Summary
Progress through mitosis requires that the right protein be degraded at the right time. One ubiquitin ligase, the Anaphase Promoting Complex or Cyclosome (APC-C) targets most of the crucial mitotic regulators by changing its substrate specificity throughout mitosis. The Spindle Assembly Checkpoint (SAC) acts on the APC-C co-activator, Cdc20 to block the degradation of metaphase substrates, e.g.: Cyclin B1 and securin, but not others, e.g.: Cyclin A. How this is achieved is unclear. Here we show that Cdc20 binds to different sites on the APC-C depending on the SAC. Cdc20 requires APC3 and APC8 to bind and activate the APC-C when the SAC is satisfied, but only requires APC8 when the SAC is active. Moreover, APC10 is crucial for Cyclin B1 and securin but not Cyclin A destruction. We conclude that the SAC causes Cdc20 to bind to different sites on the APC-C and this alters APC-C substrate specificity.
Introduction
The cell cycle uses ubiquitin-mediated proteolysis to ensure that the two daughter cells inherit an identical complement of chromosomes, and coordinate mitosis with cytokinesis. Specific mitotic regulators are degraded at specific times to allow the next step in cell division 1. One ubiquitin ligase, the Anaphase Promoting Complex-Cyclosome (APC-C), targets many essential mitotic regulators for proteolysis, including Cyclins A and B1, securin, Plk1 and the Aurora A and B kinases 2.
One of the most important questions is how the same ubiquitin ligase targets different sets of proteins at different times in mitosis; in particular how this is regulated by the Spindle Assembly Checkpoint (SAC). Part of the answer is likely to be found in the structure of the APC-C, a large (1.5 MDa) multiprotein complex composed of at least 13 subunits 2. The APC-C is organised into two main sub-complexes held together by APC1 3. One sub-complex contains the ‘catalytic’ subunits, APC11 and APC2, plus APC10 3-5. The other has several subunits with TPR motifs: APC3, 6, 7 and 8, plus APC4 and 5 that form the link to APC1. Despite this complexity, no subunit has been directly implicated in selecting substrates with the exception of APC10 (see below). Instead, most attention has focused on the APC-C co-activators.
There are two co-activators in mitotic cell cycles, Cdc20-fizzy and Cdh1-fizzy-related. It has been suggested that either Cdc20 and Cdh1 recognise substrates and recruit them to the APC-C 6-13, or substrates are recognised by a complex of the APC-C bound to Cdc20 or Cdh1 14, 15. It was originally proposed that the change in substrate specificity of the APC-C through mitosis was driven by exchanging Cdc20 for Cdh1 16 17, but Cdh1 is not essential in the yeasts 18, 19 and the majority of Drosophila, mice and chicken DT40 cells lacking Cdh1 can divide correctly 20-23. Depleting Cdh1 in human cells stabilises the Aurora kinases but other late mitotic substrates, such as Plk1, are still degraded 24. Thus, there must be other means by which the APC-C alters its substrate specificity.
One model for how APC-C complexes can discriminate between substrates is that ubiquitylation is more processive on some substrates than others 25. Processively ubiquitylated substrates should be degraded first since they can be polyubiquitinated in one round, making them less likely to be deubiquitinated by antagonistic deubiquitinases than ‘distributive’ substrates that require several rounds of binding and release 25. But this model took Cyclin A as a model distributive substrate and securin as a processive substrate, yet in mitosis Cyclin A is degraded before securin 1. The UbcH10 E2 enzyme has also been suggested to regulate APC-C substrate selection 26, 27 but we find that both mitosis and Cyclin degradation are unperturbed when UbcH10 levels are depleted by more than 90% 28.
The SAC clearly regulates the selection of some substrates over others. Some substrates are degraded when the SAC is active (prometaphase), such as Cyclin A and Nek2A, but others are degraded only when the SAC is satisfied (metaphase) such as Cyclin B1 and securin 29-32. Henceforth we refer to Cyclin A and Nek2A destruction as SAC-insensitive, and Cyclin B1 and securin destruction as SAC-sensitive.
Recently we showed that Cyclin A is degraded when the SAC is active because it binds directly to Cdc20, and is recruited by its Cks1 subunit to the APC-C 33, 34. Nek2A also binds directly to the APC-C through its carboxyl terminus 35, which has the dipeptide MR that resembles the IR dipeptide at the carboxyl terminus of Cdc20, Cdh1 and APC10 36, and is required for Cdh1 to interact with the APC3 subunit 3, 36. Whether all APC-C substrates bind to the same binding site, and it is the timing of their recruitment that regulates when they are ubiquitylated, or whether different substrates are bound to different binding sites on the APC-C, is unknown.
Here we have begun to analyse the contribution that APC-C subunits make to the targeting of specific mitotic substrates. We have used siRNA to deplete particular subunits and assayed the binding of substrates and Cdc20 to the APC-C, and in parallel the effect on the degradation of specific substrates using a live cell assay. We present evidence that the APC-C does indeed recognise its SAC-insensitive substrate Cyclin A in a different manner from its SAC-sensitive substrates, Cyclin B1 and securin, and this is determined by the manner in which Cdc20 binds to the APC-C. We show that when the SAC is active Cdc20 primarily requires APC8 to bind to the APC-C, whereas it requires APC3 and APC8 when the SAC is satisfied. The interaction with APC8 is important for APC-C activity in the presence and absence of the SAC. Furthermore, once the SAC is satisfied Cdc20 works together with APC10 to provide a binding site for SAC-sensitive substrates. Thus our results give insights into how the SAC regulates Cdc20 and the APC-C, and consequently how this alters APC-C substrate specificity.
Results
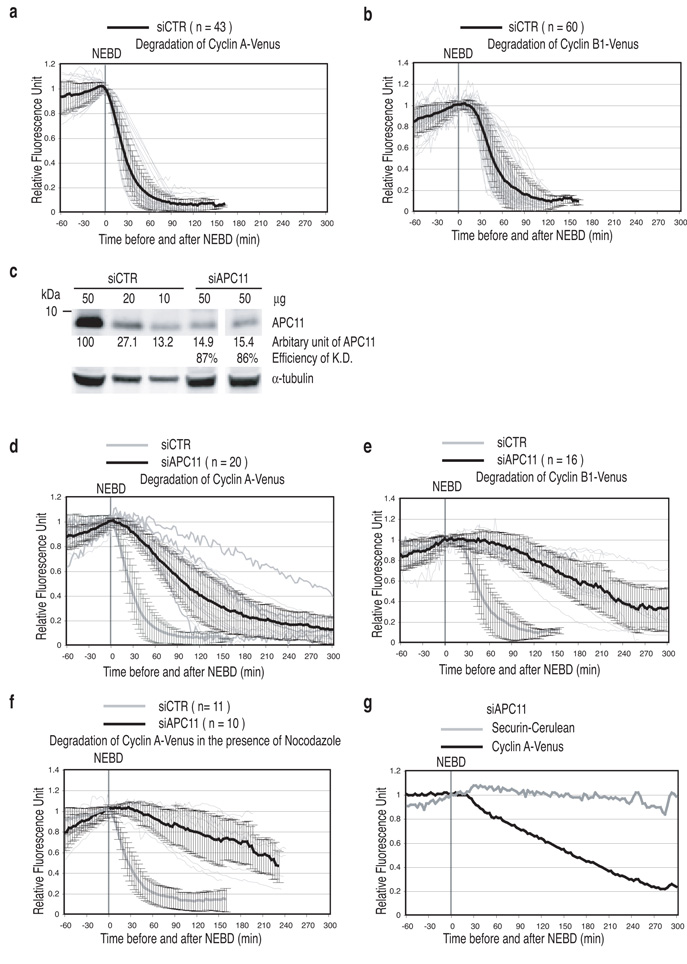
To determine how the APC-C and the SAC work together to target different substrates for degradation when the SAC is active in prometaphase, compared to when the SAC is satisfied in metaphase, we depleted individual APC-C subunits by siRNA in human cells and assayed the effect on fluorescent protein (FP)-tagged substrates using a live-cell assay 29. This allowed us to determine whether depletion affected the timing and the rate of destruction of specific substrates. We chose Cyclin A as a model SAC-insensitive substrate and Cyclin B1 and securin as SAC-sensitive substrates. Cyclin A-FP is degraded rapidly in prometaphase as soon as the nuclear envelope breaks down (NEBD) 30, 31, whereas Cyclin B1-FP and securin-FP only begin to be degraded once all the chromosomes have attached to the spindle and satisfied the SAC (Figs. 1a & b & S1) 29, 30, 37.
