strand breaks in the Apc gene could be due to hypomethylation of the gene or genomic instability induced by misincorporation of uracil into genomic DNA. In the present study, we have modified the expression of SHMT1 in mice to determine the role(s) played by de novo purine, thymidylate and methionine synthesis in the development of Apcmin-mediated intestinal cancer.
Materials and Methods
Mice
SHMT1 null (Shmt1−/+) mice were generated as previously described (19) and backcrossed a minimum of 10 generations onto the C57BL/6J strain. Mice were genotyped using the forward primer 5'-GACACTGTTCACATCCCTC-3' and the reverse primer 5'-CAAAACATTCGGGAGCCTC-3'. The forward primer corresponds to an intron 6 sequence located 5' to a loxP site and exon 7 and the reverse primer corresponds to an intron 7 sequence located downstream of a 3' loxP site (19). C57BL/6J-ApcMin/J (Apcmin/+) mice were obtained from The Jackson Laboratory (Bar Harbor, ME). Genotyping of Apcmin/+ mice was performed using the following primers recommended by The Jackson Laboratory protocol: wildtype forward primer 5'-GCCATCCCTTCACGTTAG-3', Min forward primer 5'-TTCTGAGAAAGACAGAAGTTA-3' and a common reverse primer 5'-TTCCACTTTGGCATAAGGC-3'. Shmt1−/+ mice were mated to Apcmin/+ mice. Double heterozygous offspring were intercrossed to achieve Apcmin/+ Shmt1+/+, Apcmin/+ Shmt1−/+ or Apcmin/+ Shmt1−/− mice.
Mice were maintained under specific-pathogen free conditions at the Cornell University Transgenic Mouse Core Facility (Ithaca, NY) in accordance with standard use protocols and animal welfare regulations. All study protocols were approved by the Institutional Animal Care and Use Committee of Cornell University and conform to the NIH Guide for the Care and Use of Laboratory Animals.
Diets
Mice were randomly weaned onto either a control (AIN-93G; Dyets, Inc., Bethlehem, PA) diet or a modified AIN-93G diet lacking folic acid and choline (Dyets, Inc., Bethlehem, PA) at three weeks of age. The control diet contained 2 mg/kg folic acid and 2.5 g/kg choline bitartrate and the folate/choline deficient diet contained 0 mg/kg folic acid and 0 g/kg choline bitartrate. Mice were maintained on the diet for five weeks (Apc+/+) or 11 weeks (Apcmin/+).
Immunohistochemistry
The colon from wildtype FVBN mice were fixed as Swiss rolls in 10% neutral-buffered formalin. Samples were embedded in paraffin and sectioned (5 µm). Polyclonal sheep anti-SHMT1 antibody was used to probe the sections for cellular localization of SHMT1 (20). Non-immune sheep serum was used as a negative control. Biotinylated rabbit anti-sheep IgG was used as the secondary antibody. SHMT1 was visualized with AEC (3-amino-9-ethylcarbazole) developer (Dako, Carpinteria, CA). Sections were counterstained using Gill’s hematoxylin #2 (Sigma, St. Louis, MO). Coverslips were mounted using Fluoromount-G (Sigma).
Tumor assessment
The small intestine and colon were removed, flushed with cold phosphate-buffered saline, opened longitudinally, and laid flat lumen side up for examination using a dissecting microscope as previously described (21). Tumors were counted according to intestinal location (small intestine or colon) and their diameter measured by a pathologists-trained investigator who was blinded to the genotype of the intestinal specimen. Tumor load is a function of tumor number and area and was calculated as the total tumor area per mouse.
Plasma and tissue folate determination
Folate concentration of plasma and tissues was quantified using the Lactobacillus casei microbiological assay (14). Protein concentration was quantified (22).
Detection of uracil in nuclear DNA
Uracil content in hepatic nuclear DNA was determined by gas chromatography/mass spectrometry as previously described (19). Uracil content in liver nuclear DNA has been shown to correlate uracil content in colonic nuclear DNA in Shmt1−/+ mice (MacFarlane and Stover, in preparation)
Tissue AdoMet and AdoHcy determination
The animal feeding cycle was synchronized prior to tissue harvest to ensure AdoMet levels reflected homocysteine remethylation capacity with minimal contributions from dietary methionine. Food was removed 24h prior to killing the animals. After 12 hours, each animal was given one food pellet and the animals were killed 12h later by cervical dislocation. Tissues were harvested and immediately flash frozen and stored at −80°C until analysis. Frozen tissues were sonicated in 500 µl of 0.1 M NaAcO buffer (pH 6), and protein was precipitated by adding 312 µl of 10% perchloric acid to each sample. After vortexing, samples were centrifuged at 2000 × g for 10 min at 4°C. AdoMet and AdoHcy were determined by high performance liquid chromatography as described previously (14). AdoMet and AdoHcy values were normalized to cellular protein content (22).
Western blot analysis
Total protein was extracted and quantified from frozen tissue (22). Immunoblotting was performed as previously described (19). The membranes were visualized using the SuperSignal® West Pico chemiluminescent substrate system (Pierce, Rockford, IL). For actin detection, polyclonal rabbit anti-actin antibody conjugated to horseradish peroxidase (HRP; Abcam, Cambridge, MA) was diluted 1:40,000. For SHMT1 detection, sheep anti-mouse SHMT1 antibody (20) was diluted 1:10,000, and rabbit anti-sheep IgG secondary antibody conjugated to HRP (Pierce) was diluted 1:20,000. For TYMS detection, monoclonal mouse anti-TYMS antibody (Invitrogen, Carlsbad, CA) was diluted 1:3,000, and goat-anti-mouse IgG secondary antibody conjugated to HRP (Pierce) was diluted 1:5,000. For cytoplasmic thymidine kinase (TK1) detection, monoclonal mouse anti-TK1 antibody (Abcam) was diluted 1:1,000, and horse-anti-mouse IgG secondary antibody conjugated to HRP (Cell Signaling Technology, Danvers, MA) was diluted 1:1,000. Densitometric analysis of Western blots was performed using Scion Image for Windows (Scion Corporation, Frederick MD). Three animals per group were included in the analysis. Values were normalized to GAPDH. Densitometric data are presented relative to the mean of wildtype animals fed the control diet.
