Abstract
Purpose
Multiple developmental pathways including Notch, Hedgehog, and Wnt are active in malignant brain tumors such as medulloblastoma and glioblastoma (GBM). This raises the possibility that tumors might compensate for therapy directed against one pathway by upregulating a different one. We investigated whether brain tumors show resistance to therapies against Notch, and whether targeting multiple pathways simultaneously would kill brain tumor cells more effectively than monotherapy.
Experimental Design
We used GBM neurosphere lines to investigate the effects of a gamma-secretase inhibitor (MRK-003) on tumor growth, and chromatin immunoprecipitation (ChIP) to study the regulation of other genes by Notch targets. We also evaluated the effect of combined therapy with a Hedgehog inhibitor (cyclopamine) in GBM and medulloblastoma lines, and primary human GBM cultures.
Results
GBM cells are at least partially resistant to long-term MRK-003 treatment, despite ongoing Notch pathway suppression, and show concomitant upregulation of Wnt and Hedgehog activity. The Notch target Hes1, a repressive transcription factor, bound the Gli1 first intron, and may inhibit its expression. Similar results were observed in a melanoma-derived cell line. Targeting Notch and Hedgehog simultaneously induced apoptosis, decreased cell growth, and inhibited colony-forming ability more dramatically than monotherapy. Low-passage neurospheres isolated from freshly resected human GBMs were also highly susceptible to co-inhibition of the two pathways, indicating that targeting multiple developmental pathways can be more effective than monotherapy at eliminating glioblastoma-derived cells.
Conclusion
Notch may directly suppress Hedgehog via Hes1 mediated inhibition of Gli1 transcription, and targeting both pathways simultaneously may be more effective at eliminating GBMs cells.
Introduction
Glioblastoma (GBM) is the most common malignant primary central nervous system tumor in adults and is characterized by resistance to chemo- and radiotherapy (1). Prognosis remains very poor, with most patients surviving less than two years (2) despite recent advances in surgery and chemotherapy. It has become clear that GBMs are a diverse group of tumors, with different subtypes activating distinct sets of oncogenes and signaling pathways (3). Because of this, no single therapy is likely to be effective against all GBMs, and a number of pharmacologic agents with activity against specific targets such as EGFR, Akt, Hedgehog, mTOR, PI3K, PDGFR, Raf, TGF-β are being developed (4). However, even the use of targeted therapies can be limited by the emergence of resistant tumor cells, and resistance to EGFR inhibitors (5) and Hedgehog inhibitors (6) has already been documented.
An important developmental pathway required in at least a subset of GBMs is Notch. Aberrant Notch signaling was implicated in the initiation of T-cell lymphoblastic leukemia in the early 1990s (7), and has since been demonstrated in many different hematopoietic and epithelial tumors (8-10). Upregulation of Notch pathway components has been demonstrated in GBM (11-13) as well as the malignant embryonal tumor medulloblastoma (14, 15), and Notch pathway inhibition has emerged as a potential therapy for malignant brain tumors. The four Notch receptors (Notch 1-4) bind ligands (Jagged and Delta) expressed on adjacent cells, permitting cleavage of Notch via ADAM metalloprotease and then gamma-secretase (16). The released intracellular domain of Notch (ICD) translocates to the nucleus, where it binds CBF-1/RBP-J and promotes transcription of the Hes/Hey genes which help maintain a progenitor-like state by repressing transcription of pro-differentiation genes during development (17, 18).
Many different techniques for Notch blockade have been attempted, including gamma-secretase inhibitors (GSI) (19), siRNA (12), monoclonal antibodies (20-22), and small inhibitory molecules directly affecting the transcriptional complex (23). siRNA and GSIs have been tested in the context of malignant brain tumors (12, 13, 19, 24) with promising results in vitro and in xenograft models. Over twenty Phase I/II clinical trials investigating the efficacy of GSIs in tumors are actively recruiting or awaiting activation (www.clinicaltrials.gov), but it is uncertain whether inhibition of Notch signaling alone will be sufficient to prevent tumor growth as cancer adaptation is well-documented. We assessed the effects of Notch inhibition on malignant brain tumor cells and the potential emergence of therapeutic resistance. Some GBM neurosphere lines that survived long-term Notch inhibition upregulated Wnt and Hedgehog, with the latter effect due potentially to Hes1 binding and inhibiting Gli1 at the transcriptional level. We found that inhibiting Notch and Hedgehog simultaneously dramatically decreased growth of neurosphere cultures and primary human GBM cells, suggesting this regulatory mechanism may contribute to resistance.
Materials and Methods
Cell Culture
DAOY, PFSK, U87, 22RV1, H157, KMS12, L428, Mel10, Reh, TOV-112D, and U937 were maintained in the recommended media with 10% fetal bovine serum (FBS) unless otherwise specified. HSR-GBM1 and HSR-GBM2 were maintained as neurosphere cultures in serum-free neurosphere media (25). Cell line identity was verified using SNP analysis. For all assays, cells were counted using GUAVA Viacount reagent as per manufacturer’s instructions (#4000-0040, Millipore, Billerica, MA) and equal numbers of viable cells were used for all experiments.
For drug treatment assays, adherent cell lines were plated overnight in 6- or 96-well plates (BD Falcon; BD Biosciences, Bedford, MA) with media containing 10% FBS. The next day media was changed to low-serum (0.5% FBS) and MRK-003 (26), cyclopamine (Infinity Pharmaceuticals, Cambridge, MA), or vehicle (DMSO or ethanol, respectively) was added to each well as specified. Media was changed to every 2-3 days as necessary. For neurosphere lines, cells were treated immediately upon plating with drugs as specified. Cell biomass was measured using CellTiter96 (Promega, Madison, WI) at regular intervals after treatment. Anchorage independent growth assays measuring colony forming ability were performed as previously described (19). Colonies were stained and counted 21-28 days after plating.
Neurosphere nucleofection assays were performed using the AMAXA Mouse NSC Nucleofector Kit (VPG-1004, Lonza, Basel, Switzerland) per manufacturer’s instruction, using program A-033 with 2×106 cells per condition. Cells were nucleofected with Hes1 (27) or a control plasmid and allowed to recover for 24 hours in normal media before treatment with MRK-003 or vehicle. Transfection efficiency was quantified by cotransfection with CAG-GFP and microscopic quantification of the percentage of GFP-expressing cells. Cells were harvested after 48 hours for analysis.