Figure 1.
APC11 is required to degrade all model substrates
(a and b) Degradation of model substrates in control cells. Cells were treated with siRNA oligos against GAPDH, injected with plasmids encoding Cyclin A-Venus (a) or Cyclin B1-Venus (b) in G2 phase and analysed by time-lapse DIC and fluorescence microscopy at 3 min intervals. The fluorescence of individual cells was measured, the value at NEBD set to 1 and plotted as thin light grey lines. The mean +/− s.d. for all cells is plotted as thick black lines. n = number of cells analyzed from 3 independent experiments.
(c) Depletion of human APC11. Cells were treated with siRNA oligos against GAPDH (siCTR) or APC11 for 72 hrs before assaying the indicated amounts of extract by quantitative immunoblotting. The extent of depletion was calculated from a standard curve using diluted control extract and normalisation to the level of tubulin. Results are representative of at least two experiments per siRNA treatment.
(d and e) APC11 is required to degrade both SAC–sensitive and SAC-insensitive substrates. Cells were treated with siRNA oligos against APC11, injected with a plasmid encoding Cyclin A-Venus (d) or Cyclin B1-Venus (e) and analyzed as in a. The mean +/− s.d. for all cells are plotted as the thick black lines. The experimental values are plotted in black (siAPC11) and the controls are plotted in grey (siCTR). n = number of cells analyzed from 3 independent experiments.
(f) The SAC stabilises Cyclin A-Venus in cells depleted of APC11.
Cells were treated with siRNA oligos against GAPDH (grey) or APC11 (black), injected with a plasmid expressing Cyclin A-Venus, treated with 100 ng-ml nocodazole and analysed as in a. n = number of cells analyzed from 2 independent experiments.
(g) Cyclin A is preferred over securin as a substrate when APC-C activity is limiting.
Cells were treated with siRNA oligos against APC11, injected with a plasmid expressing Cyclin A-Venus (black) and securin-Cerulean (grey) and analyzed as in a. Representative of 11 cells in 2 independent experiments.
Cyclin A is preferred to securin in cells with limiting levels of APC-C-depleted activity
We first assayed the effect of reducing overall APC-C activity in cells by depleting APC11, the catalytic subunit that recruits the E2 protein 4, 38. Depleting APC11 by >85% (Fig. 1c) delayed cells in mitosis (Fig. S2b) and prevented or grossly impaired the degradation of all the substrates we analysed: Cyclin A (Fig. 1d), Cyclin B1 (Fig. 1e & quantified in Table 1), and Securin (Fig S1b). The effect of the siRNA was specific because we could rescue the phenotype by expressing a siRNA-insensitive version of the subunit, as we could rescue for all the other APC subunit depletions in our study (Fig. S2).
Table 1.
Quantification of Cyclindegradation rates in siRNA experiments
| Cyclin A degradation | ||||
|---|---|---|---|---|
| Time NEBD to start of degradation (min) |
slope | P value | Figure | |
| siCTR | 0.81 ± 2.46 | −0.0567 ± 0.0222 | 1a | |
| siAPC11 | 21.12 ± 18.69 | −0.0160 ± 0.0087 | <0.0001 | 1d |
| siAPC3 | 24.78 ± 20.82 | −0.0167 ± 0.0066 | <0.0001 | 2b |
| siAPC10 | 0.72 ± 1.77 | −0.0578 ± 0.0276 | <0.87 | 4b |
| siAPC8 +Flag-APC8WT | 3.00 ± 4.06 | −0.0557 ±0.0199 | 6a | |
| siAPC8 +Flag- APC8N338A |
12.4 ± 0.934 | −0.0523 ±0.0253 | 0.54 | 6b |
| siAPC8 +Flag-APC8WT + Nocodazole |
−0.0442 ± 0.0167 | 6c | ||
| siAPC8 +Flag- APC8N338A + Nocodazole |
−0.0237 ± 0.0093 | <0.0001 | 6d | |
| Cyclin B1 degradation | ||||
|---|---|---|---|---|
| Time NEBD to start of degradation (min) |
slope | P value |
Figure | |
| siCTR | 68.64 ± 25.59 | −0.0642 ± 0.0289 | 1b | |
| siAPC11 | 145.38 ± 91.17 | −0.0293 ± 0.0249 | <0.0001 | 1e |
| siAPC3 | 147.33 ± 67.59 | −0.0227 ± 0.0136 | <0.0001 | 2c |
| siAPC10 | 139.38 ± 61.14 | −0.0372 ± 0.0148 | 0.0020 | 4d |
| siAPC8 +Flag-APC8WT | −0.0408 ± 0.0268 | 7a | ||
| siAPC8 +Flag- APC8N338A |
−0.0249 ± 0.0254 | 0.0003 | 7b | |
The maximal slope of the cyclin degradation was calculated by using the curve-fit function in Prism software and the standard deviation calculated. The P value compared to control cells was calculated by a student’s t-test.
We noticed that Cyclin A degradation did begin slowly to be degraded when cells reached metaphase, indicating that degradation might now be more sensitive to the SAC (Fig. S3), we provided direct support for this interpretation by adding nocodazole to maintain the SAC, which stabilised Cyclin A (Fig. 1f). Since Cyclin A degradation now began at the same time as the SAC-sensitive substrates we could directly test the ‘processivity model’ for substrate selection. Therefore, we co-expressed Cyclin A-Venus and securin-CFP in APC11-depleted cells and, in contradiction to the model, Cyclin A (a ‘distributive’ substrate) was preferentially degraded over securin (a ‘ ‘processive’ substrate) (Fig. 1g).
APC3 is required to degrade both SAC-sensitive and SAC–insensitive substrates
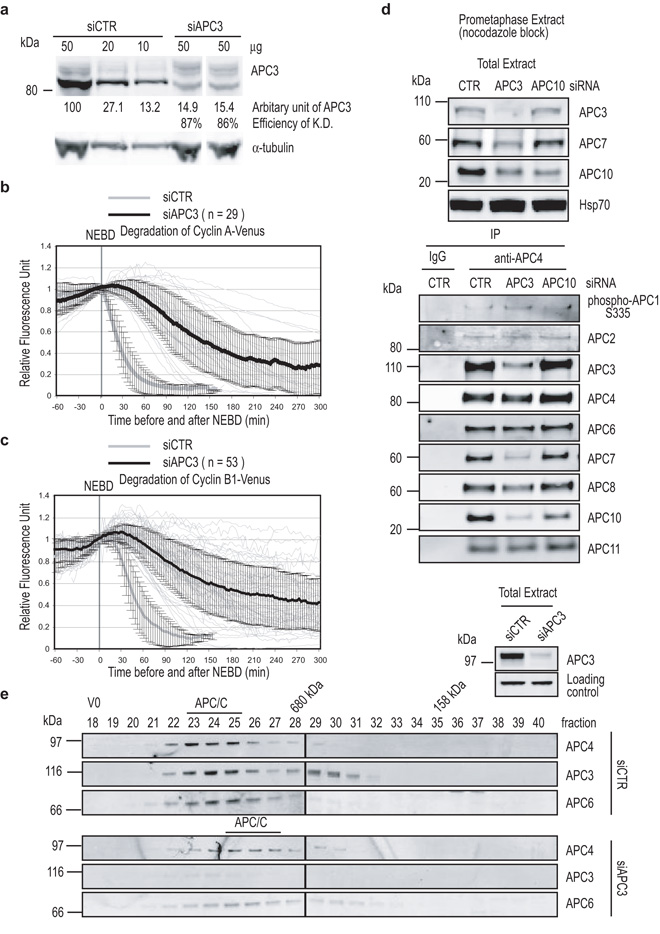
To address the question of whether APC-C subunits contribute to the change in APC-C substrate selection we depleted the APC3 subunit that binds to the IR tail of APC-C co-activators 3, 36. Mutations in the APC3 gene in yeasts 39 and in Drosophila 40 showed that it was required to degrade mitotic cyclins and, in agreement with this, depleting APC3 by siRNA to ~90% (Fig. 2a) severely delayed and inhibited the degradation of human Cyclin A (Fig. 2b), Cyclin B1 (Fig 2c) and securin (Fig. S1c) (quantified in Table 1).
