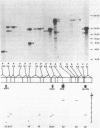
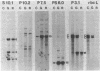
Abstract
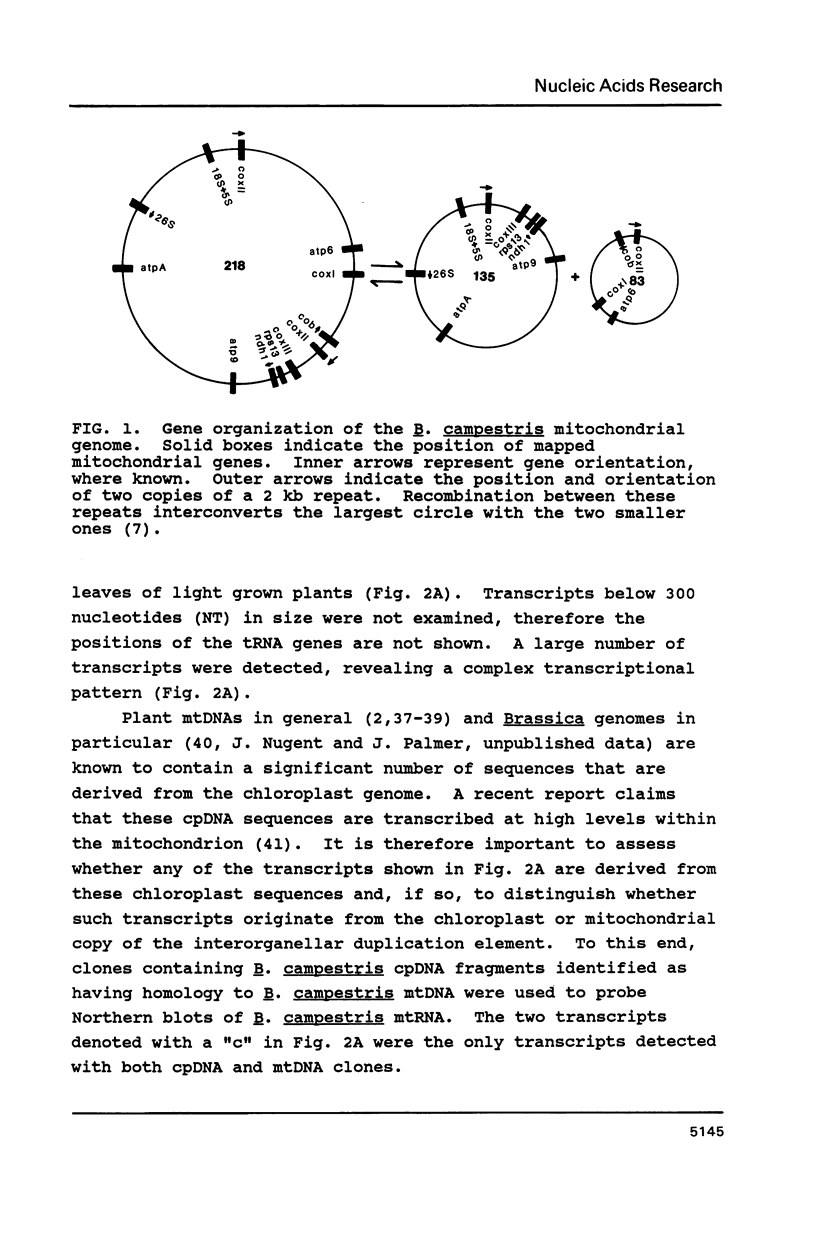
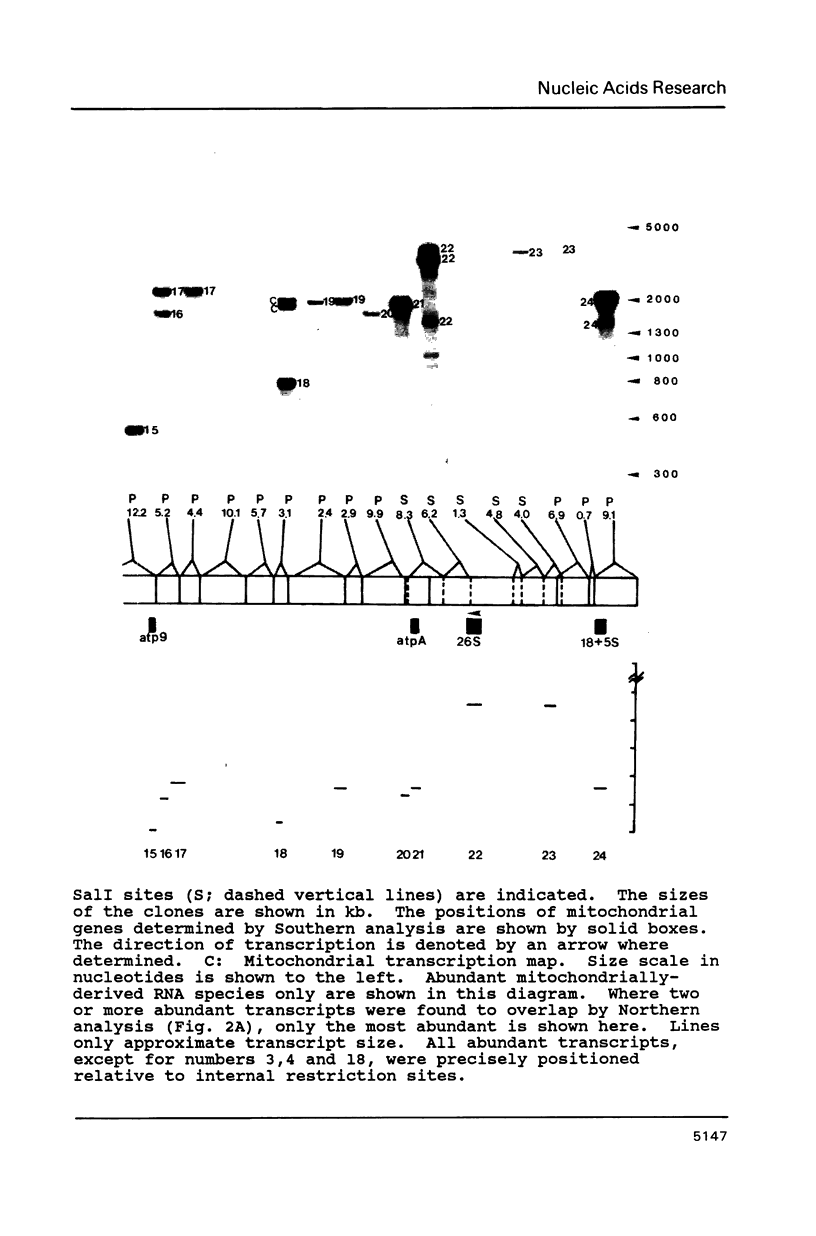
We constructed a complete transcriptional map of the 218 kb Brassica campestris (turnip) mitochondrial genome. Twenty-four abundant and positionally distinct transcripts larger than 500 nucleotides were identified by Northern analyses. Approximately 30% (61 kb) of the genome is highly transcribed. In addition, a number of less abundant transcripts, many of which overlap with each other and with the major transcripts, were also detected. If each abundant transcript represents a distinct rRNA or protein species, then plant mitochondria would appear to encode a significantly larger number of proteins than do animal mitochondria. Although B. campestris mitochondrial DNA contains a number of chloroplast DNA-derived sequences, none of these chloroplast sequences appear to be transcribed within the mitochondrion. We determined the positions of 12 genes in the B. campestris mitochondrial genome. The order of these genes in B. campestris is completely different than in maize (1) and spinach (2).