Mouse embryonic fibroblast isolation and maintenance
Mouse embryonic fibroblasts (MEFs) were freshly isolated as described in (19). Cells were cultured in alpha-minimal essential medium (alpha-MEM; Hyclone Laboratories, Logan, UT) supplemented with 10% fetal calf serum (FCS; Hyclone Laboratories), 0.1 mM nonessential amino acids (Invitrogen), 1mM sodium pyruvate (Invitrogen) and penicillin/streptomycin (Invitrogen) and incubated at 37 °C in a 5% CO2 atmosphere.
Purine synthesis assay
[14C]formate and [3H]hypoxanthine are precursors for purine nucleotide biosynthesis through the de novo and salvage pathways, respectively (23). For tracer experiments, FCS was dialyzed against PBS for over 24 h with six buffer changes to remove folate and other small molecules and then charcoal treated to remove any residual folate. The tracer medium was Defined Minimal Essential Medium (Hyclone Laboratories) that lacked glycine, serine, methionine, hypoxanthine, and folate but was supplemented with 10% dialyzed and charcoal treated FCS, 1 mg/L pyridoxyl-L-phosphate, 200 µM methionine, 20 nM leucovorin, 2 nM [3H]hypoxanthine (Moravek, Brea, CA), and 20 µM [14C]formate (Moravek). MEFs were plated 2 × 104 cells/well in 6-well plates and grown at 37 °C, 5% CO2 in tracer medium until confluent and harvested. The cell pellets were stored at −80 °C. Nuclear DNA was isolated using a DNA blood kit from Qiagen (Valencia, CA), RNase A treated and isotope levels quantified on a Beckman LS6500 scintillation counter in dual dpm mode.
Thymidylate synthesis assay
[14C]deoxyuridine and [3H]thymidine are precursors for thymidylate nucleotide biosynthesis through the de novo and salvage pathways, respectively. The tracer medium was Defined Minimal Essential Medium (Hyclone Laboratories) that lacked glycine, serine, methionine, hypoxanthine, and folate but was supplemented with 10% dialyzed and charcoal treated FCS, 1 mg/L pyridoxyl-L-phosphate, 200 µM methionine, 20 nM leucovorin, 500 nM [3H]thymidine (Moravek), and 10 µM [14C]deoxyuridine (Moravek). MEFs were plated 2 × 104 cells/well in 6-well plates and grown at 37 °C, 5% CO2 in tracer medium until confluent and harvested. The cell pellets were stored at −80 °C. Nuclear DNA was isolated using a DNA blood kit from Qiagen with RNase A treatment and isotope levels quantified on a Beckman LS6500 scintillation counter in dual dpm mode. Data were normalized to the wildtype MEF line in each independent experiment.
Microarray analysis
Microarray analysis was performed on colon tissue from Shmt1+/+, Shmt1−/+ and Shmt1−/− mice fed the AIN-93G minus folate and choline diet. Total RNA was isolated from colons and 50 ng was amplified using the Nugen Ovation V2 kit (Nugen, San Carlos, CA). Amplified RNA from nine mice (three independently processed samples from each genotype) was hybridized to Affymetrix 430A mouse gene expression chips (Affymetrix, Santa Clara, CA) and scanned using an Affymetrix GeneChip 3000 scanner. The raw array data was processed using GCOS software to obtain signal values scaled to a default target of 500. Further statistical data analysis was performed using R (24). The signal values were log2-transformed after offset by 8, and gene filtering was applied to include only the 19871 probe sets having at least one present call in the dataset. A two sample t-test was applied on the normalized log ratio of each probe set. At a p value cutoff of 0.01, 134 differential genes between wildtype and null mice were selected, while 229 differential genes were selected between wildtype and heterozygous mice (Table S1). Because only three replicates were performed for each sample, multiple testing correction was not performed. The microarray data have been deposited in NCBI's Gene Expression Omnibus (25) and are accessible through GEO Series accession number GSE14645.
Quantitative RT-PCR
Gene expression levels of candidate genes identified in the microarray screen were tested by quantitative RT-PCR using the Taqman-based Universal Probe Library (Roche). Colon RNA used in the microarray screen was primed with oligo-dT and reverse transcribed with Superscript III (Invitrogen) to generate cDNA template. Quantitative RT-PCR primers were designed using the Universal Probe Library Assay Design Center, Roche (26) and validated on an Applied Biosystems 7500 machine by comparison against a standard curve generated with rodent GAPDH control primers (Applied Biosystems, Foster City, CA). Relative quantitation analysis was then performed in triplicate for all nine samples. Genes tested, Affymetrix ID, primer sequences, and universal probe library numbers used are in Table S2.
Statistical analyses
Differences between two groups were determined by Student’s t-test analysis. Differences among more than two groups were analyzed by two-way ANOVA and Tukey’s HSD post-hoc test. Non-normally distributed data was normalized by log transformation for analysis. Groups were considered significantly different when the P value ≤ 0.05. All statistics were performed using JMP IN software, release 5.1.2 (Copyright © 1989–2004 SAS Institute Inc.).