Figure 2.
APC3 is required to degrade both SAC-sensitive and SAC–resistant substrates.
(a) Depletion of human APC3. Cells were treated with siRNA oligos against GAPDH or APC3 for 72 hrs before analyzing. Protein levels were analyzed as in Fig 1c.
(b and c) APC3 is required to degrade both SAC–insensitive and SAC-sensitive substrates. Cells were treated with siRNA oligos against APC3, injected with a plasmid encoding Cyclin A-Venus (b) or Cyclin B1-Venus (c) and analyzed as in Fig 1a. The mean +/− s.d. is plotted as thick black lines. The experimental values are plotted in black (siAPC3) and the values from the control GAPDH-depleted cells are plotted in grey (siCTR). n = number of cells analyzed from 3 independent experiments.
(d) APC-C composition in the depletion of APC3 or APC10. HeLa cells were treated with siRNA against GAPDH, APC3 or APC10 for 72 hrs before harvesting prometaphase cells in the presence of 100 ng-ml nocodazole by mitotic shake off. The APC-C was immunoprecipitated using anti-APC4 antibodies and the immunoprecipitates blotted with antibodies against phospho-APC1 (serine 355), APC2, APC3, APC4, APC6, APC7, APC8, APC10 and APC11, and the extent of depletion measured by quantitative immunoblotting for the cell extracts (top) and immunoprecipitates (bottom).
(e) Analysis of APC-C in the depletion of APC3 by size-exclusion chromatography. Extracts of control (top) or APC3-depleted (bottom) cells were separated on a Superose 6 column and fractions were blotted with antibodies against APC3, APC4 and APC6. The total cell extract is also shown (top right). Loading control refers to a cross-reacting protein recognised by the anti-APC3 antibody. The peak of APC-C migration is indicated by the black bar.
APC3 is only required for the APC-C to bind Cdc20 when the SAC is satisfied
To understand the requirement for APC3 we assayed APC-C composition and the binding of co-activator and substrates in APC3-depleted cells. Depleting APC3 destabilised APC10 and APC7 (Fig. 2d) and reduced the amount of APC10 and APC7 bound to the complex (Fig. 2d), but the composition of the rest of the APC-C was largely unaltered, as assayed by co-immunoprecipitation and size exclusion chromatography (Fig. 2d & Fig. 2e). These minimal effects on the overall APC-C structure agree with the analysis of deleting APC3 (Cdc27) in budding yeast 3.
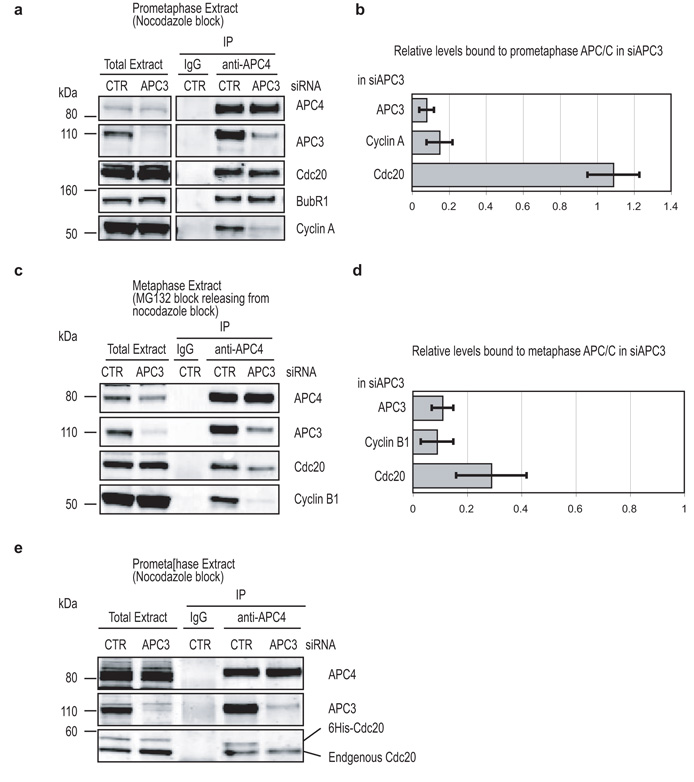
Depleting APC3 markedly reduced the binding of both Cyclin A (Fig 3a & b) and Cyclin B1 (Fig. 3c & d), most likely because Cyclin A is recruited via its Cks1 subunit to phospho-APC3, whereas Cyclin B1 requires Cdc20 to bind to the APC-C (see below). When we assayed the binding of Cdc20, however, we noted a striking difference between the APC-C immuno-purified from cells with an active SAC (prometaphase, Fig 3a) and those in which the SAC was satisfied (metaphase, Fig 3c). Depleting APC3 had no apparent effect on the amount of Cdc20 bound to the APC-C when the SAC was active, but significantly reduced the amount bound when the SAC was satisfied (compare Figs 3a & c, quantified in Figs 3b & d). That these effects were specific to the depletion of APC3 was confirmed by rescuing the depletion with siRNA-resistant APC3, which also restored APC10 binding (Fig. S4). We reasoned that this difference might be because in prometaphase Cdc20 bound to the APC-C as part of a SAC-complex but as a free protein in metaphase, and consistent with this exogenous Cdc20 could not bind to the APC-C in prometaphase in the absence of APC3 (Fig 3e). Thus we conclude that Cdc20 binds to a different site on the APC-C depending on whether the SAC is active or not.
Figure 3.
APC3 is only required to bind Cdc20 when the SAC is satisfied
(a - d) APC3 is only required to bind Cdc20 when the SAC is satisfied. HeLa cells were treated with siRNA against GAPDH (CTR) or APC3 and synchronised in prometaphase by treating with 100 ng-ml nocodazole plus 10 μM MG132 to stabilise Cyclin A (a & b), or synchronised in prometaphase with 100 ng-ml nocodazole, released into medium containing 10 μM MG132 and incubated for a further 3 hrs to obtain metaphase cells (c & d). The APC-C complex was immunoprecipitated with anti-APC4 antibodies and samples blotted for APC3, APC4, Cdc20, BubR1 and Cyclin A (a) or Cyclin B1 (b).
(b and d) Bar diagrams show the remaining amount of APC3 and the amount of Cdc20 and Cyclin A or B1 bound to the prometaphase APC-C (b) or the metaphase APC-C (d), quantified using a LI-COR Odyssey scanner and normalised to the level of APC4. Levels of the proteins bound to control APC-C were set to 1. Error bars shown are mean+/− s.d. of 3 experiments.
(e) APC3 is required for free Cdc20 to bind to the APC-C. Cells were treated with siRNA oligos against GAPDH or APC3 and synchronized as in (a). Purified recombinant His6-tagged Cdc20 was added to the cell extracts and the APC-C was immunoprecipitated with an anti-APC4 antibody. Samples were blotted for APC3, APC4 and Cdc20. Recombinant Cdc20 runs at a higher molecular mass than endogenous Cdc20.