Notch2 overexpression was achieved by incubating 4×105 dissociated cells in a 12-well plate with neurosphere media and 8ug/ml Polybrene (107689, Sigma-Aldrich, St. Louis, MO). Concentrated retrovirus designed to express Notch2 ICD with a truncated PEST domain (aa1703-2146) was added to the cells and the dish was rotated every 20 minutes for two hours, at which time 2mls media was added to the cells. Cells were harvested 48 hours later. In some assays, infected cells were treated with MRK-003 24 hours after infection and were harvested 48 hours later.
shRNAs
Lentivirus was produced as previously described (28) from shRNA constructs against human Notch1 (TRCN0000003359 and TRCN0000003360) and Notch2 (TRCN0000004895 and TRCN0000004896). Neurosphere lines were infected as described above. Cells were harvested 72 hours after infection, RNA was isolated, and target levels were assayed by QPCR.
Primary Tumor-Derived Cell Culture
JHH-GBM4, JHH-GBM10, JHH-GBM11, JHH-GBM14, JHH-GBM17, JHH-GBM18, JHH-GBM20, and JHH-GBM23 were generated from primary GBM surgical specimens at Johns Hopkins Hospital as previously described (25, 28). JHH-GBM4, JHH-GBM17, JHH-GBM18, JHH-GBM20, and JHH-GBM23 were used as primary or very low passage cultures (passage 0-2), while JHH-GBM10 and JHH-GBM11 were analyzed at passage 12-20. JHH-GBM14 was used both as a primary culture and at later passages (10-15) as indicated in the text.
Quantitative PCR
RNA was extracted using an RNeasy kit (#74104, Qiagen,Valencia, CA) with on-column DNase treatment (#79254, Qiagen) according to manufacturer’s instructions. Reverse transcription was performed, and quantitative PCR was done using SYBR Green PCR Master Mix (#4309155, Applied Biosystems, Foster City, CA) on an I-Cycler IQ Real-Time detection system (Bio-Rad, Hercules CA) according to manufacturer’s instructions. The following primers were obtained from published literature: hGli1, hPtc1B and Beta-Actin (29). hHes1, hHes5, hHey1, and hHey2 primers were designed using Primer3 (30) hHes1: Forward (F) 5′-AGTGAAGCACCTCCGGAAC-3′ Reverse (R) 5′-TCACCTCGTTCATGCACTC-3′ hHes5: Forward (F) 5′-CCGGTGGTGGAGAAGATG-3′ Reverse (R) 5′-TAGTCCTGGTGCAGGCTCTT-3′ hAxin2 primers were a generous gift from Brian Simons: Forward (F) 5′- CTGGTGGCTGGTGCAAAGAC-3′ Reverse (R) 5′-CGAGTGTGAGGTCCACGGAA-3′. The standard curve technique was used to determine expression levels and values were normalized to beta actin.
Protein Analysis
Protein was extracted from cell pellets using RIPA buffer (R0278, Sigma-Aldrich) and 30ug was run on each lane of a NuPage 4-12% Bis-Tris gel (NP0321, Invitrogen) according to manufacturer’s instructions. The antibodies used were as follows: rabbit anti-Hes1 (1:400, AVIVA Systems Biology, San Diego CA, ARP32372; 1:1000, Toray Industries, Tebiro, Kamakura, Japan), rabbit monoclonal anti-Cleaved Notch1 (Cell Signaling, Danvers, MA, #2421 and #4147), and mouse monoclonal anti-GAPDH (1:50,000, Research Diagnostics Inc., Flanders NJ, RDI-TRK564-6C5).
Chromatin Immunoprecipitation (ChIP)
ChIP was performed using two different techniques and antibodies. For the Magna ChIP kit (17-610, Millipore), cells were grown in the appropriate media, harvested during log-phase growth, crosslinked using 1% formaldehyde, and processed according to the manufacturer’s instructions. The positive control was anti-Acetyl Histone H3, negative control was rabbit IgG, and Rabbit anti-Hes1 was used for pulldown (AVIVA Systems Biology). Non-quantitative PCR was performed using the primers in Supplemental Table 1. GAPDH and Hes1 primer sets were used as negative and positive controls, respectively, as Hes1 has previously been shown to bind its own promoter (31, 32). Some samples were run using quantitative PCR as well.
The other technique used for ChIP has been previously described (33). Briefly, cells were grown in the appropriate media, harvested during log-phase growth, and crosslinked with formaldehyde. 5 μg of Hes1 antibody (AB5702, Millipore) or 5 μg control (rabbit IgG) was added to the sample and incubated for 12 hrs at 4°C. After washing, crosslinking was reversed and quantitative PCR was performed as described above using the primers in Table 1. p63 and p27Kip primer sets were used as negative and positive controls, respectively (34). Linear amplification of each primer set used for qPCR was verified by a standard curve. qPCR calculations were done as previously described (35). Briefly, the average of the cycle threshold values (CTs) was calculated for each input, sample, and control. The input CT was subtracted from the corresponding sample and control CTs. The following formula was then applied: power (1.9, negative ln (subtracted value)). This value was used for further calculations. Each sample and control was normalized by dividing both numbers by the highest value so that each ChIP experiment was scaled from 0 to 1 and outliers were removed. For sample minus control values, a negative number was replaced with a zero.
Table 1.
Top Differentially Expressed Genes in Microarray. Bold are genes altered in “Top 100” gene list
| Notch Pathway | Up: CCND1, CREBBP, DLL1, DLL3, ELAVL4,IGF1R, KLF4, TFRC, Down: EFEMP1, HES1, HES5, HEY1, HEY2, HOXA5, HOXA7, HOXA9, |
| Wnt Pathway | Up: CCND1, CREBBP, NRFA1, SFRP1, WNT5B, WNT7A, Down: ERBB2, FRZB, FZD8, TCF7L1 |
| Hedgehog Pathway | Up: MYCN, GLI2, NR4A1, Down: GLI1, GLI3, SUFU, |
| Other Pathways | Up: ABCA1, VEGFA Down: HOXA4, HOXC9, PDPN, PDGFRA, RB1, S100B, TGFB1, |
Gene Expression Analysis
Gene expression was measured using Agilent’s 44K whole human genome microarrays at the Johns Hopkins Oncology Microarray Core, with labeling, hybridization, and detection performed according to manufacturer’s instructions (Agilent Technologies, Santa Clara, CA). Differential gene expression, gene set enrichment analyses, and Analysis of Functional Annotation (AFA) were performed as previously described (36, 37), using statistical packages from the R/Bioconductor project (38, 39). Gene annotation for the microarray used in this study was obtained from the corresponding R-Bioconductor metadata packages. Raw expression data along with MIAME required information is located in the GEO database (40).