Results
Localization of SHMT1 in colonic epithelial cells
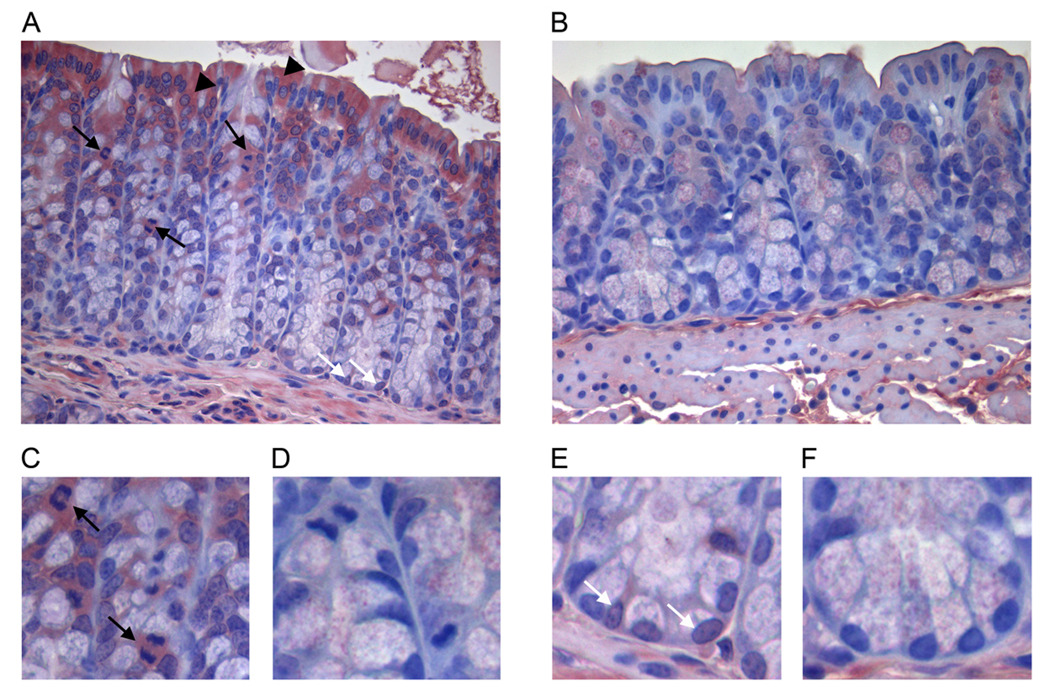
SHMT1 exhibits tissue-specific expression and is present in all tissues associated with folate-mediated pathologies, including the colon (19, 27, 28). Here we show that SHMT1 is specifically expressed in the cytoplasm of mature colonic enterocytes (Fig. 2A), in mitotically active epithelial cells in the proliferative zone of colonic crypts (Fig. 2A and C), and in the nucleus of cells residing at the base of colonic crypts (Fig. 2A and E).
Figure 2. SHMT1 localization in mouse colonocytes.
SHMT1 was detected in histologic sections of colon from wildtype FVBN mice probed with polyclonal sheep anti-SHMT1 antibody. A, SHMT1 is expressed in the cytoplasm of colonic enterocytes (black arrowheads), in mitotically active epithelial cells in the proliferative zone of the colonic crypts (black arrows), and in the nucleus of cells residing in the base of the crypts (white arrows). C, magnification of mitotically active cells expressing SHMT1. E, SHMT1 is expressed in the nucleus of cells residing at the base of the colonic crypts, indicated by white arrows. B, D and F, mouse colon probed with non-immune sheep serum.
SHMT1 and diet interact to influence intestinal tumor development
To determine the effect of Shmt1 on intestinal tumor development, Shmt1−/+ mice (19), were mated to Apcmin/+ mice, a model of spontaneous intestinal neoplasia (29). Double heterozygous offspring were intercrossed to achieve Apcmin/+ Shmt1+/+, Apcmin/+ Shmt1−/+ or Apcmin/+ Shmt1−/− mice. Mice were weaned at 21 days and randomly assigned to either a control (C) or a folate/choline deficient (FCD) diet. Plasma and colon folate concentrations were determined in Apc+/+ littermates after 5 weeks on diet to estimate folate status at the mid-point of the experiment. Folate in colon was significantly reduced by the FCD diet after 5 weeks (Table 1). This confirms that the colon is sensitive to dietary folate depletion (30). Liver folate concentrations were significantly reduced in mice only after 11 weeks on diet (Table 1).
Table 1.
Tissue folate concentrations in Apc+/+ Shmt1−/+ mice on diet for five weeks and Apcmin/+ Shmt1−/+ mice on diet for 11 weeks. Data are presented as mean ± SEM. n = 3–4.
| Apc+/+ mice on diet for 5 weeks |
Apcmin/+ mice on diet for 11 weeks |
|||||
|---|---|---|---|---|---|---|
| Diet |
Shmt1 Genotype |
Plasma (ng/ml) |
Liver (fmol/µg protein) |
Colon (fmol/µg protein) |
Plasma (ng/ml) |
Liver (fmol/µg protein) |
| Control | Shmt1+/+ | 58.56 ± 10.75 | 51.80 ± 2.94 | 35.14 ± 10.32 | 24.68 ± 7.50 | 45.72 ± 9.50 |
| Shmt1−/+ | 58.34 ± 7.05 | 56.65 ± 8.39 | 21.46 ± 4.02 | 20.91 ± 5.01 | 40.00 ± 3.24 | |
| Shmt1−/− | 40.82 ± 7.78 | 50.77 ± 8.80 | 17.09 ± 2.10 | 26.44 ± 3.08 | 41.50 ± 2.69 | |
| FCD | Shmt1+/+ | 20.60 ± 1.80 | 47.26 ± 5.77 | 9.15 ± 2.02 | 11.79 ± 2.93 | 28.49 ± 2.68 |
| Shmt1−/+ | 38.95 ± 2.84 | 44.30 ± 5.59 | 18.04 ± 2.25 | 8.37 ± 1.56 | 23.25 ± 1.85 | |
| Shmt1−/− | 8.52 ± 8.52 | 48.88 ± 3.06 | 14.89 ± 4.24 | 9.97 ± 2.32 | 29.44 ± 2.71 | |