APC10 and Cdc20 are both required to recruit SAC-sensitive substrates
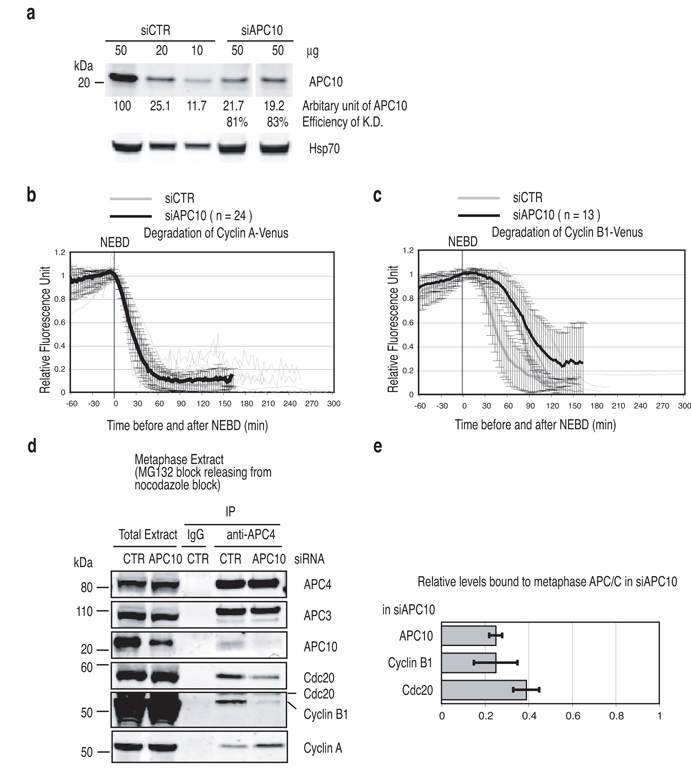
Our observation that depleting APC3 also caused a reduction in APC7 and APC10 levels meant that these proteins might also have important roles in recognising substrates. To test this we depleted APC7 (Fig. S5) or APC10 by siRNA (Fig. 4a) and assayed the destruction of Cyclin A and Cyclin B1 (Fig. 4b and c) (quantified in Table 1). Depleting APC7 had no visible effect on Cyclin B1 degradation, and caused only a minor delay in the destruction of Cyclin A, which was still degraded in nocodazole (Fig. S5). To our surprise, although depleting APC10 had no effect on the degradation of Cyclin A, which began at the correct time and proceeded at its normal rate (Fig. 4b), it delayed the timing and slowed the rate of Cyclin B1 (and securin) destruction (Fig. 4c and S1d). Consistent with this, depleting APC10 perturbed the binding of Cyclin B1 but not Cyclin A to the APC-C (Fig. 4d). Cyclin B1 primarily bound to the APC-C as a substrate because mutating the Destruction box of Cyclin B1 severely reduced its binding (Fig. S6).
Figure 4.
APC10 is required to recruit SAC-sensitive substrates
(a) Depletion of APC10. Cells were treated with siRNA oligos against GAPDH (control) or APC10 and the results from 2 independent experiments analyzed as in Fig 1c. Hsp70 was used as a loading control.
(b and c) APC10 is required for Cyclin B1 but not for Cyclin A degradation. Cells were treated with siRNA oligos against APC10, injected with a plasmid encoding Cyclin A-Venus (b) or Cyclin B1-Venus (c) and analysed by time-lapse as Fig 1a. The mean +-− s.d. for all cells is plotted in black (siAPC10) and the values from the control cells are plotted in grey (siCTR). n = number of cells analyzed from 2 independent experiments.
(d and e) APC10 mediates Cyclin B1 and Cdc20 but not Cyclin A binding to APC-C. HeLa cells were treated with siRNA against GAPDH (CTR) or APC10 and synchronised in metaphase as in Fig 3c. The APC-C complex was immunoprecipitated with anti-APC4 antibodies and samples blotted for APC3, APC4, APC10, Cdc20, Cyclin A and Cyclin B1 (d). (e) Bar diagrams show the remaining amount of APC10 and the amount of Cyclin B1 and Cdc20 bound to the metaphase APC-C quantified using a LI-COR Odyssey scanner and normalised to the level of APC4. Levels of the proteins bound to control APC-C were set to 1. The mean +/− s.d. of 3 experiments is shown.
Depleting APC10 also reduced the amount of Cdc20 bound to the APC-C in metaphase (Fig. 4d & e) but not prometaphase (data not shown). In yeast, APC10 is important for the APC-C to recognise substrates with a Destruction box 15, 41 by forming a bipartite receptor with Cdc20 42, therefore we tested whether depleting Cdc20 might have a similar effect on substrate binding. In accord with this idea, depleting Cdc20 drastically reduced the binding of Cyclin B1 to the APC-C in metaphase (Fig. S7a). By contrast, depleting Cdc20 increased the amount of Cyclin A bound to the APC-C when the SAC was active (Fig. S7b), indicating that Cdc20 was not required to recruit Cyclin A to the APC-C. Thus we conclude that, as in budding yeast 42, 43, APC10 and Cdc20 stabilise each other’s binding to the APC-C and are both required to bind canonical Destruction box substrates to the APC-C when the SAC is satisfied. However, they are not required to recruit Cyclin A to the APC-C when the SAC is active. Instead, Cyclin A is recruited by its Cks subunit 33 and we infer that Cdc20 must activate the APC-C 34, as suggested for Nek2A destruction 44.
To bind Cdc20 and degrade Cyclin A when the SAC is active the APC-C requires APC8
Since Cdc20 did not require APC3 to bind to the APC-C when the SAC was active we sought to identify its prometaphase binding site. By siRNA phenotypes we identified APC6 and APC8 as particularly important to bind Cdc20 in prometaphase (Fig. S8). Analyses in budding yeast, however, showed that eliminating APC6 (Cdc16) and APC8 (Cdc23) perturbed APC-C structure 3. To eliminate the possibility that depleting APC6 or APC8 reduced Cdc20 binding by disrupting the APC-C we sought a point mutation to perturb Cdc20-binding.
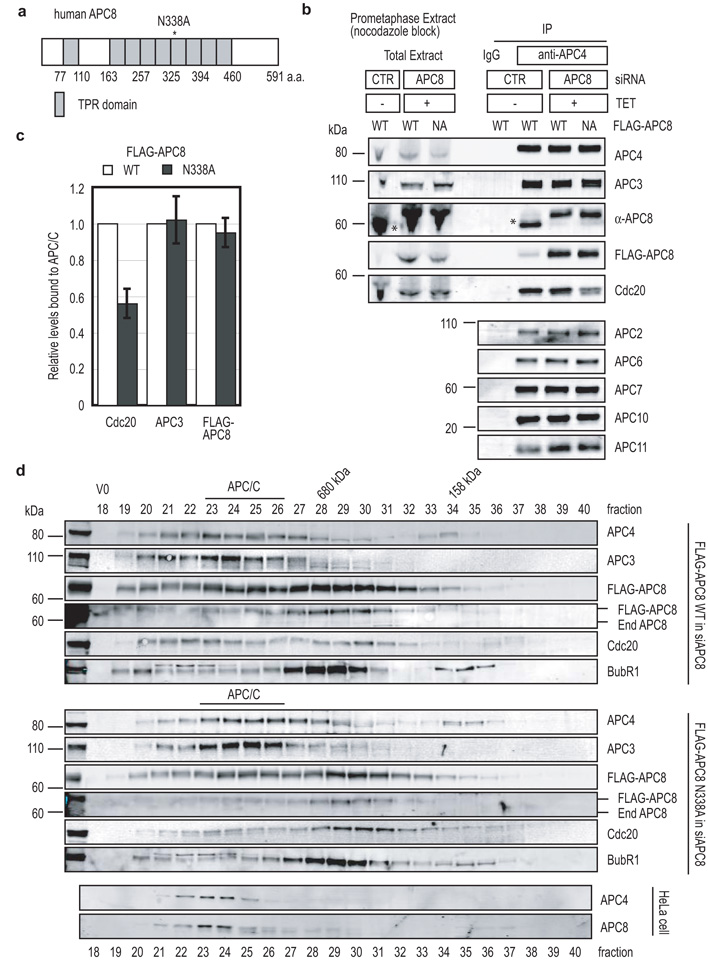
Matyskiela and Morgan identified a point-mutation in the TPR motifs of APC8 (Cdc23) that reduced binding to Cdh1 42. Therefore, we made the analogous mutation (N338A) in human APC8 (Fig. 5a) and expressed this in cells depleted of APC8 by siRNA. Compared to cells rescued with wild type APC8, the APC8N338A mutant reduced the amount of Cdc20 bound to the APC-C in prometaphase (Fig. 5b & c) but had no effect on the composition of the APC-C (Fig. 5b). Size exclusion chromatography confirmed that less Cdc20 bound to the APC-C incorporating the APC8N338A point mutant (Fig. 5d) and showed both that the APC-C migrated at its correct size (1.5 MDa) and incorporated the Flag-tagged wild type and mutant APC8 constructs to a similar extent (Fig. 5d). Consistent with the reduction in Cdc20 binding, the APC8N338A mutant delayed cells in mitosis (Fig. S9) and notably delayed the destruction of Cyclin A by around 12 min (Fig. 6a & b & quantified in Table1).
Figure 5.
A point mutation in APC8 is sufficient to reduce the binding of Cdc20 in prometaphase
(a) Schematic structure of human APC8. TPR domains are shown as grey boxes.