Results
GBM Neurosphere Cultures Survive Long-term Notch Inhibition
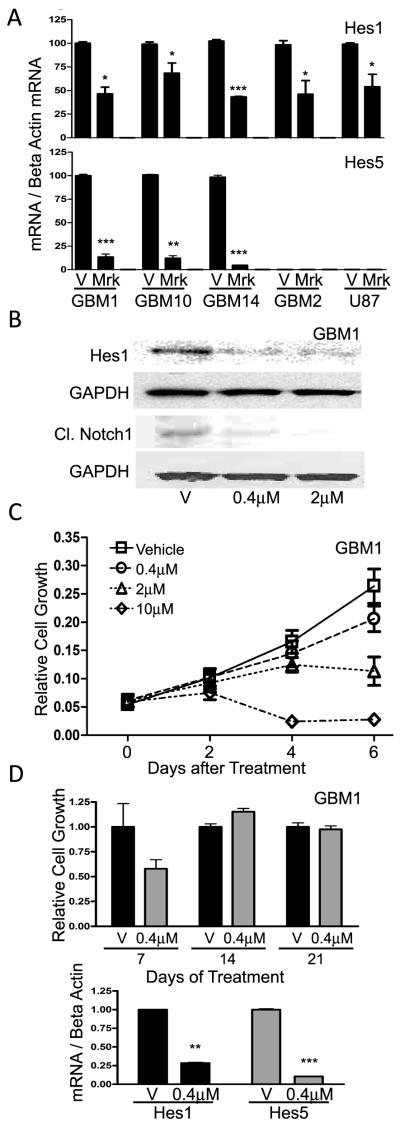
To confirm previous reports (24, 41) and examine Notch suppression in the newly derived GBM neurosphere lines JHH-GBM10 and JHH-GBM14, we treated dissociated neurospheres with MRK-003 and evaluated expression of the Notch pathway targets Hes1 and Hes5. We also looked at an adherent, serum-cultured GBM-derived line, U87. We observed that MRK-003 treatment significantly decreased expression of Hes1 in all lines, with inhibition ranging from 32% (JHH-GBM10) to 57% (JHH-GBM14; Fig. 1A). Significant reductions in the expression of Hes5 ranging from 87% (HSR-GBM1) to 95% (JHH-GBM14; Fig. 1A) were also noted in three GBM neurosphere lines. Interestingly, U87 and HSR-GBM2 did not express Hes5 at sufficient levels for quantitation, indicating some heterogeneity in Notch target levels between tumors. MRK-003 treatment also inhibited Hes1 expression by 40% in the medulloblastoma cell line DAOY (Supplemental Fig. 1A). In addition to downregulation of Hes1 transcripts, Hes1 protein levels were suppressed by MRK-003 treatment in HSR-GBM1 (Fig. 1B) and DAOY (Supplemental Fig. 1B), with 70-80% inhibition following 2 μM doses (P<0.0001). Cleaved Notch1 levels also decreased dramatically with MRK-003 treatment, dropping to almost undetectable levels (Fig. 1B) as measured by two different antibodies. We confirmed dose-dependent inhibition of HSR-GBM1 and DAOY cell growth following application of MRK-003 in relatively short term six-to-nine day growth assays (Fig. 1C and Supplemental Fig. 1C).
Figure 1.
Long-term pharmacologic Notch inhibition causes resistance. A, Hes1 and Hes5 levels measured by quantitative PCR in response to 0.4 μM MRK-003. B, Western blot and quantification showing Hes1 and cleaved Notch1 protein levels in response MRK-003. C, MTS assay measuring cell growth after treatment with MRK-003. Data are representative of N=5. D, MTS assay measuring cell growth after treatment with 0.4μM MRK-003 normalized to vehicle, upper panel, and Hes1 mRNA levels after three weeks of MRK-003 treatment, lower panel. Experiment was done in triplicate for N=2, data shown are representative. * P<0.02, ** P<0.001, *** P<0.0001, using Student’s t-test.
Interestingly, although neurosphere lines HSR-GBM1 and HSR-GBM2 showed a short-term response to Notch inhibition, they continued to grow even after three or more passages with continuous MRK-003 treatment (Fig. 1D and data not shown). These experiments were performed three or more times with similar results, indicating the capacity for long-term growth was a reproducible phenomenon. Notch signaling was still quite pronounced at the end of long-term MRK-003 treatment as demonstrated by very low Hes1 and Hes5 levels (Fig. 1D), suggesting that ongoing growth was not due to reactivation of the Notch pathway. Higher concentrations of MRK-003 (2-10 μM) did result in complete cessation of cell growth over the course of 10-14 days, indicating that at sufficiently high drug levels cells do not easily develop resistance.
Up-regulation of Hedgehog and Wnt Signaling Following Notch Inhibition
Given the ability of a subset of malignant brain tumor cells to survive long term MRK-003 treatment, we performed a microarray assay on four GBM-derived neurosphere lines with two different doses of MRK-003 to identify Notch targets and determine if other pathways might be upregulated to compensate for the loss of Notch activity (HSR-GBM1, N=3 each for two doses; HSR-GBM2, N=3; JHH-GBM10, N=3; and JHH-GBM14, N=2). As expected, canonical Notch targets including Hes1, Hes5, and Hey1 were suppressed 2.7-28 fold following MRK-003 treatment. A list of the top differentially regulated genes seen in common across all lines is included in Supplemental Table 2. We performed a gene set enrichment analysis (GSEA) comparing our data against literature-cited, manually curated pathway lists with genes up- and down-regulated by pathway activation and found that, as expected, Notch was significantly changed (P = 0.013). We found that two other developmental and proliferative pathways were significantly altered: Wnt (P = 0.00013) and Hedgehog (P = 0.0027; Table 1). Closer analysis of specific Hedgehog and Wnt targets indicated that there was an overall trend towards upregulation of canonical targets and pathway components in the presence of MRK-003.
Since Wnt and Hedgehog play an important role in stem cell maintenance and proliferation during normal brain development and can be dysregulated in tumors, we hypothesized that upregulation of these pathways might compensate for MRK-003 induced loss of Notch signaling. Using QPCR to validate our microarray data, we found that the Hedgehog targets Patched1B and Gli1 were both significantly upregulated by up to two-fold in neurosphere lines following MRK-003 treatment (2-5 μM) (Fig. 2A). The Wnt signaling target Axin2 also showed significant upregulation (Fig. 2A). In contrast, medulloblastoma cell line DAOY showed a 40% decrease in Ptc1B mRNA and no significant change in Gli1 or Axin2 (Supplemental Fig. 1D).