| P value, diet effect | 0.0003 | ns | 0.03 | 0.0009 | 0.002 | |
| P value, genotype effect | 0.021 | ns | ns | ns | ns | |
| P value, Diet × genotype effect | ns | ns | ns | ns | ns | |
Shmt1−/+ vs. Shmt1−/− are significantly different independent of diet, p<0.05
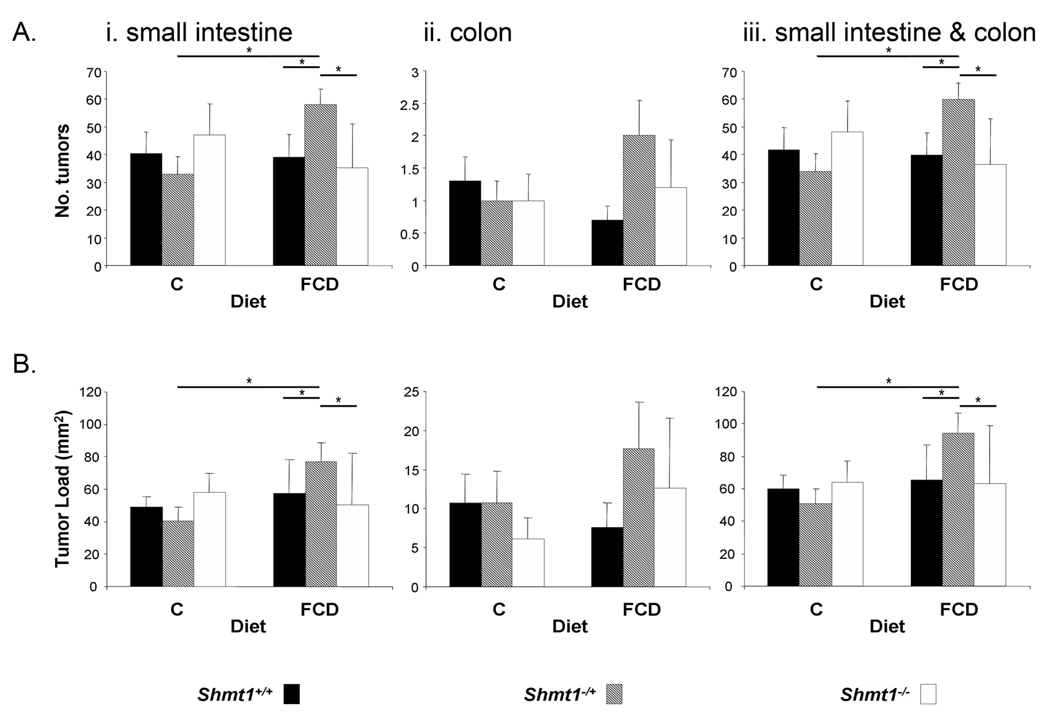
The small and large intestines from Apcmin/+ mice were examined for neoplasia at 14 weeks of age, after 11 weeks on diet. In mice fed the control diet, decreased SHMT1 expression had no effect on tumor number or load (Fig. 3A and B). The FCD diet was not associated with increased tumor number or load in Apcmin/+ Shmt1+/+ or Shmt1−/− mice. However, the FCD diet was associated with more than a 50% increase in total tumor number and load in Apcmin/+ Shmt1−/+ mice (Fig. 3, A and B). The effect in Apcmin/+ Shmt1−/+ mice on tumor number was driven by an approximate 50% increase in small intestinal tumors and a doubling of colon tumors, although the latter was not statistically significant
Figure 3. Tumor number and load in Apcmin/+ Shmt1+/+, Apcmin/+ Shmt1−/+ or Apcmin/+ Shmt1−/− mice.
A, tumor number in the small intestine (i), colon (ii) and combined small intestine and colon (iii). B, tumor load in the small intestine (i), colon (ii) and combined small intestine and colon (iii). Tumor load was calculated as the total tumor area per mouse. n = 10–13 for Shmt1+/+ and Shmt1−/+ mice, n = 4–5 for Shmt1−/− mice. Data are presented as mean ± SEM. *, p≤0.05.
De novo purine synthesis is not dependent on SHMT1 status
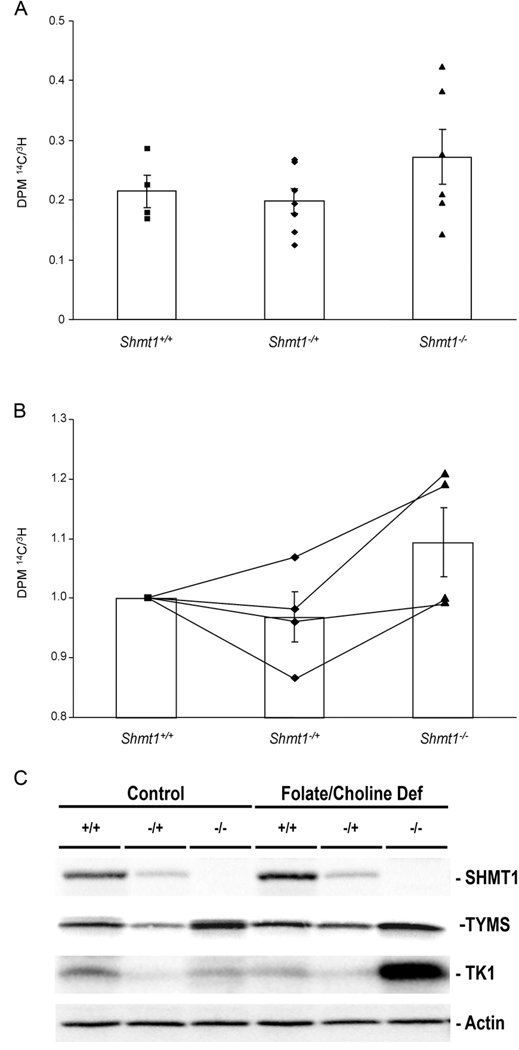
To determine the mechanism by which Shmt1 hemizygosity modifies CRC susceptibility in Apcmin/+ mice, the effects of SHMT1 depletion on de novo purine and thymidylate biosynthesis, and cellular methylation potential were determined. Mouse embryonic fibroblasts (MEFs) were employed to determine the impact of Shmt1 hemizygosity on de novo nucleotide synthesis relative to synthesis by the salvage pathway. We found that de novo purine synthesis relative to synthesis by the salvage pathway was not significantly affected by Shmt1 genotype in MEFs (Fig. 4A).
Figure 4. Purine and thymidylate synthesis.
A, [14C]formate and [3H]hypoxanthine are precursors for purine nucleotide biosynthesis through the de novo and salvage pathways, respectively. Data are presented as a ratio of de novo to salvage purine synthesis ([14C]formate/[3H]hypoxanthine). n = 4–7 independent MEF lines per genotype. B, [14C]deoxyuridine and [3H]thymidine are precursors for thymidylate nucleotide biosynthesis through the de novo and salvage pathways, respectively. Data are presented as a ratio of de novo to salvage thymidylate synthesis ([14C]deoxyuridine/[3H]thymidine). p=0.08 for a genotype effect. n = 4 independent MEF lines per genotype. Data were normalized to wildtype for independent experiments. Lines connect independent experiments. C, Representative western blots for SHMT1, TYMS, TK1 and actin in colon tissue from Apc+/+ Shmt1+/+, Shmt1−/+ and Shmt1−/− mice fed either a control or folate/choline deficient diet for five weeks from weaning.