(b) The N338A mutation reduces Cdc20 binding to prometaphase APC-C. HeLa cells with an inducible wild type Flag-APC8 (WT) or mutant Flag-APC8N338A were treated with siRNA against GAPDH (CTR) or APC8 and synchronised in prometaphase as in Fig 2d. The APC-C was immunoprecipitated using anti-APC4 antibodies and the immunoprecipitates blotted with antibodies against APC2, APC3, APC4, APC6, APC7, APC8, APC10, APC11 and Cdc20 and the extent of depletion measured by quantitative immunoblotting. The asterisk indicates endogenous APC8.
(c) Quantification of APC3, Cdc20 and Flag-APC8 bound to the APC-C by quantitative immunoblotting and normalised to the level of APC4. Levels of the proteins bound to control APC-C were set to 1. The mean +/− s.d. of 4 independent experiments is shown.
(d) Analysis of APC-C by size-exclusion chromatography. Control (bottom) and experimental HeLa cells expressing an inducible wild type Flag-APC8 (WT, top) or mutant Flag-APC8N338A (middle) were treated with siRNA oligos against APC8 and synchronised in prometaphase as in panel b and extracts prepared as in Fig. 3e. Cell extracts were fractioned on a Superose 6 column and fractions were blotted with antibodies against APC3, APC4, Flag epitope, APC8, Cdc20 and BubR1. The peak of APC-C migration is indicated by the black bar.
Figure 6.
A point mutation in APC8 prevents Cyclin A degradation in prometaphase
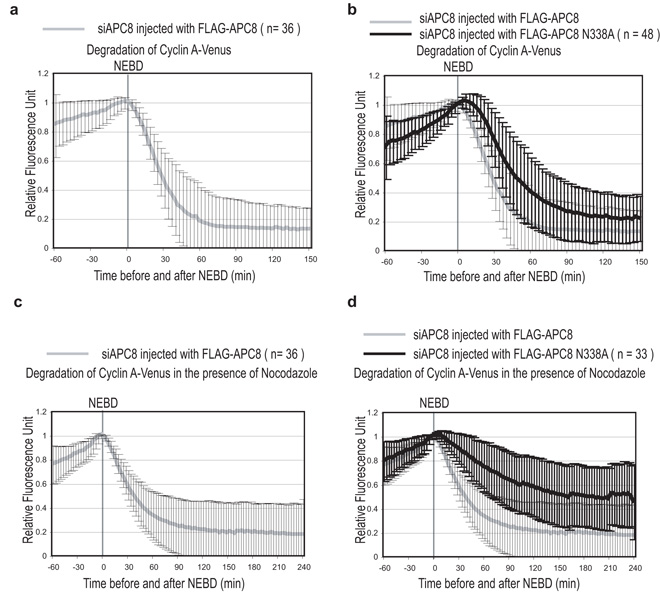
(a and b) Cyclin A degradation is delayed in cells expressing APC8N338A. Cells were treated with siRNA oligos against APC8, injected with a plasmid encoding Cyclin A-Venus and Flag-APC8 WT (a) or Flag-APC8N338A (b) and analysed by time-lapse DIC and fluorescence microscopy as in Fig 1a. The experimental mean +/− s.d. of wild type APC8 is plotted in grey in panel a and b, and of APC8 N338A plotted in black. n = number of cells analyzed in 3 independent experiments.
(c and d) Cyclin A is stabilized by nocodazole in cells expressing APC8N338A. Cells were treated with siRNA oligos against APC8, injected with a plasmid encoding Cyclin A-Venus and wild type Flag-APC8 (c) or Flag-APC8N338A (d) in G2 phase and analysed in the presence of 100 ng-ml nocodazole by time-lapse DIC and fluorescence microscopy. The experimental mean +/− s.d. of wild type APC8 is plotted in grey in panel c and d, and of APC8N338A plotted in black. n = number of cells analyzed from 3 independent experiments.
In cells rescued with the APC8N338A mutant we noticed that the destruction of Cyclin A correlated with chromosome alignment, indicating that Cyclin A might only be degraded once the SAC had been satisfied. To test this we assayed Cyclin A-FP degradation after adding nocodazole to prevent the SAC being satisfied and found that in contrast to cells expressing wild type APC8, Cyclin A was stabilised by nocodazole (Fig. 6c & d & quantified in Table1). We conclude that when the SAC is active APC8 is crucial for Cdc20 to bind the APC-C, and binding to this site is important for Cyclin A to be degraded.
The Cdc20-binding site on APC8 is required for Cyclin B1 destruction in metaphase
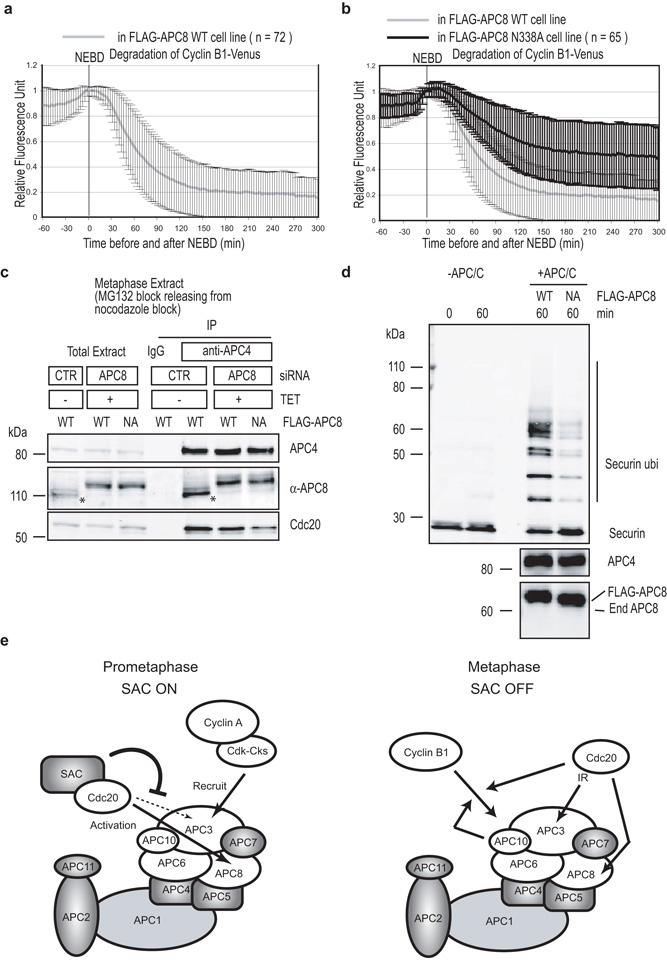
Since Cyclin A could be degraded in APC8N338A expressing cells once the SAC was satisfied, this could mean that the APC8 site was no longer required for APC-C activity. Alternatively, since Cyclin A binds directly to Cdc20 and itself binds to the APC-C through Cks1, it might be a special case. Therefore, we assayed the degradation of Cyclin B1 in cells expressing the APC8N338A mutant and found that its destruction was also inhibited (Fig. 7a & b & quantified in table1). Furthermore, the APC8N338A mutation reduced both the binding of Cdc20 when the SAC was satisfied (Fig. 7c) and APC-C activity in vitro (Fig. 7d). Thus, we conclude that APC8 has a crucial role to activate the APC-C in the presence or absence of the SAC.
Figure 7.
The Cdc20-binding site on APC8 is also required in metaphase
(a and b) Cyclin B1 degradation is inhibited in cells expressing APC8N338A. siRNA against APC8 and a plasmid encoding Cyclin B1-Venus were transfected into the HeLa cells with an inducible siRNA-resistant wild type Flag-APC8 (a) or mutant Flag-APC8N338A (b) and analysed as in Fig 1a. Mean +/− s.d. values are shown. n = number of cells analyzed from 3 independent experiments.
(c) APC8 is important for free-Cdc20 to bind to the APC-C. HeLa cells expressing an inducible wild type Flag-APC8 (WT) or mutant Flag-APC8N338A were treated with siRNA against GAPDH (CTR) or APC8 as in Fig 5b and synchronised in metaphase as in Fig 3c. The APC-C was immunoprecipitated using anti-APC4 antibodies and the immunoprecipitates blotted with antibodies against APC4, APC8 and Cdc20. The asterisk indicates endogenous APC8. (d) APC8 N338A mutation reduces APC-C activity in vitro. The APC-C incorporating Flag-APC8 or Flag-APC8N338A was prepared as in Fig 5b and its activity assayed using securin as a substrate in an in vitro ubiquitination reaction as previously described 49. (e) Model for APC-C regulation during early mitosis. In prometaphase (left), the APC-C recognizes substrates such as Cyclin A through the APC3 subunit but interacts through APC8 with Cdc20, as co-activator or as part of the SAC complex. The SAC proteins associated with Cdc20 could prevent Cdc20 accessing its APC3 binding site. In metaphase (right), the APC-C recognizes substrates such as Cyclin B1 through APC10 and Cdc20 forming a bi-partite receptor, and Cdc20 requires both APC3 and APC8 to interact with and activate the APC-C.