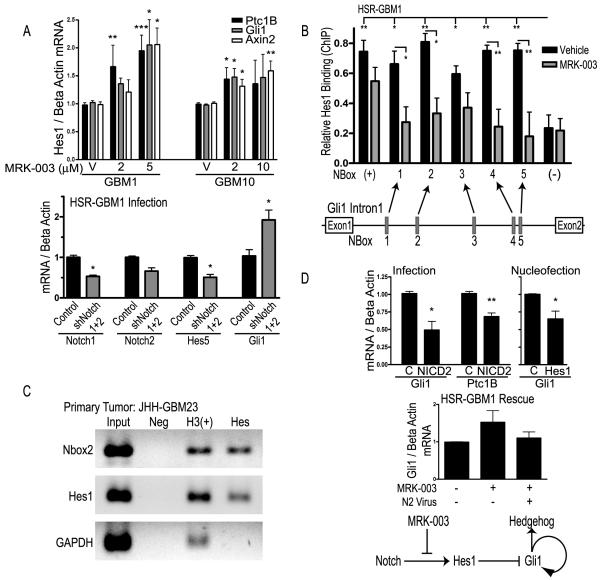
Figure 2.
Notch regulates Gli1 expression. A, mRNA levels of Hedgehog and Wnt pathway targets after treatment with MRK-003, upper panel. Expression of Notch1, Notch2, Hes5, and Gli1 after infection with shRNA, lower panel. B, ChIP of Hes1 measured via QPCR relative to IgG in HSR-GBM1. Positive and negative controls are p27 and p63, respectively. Schematic of N-Box locations in the Gli1first intron is shown below. C, ChIP of a primary GBM specimen measured via non-quantitative PCR relative to IgG. Negative and positive controls are Rabbit IgG and acetyl histone H3, respectively. D, mRNA levels of Gli1 and Ptc1B after infection with control (C) or activated Notch2 virus (NICD2), and mRNA levels of Gli1 after nucleofection with Hes1 or control plasmid, upper panel. Rescue experiment showing induction of Gli1 with MRK-003 treatment is inhibited by addition of activated Notch2 virus, lower panel. Schematic of interaction between Notch and Hedgehog signaling. * P<0.05, ** P< 0.005, *** P<0.0005, using Student’s t-test.
As GSIs are able to cleave proteins other than Notch, we used shRNA to confirm the effect of Notch inhibition on Hedgehog signaling. Infection of HSR-GBM1 neurospheres with lentivirus encoding shRNA targeting either Notch1 or Notch2 resulted in significant reductions in receptor mRNA levels (data not shown). However, inhibiting either Notch1 or Notch2 alone resulted in an overall increase in Notch signaling as evidenced by upregulation of the Notch targets Hes1 and Hes5, suggesting compensation by other Notch receptors (data not shown). We therefore simultaneously used shRNAs against both Notch1 and Notch2, and saw a decrease in mRNA encoding the two Notch receptors of approximately 30-50%, along with a decrease in Notch targets by a similar amount (Fig. 2A). Inhibition of Notch signaling using shNotch1 and shNotch2 induced Gli1 expression by approximately two-fold (Fig. 2A; P = 0.02).
Hes1 Regulates Hedgehog Signaling via Direct Binding to Gli1
As previous work has suggested the potential for crosstalk between the Notch and Hedgehog signaling cascades (14), we focused on the upregulation of Hedgehog signaling in response to MRK-003. Increased levels of Notch pathway components are seen in Hedgehog-driven medulloblastoma models (15, 42) and it was initially suggested that these tumors may be dependent on Notch signaling for survival (15). More recent studies however, indicate that Hedgehog-driven medulloblastomas can grow in the absence of canonical Notch activity (43, 44). Additionally, several groups have demonstrated that Hedgehog pathway components Gli1 and Gli2 are able to positively regulate Hes1 independently of Notch (45, 46). However, when we treated several GBM neurosphere lines with the Hedgehog inhibitor cyclopamine we did not observe decreases in Hes1 or other Notch pathway targets, suggesting that Hedgehog does not play a significant role regulating Notch targets in malignant gliomas (Supplemental Fig. 2A).
We investigated the possibility of direct Gli1 regulation by Hes1, as potential Hes1 binding sites were previously identified in the first Gli1 intron using in silico analysis (47). Hes1 negatively regulates transcription of targets, thus loss of Hes1 following Notch blockade would be predicted to relieve repression of Gli1 expression and activate the Hedgehog cascade (48). To investigate whether this mechanism occurs in brain tumors, we used chromatin immunoprecipitation (ChIP) to examine whether Hes1 bound to the five N-boxes present in the Gli1 first intron (Fig. 2B). In HSR-GBM1, we found using quantitative PCR that Hes1 interacted with all five N-boxes as strongly as with a site in the p27 promoter previously shown to be bound by Hes1 as a positive control (34). We also used an independent set of PCR primers and an alternate ChIP protocol to confirm binding to the five N-boxes in a different neurosphere line, JHH-GBM10 (Supplemental Fig. 2B). In contrast to the GBM lines, Hes1 did not bind N-box 1, 4, or 5 in DAOY cells, and showed only moderate affinity for N-boxes 2 and 3, demonstrating the degree of binding to these N-boxes is context dependent (Supplemental Fig. 2C). This is consistent with our expression data demonstrating that Hedgehog pathway targets Gli1 and Ptch1B increase in response to Notch pathway inhibition in HSR-GBM1, but show no change in response to MRK-003 in DAOY. Finally, we evaluated three primary human GBM cultures (passage 0), and found that Hes1 bound the Gli1 first intron Nboxes in one of these (Fig. 2C), suggesting that our finding can be expanded to include some primary GBMs.
When we treated neurosphere lines with MRK-003, Hes1 binding to the Gli1 first intron was reduced, suggesting decreased ability to repress Gli1 expression (Fig. 2B and Supplemental Fig. 2C). Hes1 also bound less to its own promoter at a site of auto-regulation previously documented (31, 32), indicating that Hes1 binding may be globally decreased. These findings suggest a direct negative regulatory relationship between Notch and Hedgehog signaling in some GBMs, whereby cells can upregulate Hedgehog signaling in response to pharmacological Notch pathway inhibition.
To help confirm the presence of such a regulatory mechanism, we looked at the effect of Notch pathway activation on Gli1 and other Hedgehog pathway targets. We infected HSR-GBM1 with retrovirus expressing activated Notch2 (NICD2) or control retrovirus and evaluated Notch and Hedgehog pathway activity. Consistent with our model, we found that Notch signaling targets were upregulated (Supplemental Fig. 2D) while the Hedgehog targets Gli1 and Ptc1B were significantly downregulated by 50% and 30%, respectively (P=0.007 and 0.002; Fig. 2D). Experiments using retrovirus encoding activated Notch1 or Notch3 gave very similar results (data not shown). Next, we sought to determine if the effect of Notch overexpression could be mediated through Hes1. After nucleofecting HSR-GBM1 with Hes1 or a control plasmid, we found a significant 30% reduction (P=0.02) of Gli1 in response to Hes1 overexpression (Fig 2D). The modest decreased we observed may be due to limited transfection efficiency (30-50%), as it is difficult to introduce plasmids into neurosphere lines. We sought to further confirm that the upregulation in Gli1 with MRK-003 treatment was due to Notch pathway inhibition rather than other effects of the GSI by treating cells with MRK-003 one day after infecting them with control or NICD2-expressing retrovirus. While Gli1 levels increased in response to MRK-003 with the control virus, they remained at baseline when NICD2 was present (Fig. 2D), suggesting that Notch2 activity is able to rescue the inhibition-effect of MRK-003. Taken together, our data suggest that increased expression of Hedgehog targets following Notch blockade is due at least in part to the loss of Hes1 binding at the Gli1 locus (Fig. 2D).