SHMT1, cellular methylation potential and gene expression
Our previous data suggest that Shmt1−/− mice have increased hepatic AdoMet concentrations compared to wildtype and hemizygous mice under conditions of folate depletion (19). However, AdoMet and AdoHcy levels, as well as the AdoMet/AdoHcy ratio were not significantly affected by Shmt1 genotype or diet in this study (Table 2).
Table 2.
Liver AdoMet, AdoHcy, AdoMet/AdoHcy ratio and uracil content in nuclear DNA inApcmin/+Shmt1−/+ mice after 11 weeks on diet. Data are presented as mean ± SEM values. n = 3–7 per group.
| Diet |
Shmt1 Genotype |
AdoMet (pmol/µg protein) |
AdoHcy (pmol/µg protein) |
AdoMet/ AdoHcy |
Uracil (pg/µg DNA) |
|---|---|---|---|---|---|
| AIN-93G | Shmt1+/+ | 1.4 ± 0.4 | 0.9 ± 0.2 | 1.9 ± 0.4 | 0.1 ± 0.1 |
| Shmt1−/+ | 0.8 ± 0.2 | 0.6 ± 0.0 | 1.5 ± 0.3 | 0.3 ± 0.1 | |
| Shmt1−/− | 1.5 ± 0.3 | 0.9 ± 0.1 | 1.7 ± 0.4 | 0.1 ± 0.0 | |
| AIN-93G minus folate & choline | Shmt1+/+ | 1.4 ± 0.4 | 0.7 ± 0.1 | 1.9 ± 0.4 | 0.2 ± 0.0 |
| Shmt1−/+ | 0.8 ± 0.1 | 0.6 ± 0.1 | 1.3 ± 0.1 | 0.6 ± 0.2 | |
| Shmt1−/− | 1.9 ± 0.8 | 0.8 ± 0.2 | 2.0 ± 0.6 | 0.2 ± 0.1 | |
| P value, diet effect | ns | Ns | ns | 0.06 | |
| P value, genotype effect | 0.10 | Ns | ns | 0.011 | |
| P value, Diet × genotype effect | ns | Ns | ns | ns | |
Shmt1−/+ vs. Shmt1+/+ and Shmt1−/− are significantly different independent of diet, p<0.05
In addition, we performed a microarray analysis on colons from mice fed the FCD diet to determine the effect of Shmt1 on gene expression and its relationship to tumor susceptibility. Using the NuGen Ovation amplification approach, a Present Call of between 78–81% was achieved, indicating high sensitivity in transcript detection. However, at a p-value of 0.01, less than ten genes in Shmt1 hemizygous mice and less than 15 genes in null mice of the possible ~19,000 Present genes had a two-fold or greater change in gene expression. These minimal differences strongly suggest that gene expression changes in Shmt1 mutants would be difficult to detect (Table S1). The data were confirmed by quantitative RT-PCR for 9 genes of interest (Tables S2). Only one gene, Upp1, demonstrated consistent changes in expression.
SHMT1 modifies thymidylate synthesis capacity
Previously, we have shown that uracil content in hepatic nuclear DNA was significantly affected by Shmt1 genotype (19). Here we have found that uracil content in hepatic nuclear DNA, which correlates with colon uracil content in Shmt1 knock-out mice (MacFarlane and Stover, in preparation), was significantly increased in Apcmin/+ Shmt1−/+ mice in comparison with Apcmin/+ Shmt1+/+ mice, and uracil content doubled when the Apcmin/+ Shmt1−/+ mice were fed the FCD diet (Table 2). While uracil content also doubled in Apcmin/+ Shmt1+/+ fed the FCD diet, the absolute concentration was consistently one third that observed in Apcmin/+ Shmt1−/+ regardless of diet. In addition, MEFs derived from Shmt1−/+ mice demonstrated a tendency for decreased de novo thymidylate synthesis relative to the salvage pathway in comparison with Shmt1+/+ MEFs as determined by a modified deoxyuridine suppression assay (31) (Fig. 4B). Furthermore, the protein expression of thymidylate synthase (TYMS), which functions in de novo thymidylate synthesis was decreased in the colon of Shmt1−/+ mice in comparison with wildtype mice fed the control diet (Fig. 4C; 0.69 ± 0.3 vs 1.00 ± 0.04). TK1 protein expression was not different between hemizygous and wildtype mice. The data indicate that Shmt1−/+ mice experience a reduction in de novo thymidylate synthesis capacity relative to their wildtype counterparts, which likely accounts for the increased incorporation of uracil into nuclear DNA observed in these mice.
Surprisingly, Shmt1−/− mice had lower uracil levels in nuclear DNA compared to Shmt1−/+ mice (Table 2) and MEFs from Shmt1−/− mice exhibited increased de novo thymidylate biosynthesis relative to synthesis from salvage compared to those derived from Shmt1−/+ mice (Fig. 4B). Shmt1−/− mice demonstrated markedly elevated colonic TYMS (2.86 ± 1.3 vs 1.00 ± 0.04) and TK1 (2.19 ± 1.0 vs. 0.97 ± 0.06) protein expression in comparison to Shmt1 wildtype mice. While the results were unexpected, the changes in TYMS and TK1 protein expression observed in Shmt1−/− mice could explain their increased capacity for thymidylate synthesis and decreased uracil concentration in genomic DNA in comparison with Shmt1−/+ mice.