Discussion
In this paper we have begun to determine the mechanism behind the ability of the APC-C to recognise different proteins at different times (Fig. 7e), which is crucial to the proper coordination of chromosome segregation and cytokinesis. We find that the APC-C binds to Cdc20 in a different manner when the SAC is active compared to when the SAC is satisfied. Our evidence is consistent with a site on APC8 that is particularly important to bind Cdc20 when the SAC is active, whereas APC3 also becomes important when the SAC is satisfied. Our conclusion is consistent with the most recent APC-C structures obtained by cryo-EM tomography where Cdc20 appears to bind to a different part of the APC-C depending on whether the APC-C was purified from prometaphase or metaphase cells 45. Furthermore, that APC3 and APC8 might both interact with Cdc20 is consistent with recent single particle electron microscopy analysis of the budding yeast APC-C coupled to a crystal structure of the APC3-Cdc27 N-terminal dimerisation domain, which indicates that APC8-Cdc23 and APC3-Cdc27 are homo-dimers with similar overall 3D structures (Schreiber et al., manuscript submitted and reference 46).
In agreement with previous studies in budding yeast we find that APC10 and Cdc20 are both required to bind substrates to the APC-C once the SAC is satisfied, and thus are likely to act as a bipartite receptor for substrates with canonical Destruction boxes. Just as in budding yeast 15, we were only able to detect the ternary complex of APC-C-Cdc20-Cyclin B1, and we find that APC10 and Cdc20 stabilise each other on the APC-C. At this time in mitosis Cdc20 is likely to bind to APC3, probably through its IR tail. In contrast, neither APC10 nor Cdc20 is required to form the binding site for Cyclin A when the SAC is active. Instead APC3 is required, most likely through binding the Cks protein that itself binds to the Cyclin A-Cdk complex 33.
In cells with limiting APC-C activity, Cyclin A is preferred as an APC-C substrate over Cyclin B1 and securin. This result is not consistent with the ‘processivity’ model for how the APC-C chooses its substrates 25. This model, however, was based on experiments using an in vitro ubiquitination assay 25 where it was not clear whether Cyclin A was bound to its partner Cdk and Cks proteins that are required to recruit Cyclin A to the APC-C 33, 34.
We find that APC8 but not APC3 is important to bind Cdc20 when the SAC is active, and the potential interaction site is conserved in yeast 42. Mutating this site reduces Cdc20 binding to the APC-C when the SAC is active, and prevents Cyclin A destruction even though Cyclin A is still bound to the APC-C. Once the SAC is satisfied, Cyclin A is degraded, indicating that the Cdc20 recruited by Cyclin A can activate the APC-C. However, the APC-C binds less Cdc20 and is unable to degrade Cyclin B1 in vivo, and has greatly reduced activity in vitro. Thus APC8 provides an important site for APC-C activity in prometaphase and metaphase. Consistent with this, the Cdc23N405A mutation interfered with the ubiquitylation of securin and destruction of Clb2, as well as the binding of Cdh142. Thus the interaction between APC8 and co-activator is important for APC-C activity throughout mitosis.
How Cdc20 activates the APC-C is unclear but it might change APC-C conformation as proposed for Cdh1 47. Recent evidence indicates that this activation ability lies in the amino terminus of Cdc20 44. Two motifs of Cdc20 are required for to bind and activate the APC-C: the C-box in the amino terminus, and the C-terminal IR-tail 48,49. A C-terminal peptide of Cdc20 interacts with recombinant APC3 and APC7 36 and genetically the IR motif of Cdh1 interacts with APC3-Cdc27 3, 42, whereas the C-box of Cdh1 appears to bind APC2 3. The site that interacts with APC8 is not yet known but does not appear to be either the IR domain or the C-box 42.
That APC10 is only required for to bind SAC-sensitive substrates has implications for models for how the SAC acts on the APC-C, raising the possibility that the SAC might act on APC10. Some phosphorylation sites on the APC-C specifically respond to different microtubule poisons and thus potentially depend on different SAC kinases 50. Moreover, the ability to maintain an arrest in mitosis differs in response to different microtubule poisons 51. If SAC kinases do directly regulate APC-C activity by phosphorylation then APC10 is a potential target.
The well-established target of the SAC is Cdc20, which is incorporated into a BubR1-Bub3 complex by the SAC 49, 52, 53 and we think this blocks its ability to provide a binding site (with APC10) for Cyclin B1 because we never detected an interaction between Cyclin B1 and the APC-C or Cdc20 in prometaphase extracts (data not show). In contrast, van Zon and colleagues did find Cyclin B1 associated with the prometaphase APC-C 54. Currently we cannot explain this discrepancy.
We find that the Cdc20-BubR1-Bub3 complex binds to a site requiring APC8 and not APC3. There is evidence that Mad3 (BubR1) acts as a psedusubstrate 55, therefore, BubR1-Bub3 might physically prevent Cdc20 from accessing APC3 to form the metaphase substrate binding site (with APC10). Alternatively, the BubR1-Bub3 complex might prevent Cdc20 from interacting with APC10 thereby blocking Cdc20 activity against metaphase substrates. Definitive answers to these questions will require more structural studies of the APC-C.
Supplementary Material
Acknowledgements
We thank Jan-Michael Peters for the anti-phospho APC1 antibody and Stephen Taylor for the HeLa-FRT cell line. We are grateful to David Barford for discussing results prior to publication and all the members of our laboratory for comments and criticisms. DI was supported by a fellowship from the Japanese Society for the Promotion of Science and by the Association for International Cancer Research (AICR). The work was supported by core funding to the Gurdon Institute from the Wellcome Trust and Cancer Research UK, by a project grant from the AICR and a programme grant from Cancer Research UK to JP.
Materials and Methods
Cell Culture
HeLa cells were maintained in Advanced D-MEM with 10% FBS. For synchronization at the beginning of S phase HeLa cells were treated with 2.5 mM Thymidine or 2.5 mM Thymidine followed by 2.5 μg-ml aphidicolin as previously described 56. For prometaphase, cells were released from a double thymidine block and 6 hours later treated with nocodazole at a final concentration of 0.1 ng-μl for 12 hrs. For metaphase, cells were released from the nocodazole block into medium containing 10 μM MG132 for 3 hrs.
RNAi
The following ON-TARGETplus (Dharmacon, CO, USA) oligos were used APC3-1 (GGAAAUAGCCGAGAGGUAAUU) and APC3-2 (CAAAAGAGCCUUAGUUUAAUU), APC11-1 (UCUGCAGGAUGGCAUUUAAUU), APC11-2 (AAGAUUAAGUGCUGGAACGUU), APC10-1 (GAGCUCCAUUGGUAAAUUUUU), APC10-2 (GAAAUUGGGUCACAAGCUGUU), APC6-1 (CUAUGGACCUGCAUGGAUAUU), APC6-2 (CGAGGUAACAGUUGACAAAUU), APC8-1 (GAAAUUAAAUCCUCGGUAUUU), APC8-2 (GCAGUUGCCUAUCACAAUAUU) and GAPDH (D-001830-01). Cells were transfected with 100 nM of mixture of two oligos using oligofectamine (Invitrogen, USA). Cells were transfected twice: first for 24 hr after which siRNA oligos were transfected again for a further 48 hr before harvesting cells for immunoprecipitation or analyzing by microscopy.
Microscopy
Cells were incubated on the microscope using the Delta T system (Bioptechs, PA, USA) and imaged by time-lapse fluorescence and DIC microscopy on a Leica DMIRBE or DMIR2 microscope equipped with a 40x 1.2 NA oil immersion lens. Cerulean-CFP and Venus-YFP were visualised using a JP5 filter set (Chroma, VE, USA) with excitation and emission filters in filter wheels (Lambda 10-3, Sutter Instrument Co, CA, USA) and a Cascade 512B or QuantEM CCD camera (Photometrics, AZ, USA). Multiple cell positions were captured using a Corvus (Marzhauser, Germany) or H117 (Prior, UK) stage. Shutters (Smart shutter, Sutter Instrument Co, CA, USA), filter wheels, stages, microscopes and cameras were all controlled by SlideBook software (Intelligent Imaging Innovations, CO, USA). Images were captured at 3 min intervals and analysed using Slidebook. Images were exported to ImageJ to assemble into movies.