Hedgehog Inhibition by Notch Is Seen in Other Cancers
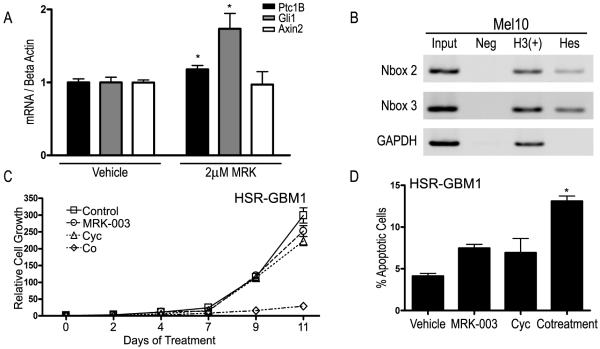
Regulation of Gli1 expression by Hes1 has not been previously reported, and we wanted to determine whether other cancer types demonstrated a similar feedback mechanism. We screened eight cell lines derived from prostate, lung, ovarian, skin, and hematopoetic cell cancers (22RV1, H157, KMS12, L428, Mel10, Reh, TOV-112D, U937) and found that Notch signaling was inhibited by MRK-003 in two. TOV-112D, an ovarian-derived line, showed a 60% decrease in Hes1 expression upon treatment with 2μM MRK-003 (P=0.0002) and Mel10, a melanoma-derived line, showed a more moderate decrease in Hes1 expression of 20% (P=0.002). Interestingly, only Mel10 showed an upregulation in Gli1 and Ptc1B with MRK-003 treatment (P=0.031 and P=0.05, respectively; Fig. 3A). Axin2 remained unchanged in both cell lines following MRK-003 introduction. Chromatin immunoprecipitation showed that Hes1 consistently (N=3) bound the Gli1 N-boxes 2 and 3 in Mel10 (Fig. 3B), along with other N-boxes to a varying degree (data not shown).
Figure 3.
Response of melanoma cell line to MRK-003. A, Gli1, Ptc1B, and Axin2 mRNA expression after MRK-003 treatment. B, ChIP using Hes1 antibody and two different N-box primer sets. C, MTS assay measuring HSR-GBM1 growth after treatment with vehicle, 0.4 μM MRK-003, 10 μM Cyclopamine, or both. D, Percent apoptosis after co-treatment for 48 hours. * P<0.05, ** P<0.005, using Student’s t-test.
Co-inhibition of Notch and Hedgehog Decreases Growth and Clonogenicity
We next evaluated whether inhibiting Hedgehog signaling in addition to Notch would augment anti-tumor effects in GBM neurosphere lines. A Hedgehog antagonist (cyclopamine) and MRK-003 were administered to HSR-GBM1 alone or together in order to detect potential interactions. We observed that cell growth over eleven days decreased slightly with MRK-003 or cyclopamine monotherapy as compared to vehicle, but decreased by approximately 90% in the presence of both drugs (Fig. 3C). Despite the absence of Hes1 binding to the Gli1 intron in DAOY, similar results were seen with co-treatment (Supplemental Fig. 3A). Because the main effect seemed to be accumulation of dead cells, rather than elongation of processes suggesting differentiation, we performed an Annexin V assay to evaluate the percentage of apoptotic cells. Doing so, we found that the percentage of apoptotic cells doubled (13%) with co-treatment as compared to either drug alone (7%; Fig. 3D).
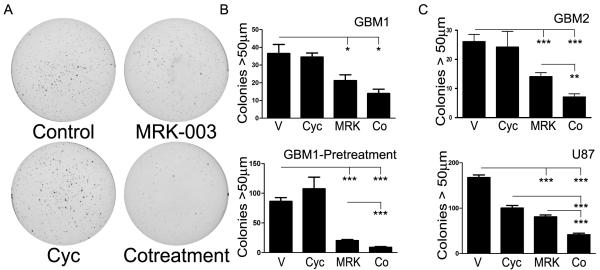
We next investigated whether co-treatment would decrease the number of clonogenic cells using an anchorage independent growth assay. HSR-GBM1, HSR-GBM2, and U87 were dissociated to single-cells, plated in soft agar, and treated continuously with vehicle, monotherapy, or co-treatment. Co-treatment was significantly more effective than monotherapy for the latter two lines, with up to 90% reduction in colony number as compared to cyclopamine alone, and up to 50% reduction as compared to MRK-003 alone (Fig. 4A-C). For HSR-GBM1, the difference between MRK-003 alone and MRK-003 plus cyclopamine was more modest and not statistically significant (P=0.077), although this trend was repeatable across independent experiments (N=3).
Figure 4.
Co-treatment inhibits GBM growth and colony forming ability. A, Sample wells from anchorage independent growth assay where HSR-GBM1 cells were treated with vehicle, 2 μM MRK-003, 5 μM cyclopamine, or both drugs. B, upper graph, Growth of HSR-GBM1 in soft agar with treatment, repeated in triplicate; lower graph HSR-GBM1 pre-treated for seven days before plating in soft agar and then grown without drug. C, HSR-GBM2 and U87 grown in soft agar with drug treatment. Performed in triplicate for N=3 for all cell lines. * P<0.02, ** P<0.001, *** P<0.0001, using Student’s t-test.
To assess our ability to specifically deplete clonogenic cells, we examined if pre-treatment would lead to a more pronounced reduction in colony formation upon co-treatment. Neurosphere cultures were grown with treatment for seven days in suspension, and then washed and equal numbers of viable cells plated in soft agar. Despite no further drug treatment, the number of colonies in the co-treatment cohort was reduced by 90% as compared with vehicle or cyclopamine, and by 50% as compared with MRK-003 alone (Fig. 4B). This shows that colony-forming cells are more sensitive to combined drug treatment and are eliminated from the population somewhat selectively. We also assessed expression of CD133, a putative tumor stem-cell marker, via FACS in HSR-GBM1 and DAOY treated for two days with vehicle, monotherapy, or both drugs. However, in these two lines co-treatment did not decrease the percentage of CD133-positive cells to a greater degree than monotherapy (data not shown). This may reflect the imperfect ability of CD133 alone to prospectively identify stem-like clonogenic cells, a fact highlighted in several recent reports (49).