Dietary folate regulation of SHMT1, thymidylate synthase and thymidine kinase
In addition to Shmt1-dependent changes to thymidylate synthesis capacity, we also observed an up regulation of members of the de novo and salvage dTMP biosynthetic pathways in response to folate deficiency. SHMT1 protein was increased by 35% (1.2 ± 0.4 vs. 0.9 ± 0.1) and TYMS protein was increased by 80% (1.8 ± 0.6 vs. 1.00 ± 0.04), as determined by densitometry, in the colons of Shmt1 wildtype mice fed the FCD diet in comparison to control fed mice (Fig. 4C), which could account for the lack of an effect of folate deficiency on uracil content in DNA (Table 2) and intestinal tumor number and load in wildtype mice (Fig. 3A and B). Although SHMT1 (0.56 ± 0.2 vs. 0.4 ± 0.06) and TYMS (1.7 ± 0.5 vs. 0.7 ± 0.3) levels were also increased in Shmt1−/+ mice fed the FCD diet, the absolute levels of these enzymes never exceeded that observed in wildtype mice (Fig. 4C). TK1 protein levels were also increased when wildtype mice were fed the FCD diet (1.0 ± 0.06 vs. 2.0 ± 0.8), which could support an increased capacity to synthesize thymidylate from the salvage pathway and also contribute to their protection from diet-mediated cancer susceptibility. TK1 also increased in Shmt1−/+ mice fed the FCD diet (1.5 ± 0.1 vs. 1.0 ± 0.6), but the induction was to a lesser extent than that observed in wildtype mice. Finally, TK1 remained high but unchanged in Shmt1−/− mice fed the FCD diet (2.0 ± 0.5 vs. 2.2 ± 1.1).
Discussion
Altered folate metabolism has been shown to be a risk factor for the development of CRC. However, the mechanism(s) involved remain unknown. Because of its metabolic properties and its cellular localization, SHMT1 is poised to regulate 1C metabolism in cells associated with neoplastic transformation and CRC risk (32). Therefore, SHMT1 expression was modified to determine the relative contributions of dTMP, purine and AdoMet synthesis, and consequently indicators of genome stability and methylation, to intestinal tumorigenesis.
Three biosynthetic pathways, purine, thymidylate and methionine synthesis, compete for 5,10-methyleneTHF, a limited cofactor in the cell. C1THF synthase can convert 5,10-methyleneTHF to 10-formylTHF, the substrate required for de novo synthesis of purines (33). While purine deprivation or inhibition of de novo purine synthesis is usually associated with G1 arrest, cytostasis and cytotoxicity (34, 35), reduced purine synthesis capacity has also been associated with abnormal DNA repair and DNA mutagenesis in mammalian cells (36, 37), and could thereby play a role in Apc-mediated intestinal tumorigenesis. However, we observed that de novo purine synthesis relative to synthesis by the salvage pathway was not significantly affected by Shmt1 genotype in mouse embryonic fibroblasts and therefore does not likely contribute to the increased tumor numbers observed in Apcmin/+ Shmt1−/+ mice.
5,10-methyleneTHF can also be irreversibly reduced by methyleneTHF reductase (MTHFR) to form 5-methyTHF, from which a methyl group is transferred to homocysteine by methionine synthase to form methionine. Methionine can then be S-adenosylated to form AdoMet, the major methyl donor in cellular methylation reactions. The balance between AdoMet and AdoHcy, the substrate and product, respectively, of transmethylation reactions is an indicator of cellular methylation potential. Cellular methylation potential can impact DNA, RNA and histone methylation patterns and has been associated with aberrant gene expression patterns in cancer. SHMT1 regulates the partitioning of methyl groups between the thymdylate and methionine synthesis pathways by preferentially shuttling its product, 5,10-methyleneTHF, towards thymidylate synthesis, as well as by tightly binding 5-methylTHF making it unavailable to the methionine cycle (14). Apcmin/+ Shmt1−/− mice tended to have increased hepatic AdoMet under folate deficient conditions, however, unlike our previous findings the difference was not statistically significant (19). We determined Shmt1-dependent changes in gene expression patterns in hemizygous mice fed the FCD diet, which were minimal, with only 10 genes identified that had significant changes in expression greater than 2 fold in Shmt1−/+ mice compared to wildtype mice. One interesting note is that the only gene that demonstrated consistent changes in expression, as confirmed by RT-PCR, was uridine phosphorylase, Upp1, which is involved in pyrimidine degradation and uridine homeostasis, indicating another mechanism by which SHMT1 status could impact on thymidylate synthesis capacity (38). A limitation to our microarray analysis was that it did not determine diet or gene-by-diet effects as mice fed the control diet were not included in the analysis. However, together the data suggest that cellular methylation potential and any associated changes in gene expression did not significantly contribute to tumorigenesis in this mouse model.
Finally, 5,10-methyleneTHF can be utilized by TYMS to methylate dUMP for the de novo synthesis of dTMP. We have shown that SHMT1 preferentially partitions one carbon units into the de novo thymidylate biosynthetic pathway (14). Consistent with our previous observations, we saw a decrease in de novo thymidylate synthesis in Shmt1−/+ mice, which was associated with an increase in uracil incorporation into nuclear DNA. Interestingly, Shmt1−/+ mice also demonstrated decreased TYMS protein expression. TK1 was induced by the FCD diet in hemizygous mice, but to a lesser extent than that observed in wiltype mice. A reduction in TK1 would result in decreased uptake of thymidine from the salvage pathway, in addition to an overall dampening of the capacity to synthesize dUMP, and subsequently dTMP (Fig. 1), while a reduction in TYMS would specifically decrease the capacity for de novo thymidylate synthesis. We conclude that impairments in de novo thymidylate biosynthesis, resulting from Shmt1 hemizygosity and its consequent effect on TYMS and TK1 protein levels, exacerbate the tumor phenotype of Apcmin/+ mice fed the FCD diet. Observed increases in SHMT1, TYMS and TK1 in wildtype mice fed the FCD diet or dramatic increases in TYMS and TK1 in Shmt1−/− mice could serve as a protective mechanism to maintain adequate dTMP synthesis thereby minimizing uracil misincorporation into nuclear DNA and consequent genome instability.