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bland M. M., Levings C. S., 3rd, Matzinger D. F. The tobacco mitochondrial ATPase subunit 9 gene is closely linked to an open reading frame for a ribosomal protein. Mol Gen Genet. 1986 Jul;204(1):8–16. doi: 10.1007/BF00330180. [DOI] [PubMed] [Google Scholar]
- Bonen L., Gray M. W. Organization and expression of the mitochondrial genome of plants I. The genes for wheat mitochondrial ribosomal and transfer RNA: evidence for an unusual arrangement. Nucleic Acids Res. 1980 Jan 25;8(2):319–335. doi: 10.1093/nar/8.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun C. J., Levings C. S. Nucleotide Sequence of the F(1)-ATPase alpha Subunit Gene from Maize Mitochondria. Plant Physiol. 1985 Oct;79(2):571–577. doi: 10.1104/pp.79.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao S., Sederoff R., Levings C. S., 3rd Nucleotide sequence and evolution of the 18S ribosomal RNA gene in maize mitochondria. Nucleic Acids Res. 1984 Aug 24;12(16):6629–6644. doi: 10.1093/nar/12.16.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomyn A., Cleeter M. W., Ragan C. I., Riley M., Doolittle R. F., Attardi G. URF6, last unidentified reading frame of human mtDNA, codes for an NADH dehydrogenase subunit. Science. 1986 Oct 31;234(4776):614–618. doi: 10.1126/science.3764430. [DOI] [PubMed] [Google Scholar]
- Clayton D. A. Transcription of the mammalian mitochondrial genome. Annu Rev Biochem. 1984;53:573–594. doi: 10.1146/annurev.bi.53.070184.003041. [DOI] [PubMed] [Google Scholar]
- Dale R. M., Mendu N., Ginsburg H., Kridl J. C. Sequence analysis of the maize mitochondrial 26 S rRNA gene and flanking regions. Plasmid. 1984 Mar;11(2):141–150. doi: 10.1016/0147-619x(84)90019-2. [DOI] [PubMed] [Google Scholar]
- Dawson A. J., Jones V. P., Leaver C. J. The apocytochrome b gene in maize mitochondria does not contain introns and is preceded by a potential ribosome binding site. EMBO J. 1984 Sep;3(9):2107–2113. doi: 10.1002/j.1460-2075.1984.tb02098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey R. E., Levings C. S., Timothy D. H. Nucleotide sequence of ATPase subunit 6 gene of maize mitochondria. Plant Physiol. 1985 Nov;79(3):914–919. doi: 10.1104/pp.79.3.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey R. E., Schuster A. M., Levings C. S., Timothy D. H. Nucleotide sequence of F(0)-ATPase proteolipid (subunit 9) gene of maize mitochondria. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1015–1019. doi: 10.1073/pnas.82.4.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dron M., Hartmann C., Rode A., Sevignac M. Gene conversion as a mechanism for divergence of a chloroplast tRNA gene inserted in the mitochondrial genome of Brassica oleracea. Nucleic Acids Res. 1985 Dec 9;13(23):8603–8610. doi: 10.1093/nar/13.23.8603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconet D., Lejeune B., Quetier F., Gray M. W. Evidence for homologous recombination between repeated sequences containing 18S and 5S ribosomal RNA genes in wheat mitochondrial DNA. EMBO J. 1984 Feb;3(2):297–302. doi: 10.1002/j.1460-2075.1984.tb01799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauron C. M., Wolstenholme D. R. Structural heterogeneity of mitochondrial DNA molecules within the genus Drosophila. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3623–3627. doi: 10.1073/pnas.73.10.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox T. D., Leaver C. J. The Zea mays mitochondrial gene coding cytochrome oxidase subunit II has an intervening sequence and does not contain TGA codons. Cell. 1981 Nov;26(3 Pt 1):315–323. doi: 10.1016/0092-8674(81)90200-2. [DOI] [PubMed] [Google Scholar]
- Harris T. In vitro mutagenesis. Nature. 1982 Sep 23;299(5881):298–299. doi: 10.1038/299298a0. [DOI] [PubMed] [Google Scholar]
- Hiesel R., Schobel W., Schuster W., Brennicke A. The cytochrome oxidase subunit I and subunit III genes in Oenothera mitochondria are transcribed from identical promoter sequences. EMBO J. 1987 Jan;6(1):29–34. doi: 10.1002/j.1460-2075.1987.tb04714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac P. G., Brennicke A., Dunbar S. M., Leaver C. J. The mitochondrial genome of fertile maize (Zea mays L.) contains two copies of the gene encoding the alpha-subunit of the F1-ATPase. Curr Genet. 1985;10(4):321–328. doi: 10.1007/BF00365628. [DOI] [PubMed] [Google Scholar]
- Isaac P. G., Jones V. P., Leaver C. J. The maize cytochrome c oxidase subunit I gene: sequence, expression and rearrangement in cytoplasmic male sterile plants. EMBO J. 1985 Jul;4(7):1617–1623. doi: 10.1002/j.1460-2075.1985.tb03828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler L. G., Avise J. C. A comparative description of mitochondrial DNA differentiation in selected avian and other vertebrate genera. Mol Biol Evol. 1985 Mar;2(2):109–125. doi: 10.1093/oxfordjournals.molbev.a040339. [DOI] [PubMed] [Google Scholar]