Primary Human-Derived GBMs Respond Strongly to Co-Inhibition of Notch and Hedgehog
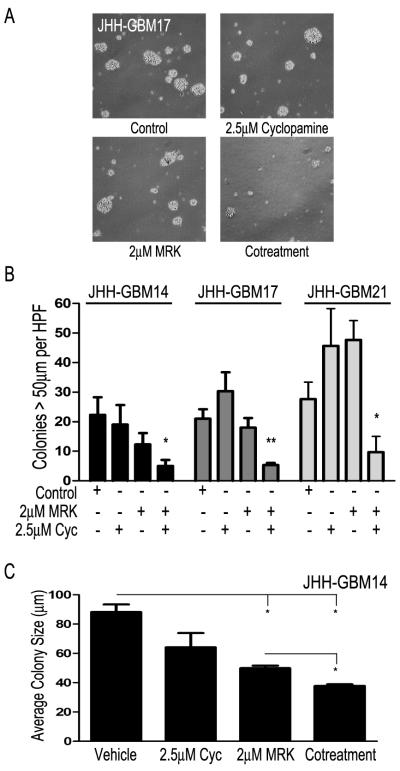
As a final test for the efficacy of combined inhibition, we treated several freshly resected primary human GBMs cultured in neurosphere media. First, we investigated whether MRK-003 was able to inhibit Notch signaling in primary GBM cultures and found that both Hes1 and Hes5 were decreased in primary GBM-derived low-passage neurosphere cultures (Supplemental Fig. 3B), and that baseline expression of Notch and Hedgehog signaling was similar to that of GBM neurosphere cultures. Next, we looked at the ability of co-treatment to prevent colony formation. We dissociated the tumors, plated them in serum-free neurosphere media, and treated the primary cultures. While some variation was observed, most likely due to the heterogeneous nature of clinical samples, overall we saw a dramatic decrease in the number and size of spheres forming over a period of 2-4 weeks in the co-treatment cohort as compared with either monotherapy or vehicle (Fig. 5A). Similar results were seen in primary passages from three GBMs (JHH-GBM17, JHH-GBM18, and JHH-GBM20) and in passage two cells from JHH-GBM14. A significant decrease of 50-80% in the number of colonies formed was seen in the lines with sufficient material for multiple replicates (Fig. 5B). The fourth line, JHH-GBM20, showed results similar to JHH-GBM14, but the assay was only performed once due to limited starting material. We also observed a decrease in the average size of tumor neurospheres formed in co-treated cultures as compared to vehicle or monotherapy, (P<0.05), suggesting a limited proliferative capacity in remaining cells (Fig. 5C).
Figure 5.
Co-treatment inhibits primary GBM growth. A, Dissociated primary GBM cells were plated and treated with vehicle, 2 μM MRK-003, 2.5 μM cyclopamine, or both. Scale bar = 300 μm. B, Quantification of sphere number after 1-4 weeks in culture. C, Sphere size in JHH-GBM14 after co-treatment. * P<0.05, ** P<0.002 ** P<0.0002, using Student’s t-test.
Discussion
GBMs are highly resistant to current treatments such as chemotherapy and radiation, and a deeper understanding of GBM biology is necessary for effective tumor elimination. In light of this, large-scale efforts characterizing genetic alterations in GBM have demonstrated that GBM subtypes exist and suggested that multiple pathways could be targeted to treat tumors more effectively (3, 50, 51). We investigated whether resistance could emerge in response to treatment with a Notch inhibitor in vitro, and found a significant number of cells were able to grow in the presence of long-term MRK-003 treatment. We found Hedgehog signaling was up-regulated in response to Notch inhibition and that direct interaction between Hes1 and Gli1 occurs in GBM neurosphere lines and primary GBM samples. This interaction could account for the increase in Hedgehog signaling during Notch suppression and is a potential mechanism of resistance. We demonstrated that targeting both Notch and Hedgehog simultaneously increases apoptosis and inhibits colony-forming ability more dramatically than either monotherapy. Moreover, freshly dissected human GBMs are also highly susceptible to co-inhibition. These findings indicate that targeting both pathways is more effective than monotherapy at eliminating GBM cells in some neurospheres in vitro, and that suggest that co-treatment should be considered in patients after additional preclinical in vivo studies are performed.
The finding that Hes1 binds to the Gli1 locus and may regulate its expression at a transcriptional level is both novel and unexpected. The fact that only a subset of cell lines, primary GBM specimens, and cancer types exhibit this relationship suggests other factors, such as binding partners, heterochromatin structure, or methylation, may also be important. We investigated whether inverse expression levels between Hes1 and Gli1 were seen in human GBMs by analyzing public databases (www.rembrant.org) and previously published data sets (52), but found no significant relationship between the two (data not shown). This may be due to the fact that this regulatory relationship is only present in a subset of human GBMs. The molecular mechanism and functional role of this feedback mechanism in cancer requires further investigation, as does the question of whether this regulatory mechanism occurs in normal development.
In summary, co-treatment with Hedgehog and Notch inhibitors clearly increases cell death and decreases colony forming ability in vitro, suggesting the potential as a combinatorial chemotherapy agent. In vivo studies will be required to evaluate whether or not this is a feasible treatment in mouse xenografts prior to any clinical trials. A study evaluating Hedgehog and Notch co-inhibition in advanced breast cancer is currently underway (NCT01071564) and may pave the road for determining the safety of co-treatment. Our data supports the increasing awareness of cell signaling complexity in tumors, and the potential for adaptation and evasion following individual pathway blockade. Further evaluation of the molecular links between the Notch, Hedgehog and Wnt pathways seems indicated, and successful tumor elimination may require broader approaches targeting several pathways.
Statement of Translational Relevance.
The emergence of therapeutic resistance is a significant concern when targeting many signaling pathways and tumor types. We found that in vitro Notch pathway blockade in glioblastoma (GBM) cells using a gamma-secretase inhibitor (GSI) led to increased activity in two other pathways important for neural development—Wnt and Hedgehog. The Notch target Hes1, a transcriptional repressor, can directly bind the first Gli1 intron, suggesting a mechanism by which Notch can inhibit Hedgehog activity. Inhibition of both Notch and Hedgehog in vitro dramatically decreased the growth of GBM cell lines, as well as low-passage neurospheres derived from primary human tumors. These findings demonstrate that Notch-targeted therapeutics can lead to alterations in other developmental signaling cascades which promote tumor survival, and suggest that combined treatment with Hedgehog pathway inhibitors may be able to increase the efficacy of GSIs in some cancer patients.