In the Apcmin/+ mouse model of CRC, Shmt1 hemizygosity did not significantly affect either de novo purine synthesis or cellular methylation potential, indicating that these pathways were unlikely to contribute to the increased tumorigenesis observed in Shmt1−/+ mice. However, changes in dTMP synthesis were significantly and inversely correlated with increased risk for intestinal cancer development in a gene-by-diet interaction. Furthermore, de novo thymidylate synthesis capacity appears to be dynamically regulated by diet and Shmt1 expression. TYMS and TK1 protein levels respond to changes in SHMT1 expression, and TYMS, TK1 and SHMT1 levels respond to dietary folate/choline deficiency. As a result of these dynamic changes, thymidylate synthesis capacity, as indicated by uracil content in nuclear DNA and metabolic flux assays in MEFs, was lowest in Shmt1−/+ mice, which exhibited the highest tumor number and load in Apcmin/+ mice under folate/choline deficient conditions.
Loss of heterozygosity of the Apc gene is an early and necessary event for intestinal neoplastic transformation in the Apcmin/+ mice (39). It has previously been shown that colonic folate 1concentrations correlate with strand breaks in the Apc gene in rats fed a severely folate deficient diet for 5 weeks and that the strand breaks were inversely correlated with steady-state transcript levels of Apc (18). In addition, mild folate depletion in combination with other B vitamins, including vitamin B6, B12 and methionine, was associated with DNA strand breaks within the Apc mutation cluster region, decreased expression of a number of genes involved in the Wnt signaling pathway, and suppression of apoptosis in colonocytes (30). The increased uracil misincorporation into DNA and subsequent genome instability associated with Shmt1 hemizygosity in combination with dietary folate deficiency accelerated the loss of heterozygosity of the Apc gene and resulted in increased cellular transformation and tumor development. Compensatory mechanisms appear to buffer thymidylate synthesis capacity under folate-restricted conditions, as demonstrated by the up regulation of SHMT1, TYMS and TK1 in the colon of FCD-fed mice, which provide protection from loss of Apc heterozygosity and tumorigenesis. Together, our data provide a plausible mechanism by which folate-dependent thymidylate synthesis and consequential changes to genomic stability contribute to Apc-mediated intestinal cancer.
Supplementary Material
Aknowledgements
We would like to thank Sylvia Allen and Rachel Slater for technical assistance. We would also like to thank the Cornell Microarray Core and Dr. Wei Wang for performing the hybridization and analysis. This work was supported by Public Health Service grant CA105440 to PJS.
Grant support: This work was supported by Public Health Service grant CA105440 to PJS.
Footnotes
Conflict of interest: None declared.
REFERENCES
- 1.Giovannucci E, Stampfer MJ, Colditz GA, et al. Folate, methionine, and alcohol intake and risk of colorectal adenoma. J Natl Cancer Inst. 1993;85:875–884. doi: 10.1093/jnci/85.11.875. [DOI] [PubMed] [Google Scholar]
- 2.Ma J, Stampfer MJ, Giovannucci E, et al. Methylenetetrahydrofolate reductase polymorphism, dietary interactions, and risk of colorectal cancer. Cancer Res. 1997;57:1098–1102. [PubMed] [Google Scholar]
- 3.Kim J, Kim DH, Lee BH, et al. Folate intake and the risk of colorectal cancer in a Korean population. Eur J Clin Nutr. 2009;63:1057–1064. doi: 10.1038/ejcn.2009.37. [DOI] [PubMed] [Google Scholar]
- 4.Cole BF, Baron JA, Sandler RS, et al. Folic acid for the prevention of colorectal adenomas: a randomized clinical trial. JAMA. 2007;297:2351–2359. doi: 10.1001/jama.297.21.2351. [DOI] [PubMed] [Google Scholar]
- 5.Logan RF, Grainge MJ, Shepherd VC, Armitage NC, Muir KR. Aspirin and folic acid for the prevention of recurrent colorectal adenomas. Gastroenterology. 2008;134:29–38. doi: 10.1053/j.gastro.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 6.Shane B. Folate Chemistry and Metabolism. In: Bailey LB, editor. Folate in Health and Disease. New York: Marcel Dekker, Inc.; 1995. pp. 1–22. [Google Scholar]
- 7.Fox JT, Stover PJ. Folate-mediated one-carbon metabolism. Vitam Horm. 2008;79:1–44. doi: 10.1016/S0083-6729(08)00401-9. [DOI] [PubMed] [Google Scholar]
- 8.James SJ, Yin L. Diet-induced DNA damage and altered nucleotide metabolism in lymphocytes from methyl-donor-deficient rats. Carcinogenesis. 1989;10:1209–1214. doi: 10.1093/carcin/10.7.1209. [DOI] [PubMed] [Google Scholar]
- 9.Branda RF, Blickensderfer DB. Folate deficiency increases genetic damage caused by alkylating agents and gamma-irradiation in Chinese hamster ovary cells. Cancer Res. 1993;53:5401–5408. [PubMed] [Google Scholar]
- 10.Duthie SJ, Hawdon A. DNA instability (strand breakage, uracil misincorporation, and defective repair) is increased by folic acid depletion in human lymphocytes in vitro. Faseb J. 1998;12:1491–1497. [PubMed] [Google Scholar]
- 11.Wainfan E, Poirier LA. Methyl groups in carcinogenesis: effects on DNA methylation and gene expression. Cancer Res. 1992;52:2071s–2077s. [PubMed] [Google Scholar]
- 12.Friso S, Choi SW, Girelli D, et al. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc Natl Acad Sci U S A. 2002;99:5606–5611. doi: 10.1073/pnas.062066299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaudet F, Hodgson JG, Eden A, et al. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 14.Herbig K, Chiang EP, Lee LR, Hills J, Shane B, Stover PJ. Cytoplasmic serine hydroxymethyltransferase mediates competition between folate-dependent deoxyribonucleotide and S-adenosylmethionine biosyntheses. J Biol Chem. 2002;277:38381–38389. doi: 10.1074/jbc.M205000200. [DOI] [PubMed] [Google Scholar]