- Kolodner R., Tewari K. K. Physicochemical characterization of mitochondrial DNA from pea leaves. Proc Natl Acad Sci U S A. 1972 Jul;69(7):1830–1834. doi: 10.1073/pnas.69.7.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver C. J., Hack E., Forde B. G. Protein synthesis by isolated plant mitochondria. Methods Enzymol. 1983;97:476–484. doi: 10.1016/0076-6879(83)97156-2. [DOI] [PubMed] [Google Scholar]
- Lonsdale D. M., Hodge T. P., Fauron C. M. The physical map and organisation of the mitochondrial genome from the fertile cytoplasm of maize. Nucleic Acids Res. 1984 Dec 21;12(24):9249–9261. doi: 10.1093/nar/12.24.9249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale D. M., Hodge T. P., Howe C. J., Stern D. B. Maize mitochondrial DNA contains a sequence homologous to the ribulose-1,5-bisphosphate carboxylase large subunit gene of chloroplast DNA. Cell. 1983 Oct;34(3):1007–1014. doi: 10.1016/0092-8674(83)90558-5. [DOI] [PubMed] [Google Scholar]
- Manna E., Brennicke A. Primary and secondary structure of 26S ribosomal RNA of Oenothera mitochondria. Curr Genet. 1985;9(6):505–515. doi: 10.1007/BF00434055. [DOI] [PubMed] [Google Scholar]
- Palmer J. D., Herbon L. A. Tricircular mitochondrial genomes of Brassica and Raphanus: reversal of repeat configurations by inversion. Nucleic Acids Res. 1986 Dec 22;14(24):9755–9764. doi: 10.1093/nar/14.24.9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J. D., Herbon L. A. Unicircular structure of the Brassica hirta mitochondrial genome. Curr Genet. 1987;11(6-7):565–570. doi: 10.1007/BF00384620. [DOI] [PubMed] [Google Scholar]
- Palmer J. D. Physical and gene mapping of chloroplast DNA from Atriplex triangularis and Cucumis sativa. Nucleic Acids Res. 1982 Mar 11;10(5):1593–1605. doi: 10.1093/nar/10.5.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozier C., Rocipon M., Mache R. Post-maturation of the plastid ribosomal RNA in the plant kingdom. J Mol Evol. 1979 Nov;13(4):271–279. doi: 10.1007/BF01731367. [DOI] [PubMed] [Google Scholar]
- Shinozaki K., Ohme M., Tanaka M., Wakasugi T., Hayashida N., Matsubayashi T., Zaita N., Chunwongse J., Obokata J., Yamaguchi-Shinozaki K. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986 Sep;5(9):2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. B., Bang A. G., Thompson W. F. The watermelon mitochondrial URF-1 gene: evidence for a complex structure. Curr Genet. 1986;10(11):857–869. doi: 10.1007/BF00418532. [DOI] [PubMed] [Google Scholar]
- Stern D. B., Newton K. J. Isolation of plant mitochondrial RNA. Methods Enzymol. 1986;118:488–496. doi: 10.1016/0076-6879(86)18095-5. [DOI] [PubMed] [Google Scholar]
- Stern D. B., Newton K. J. Mitochondrial gene expression in Cucurbitaceae: conserved and variable features. Curr Genet. 1985;9(5):395–404. doi: 10.1007/BF00421611. [DOI] [PubMed] [Google Scholar]
- Stern D. B., Palmer J. D. Extensive and widespread homologies between mitochondrial DNA and chloroplast DNA in plants. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1946–1950. doi: 10.1073/pnas.81.7.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. B., Palmer J. D. Recombination sequences in plant mitochondrial genomes: diversity and homologies to known mitochondrial genes. Nucleic Acids Res. 1984 Aug 10;12(15):6141–6157. doi: 10.1093/nar/12.15.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. B., Palmer J. D. Tripartite mitochondrial genome of spinach: physical structure, mitochondrial gene mapping, and locations of transposed chloroplast DNA sequences. Nucleic Acids Res. 1986 Jul 25;14(14):5651–5666. doi: 10.1093/nar/14.14.5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward B. L., Anderson R. S., Bendich A. J. The mitochondrial genome is large and variable in a family of plants (cucurbitaceae). Cell. 1981 Sep;25(3):793–803. doi: 10.1016/0092-8674(81)90187-2. [DOI] [PubMed] [Google Scholar]
- Young E. G., Hanson M. R., Dierks P. M. Sequence and transcription analysis of the Petunia mitochondrial gene for the ATP synthase proteolipid subunit. Nucleic Acids Res. 1986 Oct 24;14(20):7995–8006. doi: 10.1093/nar/14.20.7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski G., Whitfeld P. R., Bottomley W. Sequence of the gene for the large subunit of ribulose 1,5-bisphosphate carboxylase from pea chloroplasts. Nucleic Acids Res. 1986 May 12;14(9):3975–3975. doi: 10.1093/nar/14.9.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]