Supplementary Material
A, Hes1 and Hes5 expression after treatment with cyclopamine. B, ChIP of Hes1 in JHH-GBM10. C, ChIP of Hes1 measured via quantitative PCR in DAOY. D, Hes5 expression after infection with NICD2 virus.
A, MTS assay measuring DAOY growth after treatment with vehicle, 0.4 μM MRK-003, 10 μM Cyclopamine, or both. B, Response of low passage GBM-derived neurosphere lines to MRK-003.
Primers used for chromatin immunoprecipitation.
Effect of MRK-003 on medulloblastoma cell line DAOY. A, Hes1 mRNA in response to treatment with MRK-003. B, Western blot and quantification of Hes1 protein after MRK-003 treatment (μM). C, MTS assay measuring cell growth after MRK-003 treatment. D, QPCR of other pathway targets in response to MRK-003 treatment. * P <0.05, * P<0.005, using Student’s t-test.
List of top genes altered by treatment with MRK-003.
Acknowledgements
We would like to thank the following for reagents: Ryoichiro Kageyama and Tetso Sudo (Hes1 antibody), Madeleine Carreau (Hes1 plasmid), Angelo Vescovi (HSR-GBM1 and HSR-GBM2), Craig Peacock and Zhenhua Huang (adherent cell lines), the JHU Genetic Core (shRNAs). Thanks to Naheed Gul and Michael Coonfield for technical assistance. We would like to acknowledge grant support to Dr. Eberhart from NIH/NS55089 and the Brain Tumor Funders Collaborative, and to Dr. Golpakrishnan from the American Society Grant RSG-090273-01-DDC and the Brain Tumor Spore Grant 5P50CA127001-02. Additional thanks to Merck and its scientists for providing MRK-003.
References
- 1.Louis DN, International Agency for Research on Cancer . WHO classification of tumours of the central nervous system. 4 th ed. International Agency for Research on Cancer; Lyon: 2007. [Google Scholar]
- 2.Adamson C, Kanu OO, Mehta AI, et al. Glioblastoma multiforme: a review of where we have been and where we are going. Expert Opin Investig Drugs. 2009;18:1061–83. doi: 10.1517/13543780903052764. [DOI] [PubMed] [Google Scholar]
- 3.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–73. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 4.Kanu OO, Mehta A, Di C, et al. Glioblastoma multiforme: a review of therapeutic targets. Expert Opin Ther Targets. 2009;13:701–18. doi: 10.1517/14728220902942348. [DOI] [PubMed] [Google Scholar]
- 5.Huang TT, Sarkaria SM, Cloughesy TF, Mischel PS. Targeted therapy for malignant glioma patients: lessons learned and the road ahead. Neurotherapeutics. 2009;6:500–12. doi: 10.1016/j.nurt.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yauch RL, Dijkgraaf GJ, Alicke B, et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science. 2009;326:572–4. doi: 10.1126/science.1179386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellisen LW, Bird J, West DC, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–61. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 8.Bolos V, Grego-Bessa J, de la Pompa JL. Notch signaling in development and cancer. Endocr Rev. 2007;28:339–63. doi: 10.1210/er.2006-0046. [DOI] [PubMed] [Google Scholar]
- 9.Aster JC, Pear WS, Blacklow SC. Notch signaling in leukemia. Annu Rev Pathol. 2008;3:587–613. doi: 10.1146/annurev.pathmechdis.3.121806.154300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koch U, Radtke F. Notch and cancer: a double-edged sword. Cell Mol Life Sci. 2007;64:2746–62. doi: 10.1007/s00018-007-7164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ignatova TN, Kukekov VG, Laywell ED, Suslov ON, Vrionis FD, Steindler DA. Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro. Glia. 2002;39:193–206. doi: 10.1002/glia.10094. [DOI] [PubMed] [Google Scholar]
- 12.Purow BW, Haque RM, Noel MW, et al. Expression of Notch-1 and its ligands, Delta-like-1 and Jagged-1, is critical for glioma cell survival and proliferation. Cancer Res. 2005;65:2353–63. doi: 10.1158/0008-5472.CAN-04-1890. [DOI] [PubMed] [Google Scholar]
- 13.Kanamori M, Kawaguchi T, Nigro JM, et al. Contribution of Notch signaling activation to human glioblastoma multiforme. J Neurosurg. 2007;106:417–27. doi: 10.3171/jns.2007.106.3.417. [DOI] [PubMed] [Google Scholar]
- 14.Solecki DJ, Liu XL, Tomoda T, Fang Y, Hatten ME. Activated Notch2 signaling inhibits differentiation of cerebellar granule neuron precursors by maintaining proliferation. Neuron. 2001;31:557–68. doi: 10.1016/s0896-6273(01)00395-6. [DOI] [PubMed] [Google Scholar]
- 15.Hallahan AR, Pritchard JI, Hansen S, et al. The SmoA1 mouse model reveals that notch signaling is critical for the growth and survival of sonic hedgehog-induced medulloblastomas. Cancer Res. 2004;64:7794–800. doi: 10.1158/0008-5472.CAN-04-1813. [DOI] [PubMed] [Google Scholar]
- 16.Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- 17.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–33. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194:237–55. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 19.Fan X, Matsui W, Khaki L, et al. Notch pathway inhibition depletes stem-like cells and blocks engraftment in embryonal brain tumors. Cancer Res. 2006;66:7445–52. doi: 10.1158/0008-5472.CAN-06-0858. [DOI] [PubMed] [Google Scholar]
- 20.Li K, Li Y, Wu W, et al. Modulation of Notch signaling by antibodies specific for the extracellular negative regulatory region of NOTCH3. J Biol Chem. 2008;283:8046–54. doi: 10.1074/jbc.M800170200. [DOI] [PubMed] [Google Scholar]
- 21.Hoey T, Yen WC, Axelrod F, et al. DLL4 blockade inhibits tumor growth and reduces tumor-initiating cell frequency. Cell Stem Cell. 2009;5:168–77. doi: 10.1016/j.stem.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 22.Wu Y, Cain-Hom C, Choy L, et al. Therapeutic antibody targeting of individual Notch receptors. Nature. 464:1052–7. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