Immunoprecipitation
Protein complexes were immunoprecipitated with antibodies covalently coupled to Dynabeads (Invitrogen) using HEPES buffer (100 mM KaCl, 40 mM Hepes pH 7.8, 10 mM EDTA, 10 % Glycerol, 0.1 % NP-40, 1 mM DTT, Roche complete inhibitor cocktail tablet, 0,2 μM microcystin, 1 mM PMSF) for incubation and washing. Cells for immunoprecipitation were lysed with HEPES buffer for 10 min on ice and clarified by a 20000 x g spin for 10 min.
Gel filtration column chromatography
Cells were resuspended in buffer A (140 mM NaCl, 30 mM Hepes pH 7.8, 6 mM MgCl2, 5% glycerol, 1 mM dithiothreitol (DTT), Roche complete inhibitor cocktail tablet, 0,2 μM microcystin, 1 mM PMSF) at a 1:1 ratio of buffer to cells, and opened by nitrogen cavitation (1000 PSI, 30 min, Parr Instrument). Lysed cells were centrifuged at 20,000g for 10 min and 259,000g for 10 min before loading on Superose 6 PC 3.2-30 (GE Healthcare). The column was run at a flow of 25 μl-min–1 in buffer B (140 mM NaCl, 30 mM Hepes 7.8, 5% glycerol, 1 mM DTT) and 50 μl fractions collected.
Antibodies
The following antibodies were used at the indicated dilutions. Cdc20 (A301-180A, Bethyl laboratories) 1:200, BubR1 (A300-386A, Bethyl laboratories), 1:500, Cyclin B1 (mAb GNS-1, BD Pharmingen) 1:1000, Cyclin A (mAb AT10.3, CRUK) 1:1000, APC3 (610455, BD Transduction Laboratories) 1:500, APC4 (monoclonal antibody raised against a C-terminal peptide) 1:500, APC11 (monoclonal antibody raised against a C-terminal peptide) 1:500, APC7 (Abcam 4171) 1:500, APC8 (Biolegend) 1:500, APC6 (Santa Cruz Biotechnology), APC10 (raised against full length protein) 1:1000, Hsp70 (Sigma) 1:5000.
Secondary antibodies used for LiCor: Alexa Flour 680 rabbit anti-goat (A21088), Alexa Fluor 680 goat anti-mouse (A21057), Alexa Fluor 680 goat anti-rabbit (A21076) all used at 1:5000.
Quantitative immunoblotting
After blotting with primary antibodies, blots were incubated with fluorescently labelled secondary antibodies and the fluorescence measured using a LI-COR Odyssey CCD scanner according to the manufacturer’s instructions (LI-COR Biosciences, NE, USA).
Stable inducible cell line
The HeLa-FRT cell line (gift of Stephen Taylor, University of Manchester) was transfected using the FLIP-in system (Invitrogen), to generate stable inducible cell lines. Cells were induced with tetracycline (1 μg-ml, Calbiochem) 48 hr before harvesting. APC3 and APC8 ORF were cloned into a modified version of pCDNA5-FRT-TO (Invitrogen).
Analysis statistics of degradation curves
The fluorescence of individual cells was measured and the value at NEBD set to 1. The relative fluorescence units were transferred to Prism Software and the maximal slope of cyclin degradation was obtained by using Prism software. The maximal slope was calculated by nonlinear regression analysis (curve fit) and assuming a sigmoidal dose-response (variable slope). The standard deviation was calculated from these values. P values were calculated by a student’s t-test against the control cells.
References
- 1.Pines J. Mitosis: a matter of getting rid of the right protein at the right time. Trends Cell Biol. 2006;16:55–63. doi: 10.1016/j.tcb.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 2.Peters JM. The anaphase promoting complex-cyclosome: a machine designed to destroy. Nature reviews. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 3.Thornton BR, et al. An architectural map of the anaphase-promoting complex. Genes & development. 2006;20:449–460. doi: 10.1101/gad.1396906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gmachl M, Gieffers C, Podtelejnikov AV, Mann M, Peters JM. The RING-H2 finger protein APC11 and the E2 enzyme UBC4 are sufficient to ubiquitinate substrates of the anaphase-promoting complex. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:8973–8978. doi: 10.1073/pnas.97.16.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang Z, et al. APC2 Cullin protein and APC11 RING protein comprise the minimal ubiquitin ligase module of the anaphase-promoting complex. Molecular biology of the cell. 2001;12:3839–3851. doi: 10.1091/mbc.12.12.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwab M, Neutzner M, Mocker D, Seufert W. Yeast Hct1 recognizes the mitotic cyclin Clb2 and other substrates of the ubiquitin ligase APC. The EMBO journal. 2001;20:5165–5175. doi: 10.1093/emboj/20.18.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sorensen CS, et al. A conserved cyclin-binding domain determines functional interplay between anaphase-promoting complex-Cdh1 and cyclin A-Cdk2 during cell cycle progression. Molecular and cellular biology. 2001;21:3692–3703. doi: 10.1128/MCB.21.11.3692-3703.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burton JL, Solomon MJ. Hsl1p, a swe1p inhibitor, is degraded via the anaphase-promoting complex. Molecular and cellular biology. 2000;20:4614–4625. doi: 10.1128/mcb.20.13.4614-4625.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kraft C, Vodermaier HC, Maurer-Stroh S, Eisenhaber F, Peters JM. The WD40 propeller domain of Cdh1 functions as a destruction box receptor for APC-C substrates. Molecular cell. 2005;18:543–553. doi: 10.1016/j.molcel.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 10.Hilioti Z, Chung Y, Mochizuki Y, Hardy CF, Cohen-Fix O. The anaphase inhibitor Pds1 binds to the APC-C-associated protein Cdc20 in a destruction box-dependent manner. Curr Biol. 2001;11:1347–1352. doi: 10.1016/s0960-9822(01)00399-2. [DOI] [PubMed] [Google Scholar]
- 11.Pfleger CM, Lee E, Kirschner MW. Substrate recognition by the Cdc20 and Cdh1 components of the anaphase-promoting complex. Genes & development. 2001;15:2396–2407. doi: 10.1101/gad.918201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mallory MJ, Cooper KF, Strich R. Meiosis-specific destruction of the Ume6p repressor by the Cdc20-directed APC-C. Molecular cell. 2007;27:951–961. doi: 10.1016/j.molcel.2007.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimata Y, et al. A mutual inhibition between APC-C and its substrate Mes1 required for meiotic progression in fission yeast. Developmental cell. 2008;14:446–454. doi: 10.1016/j.devcel.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 14.Eytan E, Moshe Y, Braunstein I, Hershko A. Roles of the anaphase-promoting complex-cyclosome and of its activator Cdc20 in functional substrate binding. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2081–2086. doi: 10.1073/pnas.0510695103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Passmore LA, Barford D. Coactivator functions in a stoichiometric complex with anaphase-promoting complex-cyclosome to mediate substrate recognition. EMBO Rep. 2005;6:873–878. doi: 10.1038/sj.embor.7400482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sullivan M, Morgan DO. Finishing mitosis, one step at a time. Nature reviews. 2007;8:894–903. doi: 10.1038/nrm2276. [DOI] [PubMed] [Google Scholar]
- 17.Pfleger CM, Kirschner MW. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes & development. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- 18.Blanco MA, Sanchez-Diaz A, de Prada JM, Moreno S. APC(ste9-srw1) promotes degradation of mitotic cyclins in G(1) and is inhibited by cdc2 phosphorylation. The EMBO journal. 2000;19:3945–3955. doi: 10.1093/emboj/19.15.3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Visintin R, Prinz S, Amon A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science (New York, N.Y. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- 20.Sigrist SJ, Lehner CF. Drosophila fizzy-related down-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell. 1997;90:671–681. doi: 10.1016/s0092-8674(00)80528-0. [DOI] [PubMed] [Google Scholar]