- 15.Anderson DD, Stover PJ. SHMT1 and SHMT2 are functionally redundant in nuclear de novo thymidylate biosynthesis. PLoS ONE. 2009;4:e5839. doi: 10.1371/journal.pone.0005839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woeller CF, Anderson DD, Szebenyi DM, Stover PJ. Evidence for small ubiquitin-like modifier-dependent nuclear import of the thymidylate biosynthesis pathway. J Biol Chem. 2007;282:17623–17631. doi: 10.1074/jbc.M702526200. [DOI] [PubMed] [Google Scholar]
- 17.Fodde R, Brabletz T. Wnt/beta-catenin signaling in cancer stemness and malignant behavior. Curr Opin Cell Biol. 2007;19:150–158. doi: 10.1016/j.ceb.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 18.Kim YI, Shirwadkar S, Choi SW, Puchyr M, Wang Y, Mason JB. Effects of dietary folate on DNA strand breaks within mutation-prone exons of the p53 gene in rat colon. Gastroenterology. 2000;119:151–161. doi: 10.1053/gast.2000.8518. [DOI] [PubMed] [Google Scholar]
- 19.MacFarlane AJ, Liu X, Perry CA, et al. Cytoplasmic serine hydroxymethyltransferase regulates the metabolic partitioning of methylenetetrahydrofolate but is not essential in mice. J Biol Chem. 2008;283:25846–25853. doi: 10.1074/jbc.M802671200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu X, Szebenyi DM, Anguera MC, Thiel DJ, Stover PJ. Lack of catalytic activity of a murine mRNA cytoplasmic serine hydroxymethyltransferase splice variant: evidence against alternative splicing as a regulatory mechanism. Biochemistry. 2001;40:4932–4939. doi: 10.1021/bi002598t. [DOI] [PubMed] [Google Scholar]
- 21.Chiu CH, McEntee MF, Whelan J. Sulindac causes rapid regression of preexisting tumors in Min/+ mice independent of prostaglandin biosynthesis. Cancer Res. 1997;57:4267–4273. [PubMed] [Google Scholar]
- 22.Bensadoun A, Weinstein D. Assay of proteins in the presence of interfering materials. Anal Biochem. 1976;70:241–250. doi: 10.1016/s0003-2697(76)80064-4. [DOI] [PubMed] [Google Scholar]
- 23.Field MS, Szebenyi DM, Stover PJ. Regulation of de novo purine biosynthesis by methenyltetrahydrofolate synthetase in neuroblastoma. J Biol Chem. 2006;281:4215–4221. doi: 10.1074/jbc.M510624200. [DOI] [PubMed] [Google Scholar]
- 24.The R Project for Statistical Computing. 2010 Available from: http://www.r-project.org/ [Google Scholar]
- 25.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Universal ProbeLibrary. 2010 Available from: https://www.roche-applied-science.com/sis/rtpcr/upl/index.jsp?id=UP030000. [Google Scholar]
- 27.Girgis S, Nasrallah IM, Suh JR, et al. Molecular cloning, characterization and alternative splicing of the human cytoplasmic serine hydroxymethyltransferase gene. Gene. 1998;210:315–324. doi: 10.1016/s0378-1119(98)00085-7. [DOI] [PubMed] [Google Scholar]
- 28.Stover PJ, Chen LH, Suh JR, Stover DM, Keyomarsi K, Shane B. Molecular cloning, characterization, and regulation of the human mitochondrial serine hydroxymethyltransferase gene. J Biol Chem. 1997;272:1842–1848. doi: 10.1074/jbc.272.3.1842. [DOI] [PubMed] [Google Scholar]
- 29.Su LK, Kinzler KW, Vogelstein B, et al. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 1992;256:668–670. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 30.Liu Z, Choi SW, Crott JW, et al. Mild depletion of dietary folate combined with other B vitamins alters multiple components of the Wnt pathway in mouse colon. J Nutr. 2007;137:2701–2708. doi: 10.1093/jn/137.12.2701. [DOI] [PubMed] [Google Scholar]
- 31.Killmann SA. Effect of Deoxyuridine on Incorporation of Tritiated Thymidine: Difference between Normoblasts and Megaloblasts. Acta Med Scand. 1964;175:483–488. doi: 10.1111/j.0954-6820.1964.tb00597.x. [DOI] [PubMed] [Google Scholar]
- 32.Radtke F, Clevers H. Self-renewal and cancer of the gut: two sides of a coin. Science. 2005;307:1904–1909. doi: 10.1126/science.1104815. [DOI] [PubMed] [Google Scholar]
- 33.Tan LU, Drury EJ, MacKenzie RE. Methylenetetrahydrofolate dehydrogenasemethenyltetrahydrofolate cyclohydrolase-formyltetrahydrofolate synthetase. A multifunctional protein from porcine liver. J Biol Chem. 1977;252:1117–1122. [PubMed] [Google Scholar]
- 34.Kondo M, Yamaoka T, Honda S, et al. The rate of cell growth is regulated by purine biosynthesis via ATP production and G(1) to S phase transition. J Biochem. 2000;128:57–64. doi: 10.1093/oxfordjournals.jbchem.a022730. [DOI] [PubMed] [Google Scholar]
- 35.Bronder JL, Moran RG. Antifolates targeting purine synthesis allow entry of tumor cells into S phase regardless of p53 function. Cancer Res. 2002;62:5236–5241. [PubMed] [Google Scholar]
- 36.Mathews CK. DNA precursor metabolism and genomic stability. Faseb J. 2006;20:1300–1314. doi: 10.1096/fj.06-5730rev. [DOI] [PubMed] [Google Scholar]
- 37.Collins AR, Black DT, Waldren CA. Aberrant DNA repair and enhanced mutagenesis following mutagen treatment of Chinese hamster Ade-C cells in a state of purine deprivation. Mutat Res. 1988;193:145–155. doi: 10.1016/0167-8817(88)90045-4. [DOI] [PubMed] [Google Scholar]
- 38.Pizzorno G, Cao D, Leffert JJ, Russell RL, Zhang D, Handschumacher RE. Homeostatic control of uridine and the role of uridine phosphorylase: a biological and clinical update. Biochim Biophys Acta. 2002;1587:133–144. doi: 10.1016/s0925-4439(02)00076-5. [DOI] [PubMed] [Google Scholar]
- 39.Levy DB, Smith KJ, Beazer-Barclay Y, Hamilton SR, Vogelstein B, Kinzler KW. Inactivation of both APC alleles in human and mouse tumors. Cancer Res. 1994;54:5953–5958. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.