- 23.Moellering RE, Cornejo M, Davis TN, et al. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462:182–8. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan X, Khaki L, Zhu TS, et al. Notch Pathway Blockade Depletes CD133-Positive Glioblastoma Cells and Inhibits Growth of Tumor Neurospheres and Xenografts. Stem Cells. 2009 doi: 10.1002/stem.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galli R, Binda E, Orfanelli U, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–21. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 26.Lewis HD, Leveridge M, Strack PR, et al. Apoptosis in T cell acute lymphoblastic leukemia cells after cell cycle arrest induced by pharmacological inhibition of notch signaling. Chem Biol. 2007;14:209–19. doi: 10.1016/j.chembiol.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 27.Tremblay CS, Huang FF, Habi O, et al. HES1 is a novel interactor of the Fanconi anemia core complex. Blood. 2008;112:2062–70. doi: 10.1182/blood-2008-04-152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bar EE, Lin A, Mahairaki V, Matsui W, Eberhart CG. Hypoxia increases the expression of stem-cell markers and promotes clonogenicity in glioblastoma neurospheres. Am J Pathol. 177:1491–502. doi: 10.2353/ajpath.2010.091021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bar EE, Chaudhry A, Lin A, et al. Cyclopamine-mediated hedgehog pathway inhibition depletes stem-like cancer cells in glioblastoma. Stem Cells. 2007;25:2524–33. doi: 10.1634/stemcells.2007-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosen SaSHJ. Primer3 on the WWW for general users and for biologist programmers. Humana Press; Totowa, NJ: 2000. [DOI] [PubMed] [Google Scholar]
- 31.Takebayashi K, Sasai Y, Sakai Y, Watanabe T, Nakanishi S, Kageyama R. Structure, chromosomal locus, and promoter analysis of the gene encoding the mouse helix-loop-helix factor HES-1. Negative autoregulation through the multiple N box elements. J Biol Chem. 1994;269:5150–6. [PubMed] [Google Scholar]
- 32.Hirata H, Yoshiura S, Ohtsuka T, et al. Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science. 2002;298:840–3. doi: 10.1126/science.1074560. [DOI] [PubMed] [Google Scholar]
- 33.Aguilera DG, Das CM, Sinnappah-Kang ND, et al. Reactivation of death receptor 4 (DR4) expression sensitizes medulloblastoma cell lines to TRAIL. J Neurooncol. 2009;93:303–18. doi: 10.1007/s11060-008-9788-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murata K, Hattori M, Hirai N, et al. Hes1 directly controls cell proliferation through the transcriptional repression of p27Kip1. Mol Cell Biol. 2005;25:4262–71. doi: 10.1128/MCB.25.10.4262-4271.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aparicio O, Geisberg JV, Struhl K. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr Protoc Cell Biol. 2004 doi: 10.1002/0471143030.cb1707s23. Chapter 17:Unit 17 7. [DOI] [PubMed] [Google Scholar]
- 36.Daniel VC, Marchionni L, Hierman JS, et al. A primary xenograft model of small-cell lung cancer reveals irreversible changes in gene expression imposed by culture in vitro. Cancer Res. 2009;69:3364–73. doi: 10.1158/0008-5472.CAN-08-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaeffer EM, Marchionni L, Huang Z, et al. Androgen-induced programs for prostate epithelial growth and invasion arise in embryogenesis and are reactivated in cancer. Oncogene. 2008;27:7180–91. doi: 10.1038/onc.2008.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ihaka R, Gentleman R. R: A language for data analysis and graphics. Journal of Computational and Graphical Statistics. 1996;5:299–314. [Google Scholar]
- 40.Wheeler DL, Barrett T, Benson DA, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J, Wakeman TP, Lathia JD, et al. Notch promotes radioresistance of glioma stem cells. Stem Cells. 28:17–28. doi: 10.1002/stem.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dakubo GD, Mazerolle CJ, Wallace VA. Expression of Notch and Wnt pathway components and activation of Notch signaling in medulloblastomas from heterozygous patched mice. J Neurooncol. 2006;79:221–7. doi: 10.1007/s11060-006-9132-2. [DOI] [PubMed] [Google Scholar]
- 43.Julian E, Dave RK, Robson JP, Hallahan AR, Wainwright BJ. Canonical Notch signaling is not required for the growth of Hedgehog pathway-induced medulloblastoma. Oncogene. doi: 10.1038/onc.2010.101. [DOI] [PubMed] [Google Scholar]
- 44.Hatton BA, Villavicencio EH, Pritchard J, et al. Notch signaling is not essential in sonic hedgehog-activated medulloblastoma. Oncogene. doi: 10.1038/onc.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wall DS, Mears AJ, McNeill B, et al. Progenitor cell proliferation in the retina is dependent on Notch-independent Sonic hedgehog/Hes1 activity. J Cell Biol. 2009;184:101–12. doi: 10.1083/jcb.200805155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ingram WJ, McCue KI, Tran TH, Hallahan AR, Wainwright BJ. Sonic Hedgehog regulates Hes1 through a novel mechanism that is independent of canonical Notch pathway signalling. Oncogene. 2008;27:1489–500. doi: 10.1038/sj.onc.1210767. [DOI] [PubMed] [Google Scholar]
- 47.Katoh Y, Katoh M. Integrative genomic analyses on GLI1: positive regulation of GLI1 by Hedgehog-GLI, TGFbeta-Smads, and RTK-PI3K-AKT signals, and negative regulation of GLI1 by Notch-CSL-HES/HEY, and GPCR-Gs-PKA signals. Int J Oncol. 2009;35:187–92. doi: 10.3892/ijo_00000328. [DOI] [PubMed] [Google Scholar]
- 48.Katoh Y, Katoh M. Hedgehog signaling pathway and gastrointestinal stem cell signaling network (review) Int J Mol Med. 2006;18:1019–23. [PubMed] [Google Scholar]
- 49.Chen R, Nishimura MC, Bumbaca SM, et al. A hierarchy of self-renewing tumor-initiating cell types in glioblastoma. Cancer Cell. 17:362–75. doi: 10.1016/j.ccr.2009.12.049. [DOI] [PubMed] [Google Scholar]
- 50.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A, Hes1 and Hes5 expression after treatment with cyclopamine. B, ChIP of Hes1 in JHH-GBM10. C, ChIP of Hes1 measured via quantitative PCR in DAOY. D, Hes5 expression after infection with NICD2 virus.
A, MTS assay measuring DAOY growth after treatment with vehicle, 0.4 μM MRK-003, 10 μM Cyclopamine, or both. B, Response of low passage GBM-derived neurosphere lines to MRK-003.
Primers used for chromatin immunoprecipitation.
Effect of MRK-003 on medulloblastoma cell line DAOY. A, Hes1 mRNA in response to treatment with MRK-003. B, Western blot and quantification of Hes1 protein after MRK-003 treatment (μM). C, MTS assay measuring cell growth after MRK-003 treatment. D, QPCR of other pathway targets in response to MRK-003 treatment. * P <0.05, * P<0.005, using Student’s t-test.
List of top genes altered by treatment with MRK-003.