- 21.Sudo T, et al. Activation of Cdh1-dependent APC is required for G1 cell cycle arrest and DNA damage-induced G2 checkpoint in vertebrate cells. The EMBO journal. 2001;20:6499–6508. doi: 10.1093/emboj/20.22.6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li M, et al. The adaptor protein of the anaphase promoting complex Cdh1 is essential in maintaining replicative lifespan and in learning and memory. Nature cell biology. 2008 doi: 10.1038/ncb1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Higuera I, et al. Genomic stability and tumour suppression by the APC-C cofactor Cdh1. Nature cell biology. 2008;10:802–811. doi: 10.1038/ncb1742. [DOI] [PubMed] [Google Scholar]
- 24.Floyd S, Pines J, Lindon C. APC-C Cdh1 targets aurora kinase to control reorganization of the mitotic spindle at anaphase. Curr Biol. 2008;18:1649–1658. doi: 10.1016/j.cub.2008.09.058. [DOI] [PubMed] [Google Scholar]
- 25.Rape M, Reddy SK, Kirschner MW. The processivity of multiubiquitination by the APC determines the order of substrate degradation. Cell. 2006;124:89–103. doi: 10.1016/j.cell.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 26.Summers MK, Pan B, Mukhyala K, Jackson PK. The unique N terminus of the UbcH10 E2 enzyme controls the threshold for APC activation and enhances checkpoint regulation of the APC. Molecular cell. 2008;31:544–556. doi: 10.1016/j.molcel.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rape M, Kirschner MW. Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature. 2004;432:588–595. doi: 10.1038/nature03023. [DOI] [PubMed] [Google Scholar]
- 28.Walker A, Acquaviva C, Matsusaka T, Koop L, Pines J. UbcH10 has a rate-limiting role in G1 phase but might not act in the spindle checkpoint or as part of an autonomous oscillator. Journal of cell science. 2008;121:2319–2326. doi: 10.1242/jcs.031591. [DOI] [PubMed] [Google Scholar]
- 29.Clute P, Pines J. Temporal and spatial control of cyclin B1 destruction in metaphase. Nature cell biology. 1999;1:82–87. doi: 10.1038/10049. [DOI] [PubMed] [Google Scholar]
- 30.den Elzen N, Pines J. Cyclin A is destroyed in prometaphase and can delay chromosome alignment and anaphase. The Journal of cell biology. 2001;153:121–136. doi: 10.1083/jcb.153.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geley S, et al. APC-C-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J. Cell Biol. 2001;153:137–148. doi: 10.1083/jcb.153.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hames RS, Wattam SL, Yamano H, Bacchieri R, Fry AM. APC-C-mediated destruction of the centrosomal kinase Nek2A occurs in early mitosis and depends upon a cyclin A-type D-box. The EMBO journal. 2001;20:7117–7127. doi: 10.1093/emboj/20.24.7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Fiore B, Pines J. How Cyclin A destruction escapes the spindle assembly checkpoint. J. Cell Biol. 2010 doi: 10.1083/jcb.201001083. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolthuis R, et al. Cdc20 and Cks direct the spindle checkpoint-independent destruction of cyclin A. Molecular cell. 2008;30:290–302. doi: 10.1016/j.molcel.2008.02.027. [DOI] [PubMed] [Google Scholar]
- 35.Hayes MJ, et al. Early mitotic degradation of Nek2A depends on Cdc20-independent interaction with the APC-C. Nature cell biology. 2006;8:607–614. doi: 10.1038/ncb1410. [DOI] [PubMed] [Google Scholar]
- 36.Vodermaier HC, Gieffers C, Maurer-Stroh S, Eisenhaber F, Peters JM. TPR subunits of the anaphase-promoting complex mediate binding to the activator protein CDH1. Curr Biol. 2003;13:1459–1468. doi: 10.1016/s0960-9822(03)00581-5. [DOI] [PubMed] [Google Scholar]
- 37.Hagting A, et al. Human securin proteolysis is controlled by the spindle checkpoint and reveals when the APC-C switches from activation by Cdc20 to Cdh1. The Journal of cell biology. 2002;157:1125–1137. doi: 10.1083/jcb.200111001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohta T, Michel JJ, Schottelius AJ, Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Molecular cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- 39.Zachariae W, Nasmyth K. TPR proteins required for anaphase progression mediate ubiquitination of mitotic B-type cyclins in yeast. Molecular biology of the cell. 1996;7:791–801. doi: 10.1091/mbc.7.5.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deak P, Donaldson M, Glover DM. Mutations in makos, a Drosophila gene encoding the Cdc27 subunit of the anaphase promoting complex, enhance centrosomal defects in polo and are suppressed by mutations in twins-aar, which encodes a regulatory subunit of PP2A. Journal of cell science. 2003;116:4147–4158. doi: 10.1242/jcs.00722. [DOI] [PubMed] [Google Scholar]
- 41.Carroll CW, Enquist-Newman M, Morgan DO. The APC subunit Doc1 promotes recognition of the substrate destruction box. Curr Biol. 2005;15:11–18. doi: 10.1016/j.cub.2004.12.066. [DOI] [PubMed] [Google Scholar]
- 42.Matyskiela ME, Morgan DO. Analysis of Activator-Binding Sites on the APC-C Supports a Cooperative Substrate-Binding Mechanism. Molecular cell. 2009;34:68–80. doi: 10.1016/j.molcel.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Passmore LA, et al. Doc1 mediates the activity of the anaphase-promoting complex by contributing to substrate recognition. The EMBO journal. 2003;22:786–796. doi: 10.1093/emboj/cdg084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kimata Y, Baxter JE, Fry AM, Yamano H. A role for the Fizzy-Cdc20 family of proteins in activation of the APC-C distinct from substrate recruitment. Molecular cell. 2008;32:576–583. doi: 10.1016/j.molcel.2008.09.023. [DOI] [PubMed] [Google Scholar]
- 45.Herzog F, et al. Structure of the Anaphase-Promoting Complex-Cyclosome Interacting with a Mitotic Checkpoint Complex. Science (New York, N.Y. 2009;323:1477–1481. doi: 10.1126/science.1163300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Z, et al. Molecular structure of the N-terminal domain of the APC-C subunit Cdc27 reveals a homo-dimeric tetratricopeptide repeat architecture. Journal of molecular biology. 2010;397:1316–1328. doi: 10.1016/j.jmb.2010.02.045. [DOI] [PubMed] [Google Scholar]
- 47.Dube P, et al. Localization of the coactivator Cdh1 and the cullin subunit Apc2 in a cryo-electron microscopy model of vertebrate APC-C. Molecular cell. 2005;20:867–879. doi: 10.1016/j.molcel.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 48.Yu H. Cdc20: a WD40 activator for a cell cycle degradation machine. Molecular cell. 2007;27:3–16. doi: 10.1016/j.molcel.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 49.Nilsson J, Yekezare M, Minshull J, Pines J. The APC-C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nature cell biology. 2008;10:1411–1420. doi: 10.1038/ncb1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steen JA, et al. Different phosphorylation states of the anaphase promoting complex in response to antimitotic drugs: a quantitative proteomic analysis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:6069–6074. doi: 10.1073/pnas.0709807104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gascoigne KE, Taylor SS. Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer cell. 2008;14:111–122. doi: 10.1016/j.ccr.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 52.Kulukian A, Han JS, Cleveland DW. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Developmental cell. 2009;16:105–117. doi: 10.1016/j.devcel.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang Z, Bharadwaj R, Li B, Yu H. Mad2-Independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Developmental cell. 2001;1:227–237. doi: 10.1016/s1534-5807(01)00019-3. [DOI] [PubMed] [Google Scholar]
- 54.van Zon W, et al. The APC-C recruits cyclin B1-Cdk1-Cks in prometaphase before D box recognition to control mitotic exit. The Journal of cell biology. 2010;190:587–602. doi: 10.1083/jcb.200912084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burton JL, Solomon MJ. Mad3p, a pseudosubstrate inhibitor of APCCdc20 in the spindle assembly checkpoint. Genes & development. 2007;21:655–667. doi: 10.1101/gad.1511107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pines J, Hunter T. Isolation of a human cyclin cDNA: evidence for cyclin mRNA and protein regulation in the cell cycle and for interaction with p34cdc2. Cell. 1989;58:833–846. doi: 10.1016/0092-8674(89)90936-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.