Abstract
Purpose
Assessment of embryo viability is a key component of in vitro fertilization (IVF) and currently relies largely on embryo morphology and cleavage rate. In this study, we used receiver operating characteristic (ROC) analysis to compare the Viability Score (generated by metabolomic profiling of spent embryo culture media using near infrared (NIR) spectroscopy) to morphologic grading for predicting pregnancy in women undergoing single embryo transfer (SET) on day 5.
Methods
A total of 198 spent embryo culture media samples were collected in four IVF centers located in the USA, Europe and Australia. First, 137 samples (training set) were analyzed by NIR to develop an algorithm that generates a Viability Score predictive of pregnancy for each sample. Next, 61 samples (validation set) were analyzed by observers blinded to embryo morphology and IVF outcome, using the Day 5 algorithm generated with the training set. Pregnancy was defined as fetal cardiac activity (FCA) at 12 weeks of gestation.
Results
The Area Under the Curve (AUC) was greater for the metabolomic Viability Score compared to Morphology [Training set: 0.75 versus 0.55, p = 0.0011; Validation set: 0.68 versus 0.50, P = 0.021], and for a Composite score (obtained using a model combining Viability Score with morphologic grading), compared to morphology alone [0.74 versus 0.50, p = 0.004].
Conclusions
Our findings suggest that Viability Score alone or in combination with morphologic grading has the potential to be a better classifier for pregnancy outcome than morphology alone in women undergoing SET on day 5.
Keywords: ROC analysis, Assisted reproductive technologies, ART, In vitro fertilization, IVF, Morphologic grade, Metabolomics, Viability Score
Introduction
Infertility is estimated to affect 15% of reproductive age women [1]. Among numerous treatment options that are available to infertile couples, those involving in vitro fertilization (IVF) are associated with highest success rates [1]. Consequently, the use of IVF has been increasing steadily worldwide, accounting for more than 1% of births in the United Sates and up to 3% in European countries. It is estimated that more than 2,000,000 IVF cycles will be performed worldwide in 2015 [2]. Consequently, issues associated with the performance and outcome of IVF are becoming increasingly important.
A key aspect of IVF is the assessment of embryos generated in vitro to identify the one(s) most likely to result in pregnancy. An association between embryos’ cleavage rate and morphologic characteristics, and their ability to implant has been reported soon after the first live birth using IVF, and development of controlled ovarian hyperstimulation protocols [3–5]. Since then, a multitude of morphologic parameters associated with increased viability of pronuclear, cleavage stage, and blastocyst stage embryos were identified, and numerous embryo grading systems have been developed [6–12]. These strategies based on embryo morphology, combined with improved culture environment, have led to significant improvements in implantation and pregnancy rates [13], and currently constitute the mainstay of embryo assessment in IVF laboratories worldwide.
Despite significant improvements in IVF outcomes using morphologic embryo assessment, the ability to predict the implantation potential of an individual embryo in the IVF laboratory remains far from ideal. Indeed, even for embryos selected using best morphologic criteria, implantation rates remain at 49% for embryos transferred on day 3 [10]. Consequently, multiple embryo transfer is routinely performed in countries without strict regulations, such as the United States, where more than 80% of all transferred embryos fail to implant [14]. Multiple embryo transfer in IVF is associated with significant increase in multiple pregnancies, which, in turn result in an increased risk of preterm birth and its complications such as cerebral palsy, and infant death (see [14] for review).
Limitations of embryo assessment methodologies based on morphology and cleavage rate has led investigators to pursue additional approaches to improve the accuracy with which embryo viability is predicted in the IVF laboratory. Within this context, spent embryo culture media nutrient and metabolite content has been studied to determine an embryo’s metabolic activity as a reflection of its viability. Pyruvate, lactate, glucose, and amino acids in the embryo culture medium have all been reported to correlate with embryo development and implantation potential (see [15, 16] for review). Global assessment of the embryo culture media proteome [17] as well as specific proteins such as soluble human leukocyte antigen-G (sHLA-G) [18, 19] have also been studied as predictors of embryo viability.
Unfortunately, blinded validation of the predictive/diagnostic value of most of these approaches have not yet been reported. In addition, their use requires dedicated equipment and technical staff, which would be cost prohibitive in most embryology laboratories. Finally, these techniques frequently do not produce results quickly enough to allow the information to be used clinically in the limited window of time acceptable for embryo transfer. Therefore, the need for a technology that predicts reproductive potential of embryos through a rapid, non-invasive, and clinically applicable platform remains.
The complete array of small-molecule metabolites that are found within a biological system constitutes the metabolome and reflects the functional phenotype [20]. Metabolomics, is the systematic study of this dynamic inventory of metabolites, as small molecular biomarkers representing the functional phenotype in a biological system (see [16] for review). Using various forms of spectral and analytical approaches, metabolomics attempts to determine and quantify metabolites associated with physiologic and pathologic states (see [16] for review).
We have recently reported that non-invasive metabolomic profiling of embryo culture media using Raman and Near Infrared (NIR) spectroscopy correlates with pregnancy outcome in women undergoing IVF [21]. Our initial findings were confirmed in prospective blinded studies using multiple [22] and single embryo transfer [23, 24], which also revealed that metabolomic profiling constitutes a parameter independent of morphology [23, 24]. In the current study, we used receiver operating characteristic (ROC) analysis to compare NIR spectroscopy-based metabolomics to morphologic grading for predicting outcome in women undergoing single embryo transfer on day 5.
Materials and methods
Patient selection, treatment, and sample collection
All patients participating in the study were recruited from the Fertilitetscentrum (GSFC), Gothenburg, Sweden, Reproductive Sciences Center (RSC), Boston, USA, Shady Grove Fertility Clinic (SGFC), Maryland, USA and the Sydney IVF Centre (SYD), Sydney, Australia. Institutional Review Board approval was obtained in each center prior to the initiation of the study. All patients undergoing IVF with a Day 5 single embryo transfer (SET) from November 2007 to January 2009 were considered for participation in the study, and a total of 198 spent embryo culture media samples were collected.
In all centers standard controlled ovarian hyperstimulation protocols were used as previously described [24]. Pituitary suppression was achieved with either GnRH agonists or antagonists. Stimulation with FSH (150–600 IU per day) was monitored by measuring serum E2 levels and/or follicle growth. Human chorionic gonadotropin was administered when patients had reached the individual clinics trigger point for follicular growth. Oocytes were collected 34–36 h after hCG.
Retrieved oocytes were rinsed, graded, and placed in the clinic’s routine retrieval media. All incubations were performed at 37°C in 5% CO2, 5%O2, 90%N2 or 5–6% CO2 in air. Oocyte insemination was initiated approximately 40 h after HCG injection using standard IVF and/or ICSI procedures as previously described [24].
At 16–18 h after insemination (day 1), each oocyte was examined for evidence of fertilization and placed into either Vitrolife G1 (Vitrolife AB, Goteburg, Sweden), Sage Cleavage (Cooper Surgical, Trumbull, CT, USA), Global (IVFonline LLC, Guelph, ON, Canada) or Cook’s Cleavage media (Cook Group Inc. Bloomington, IN, USA) for culture to the cleavage stage. On Day 3, embryos were placed into individual droplets of between 15 to 25 μl of Vitrolife CCM media, Sage Blastocyst, Global or Cook’s Blastocyst Media for culture to the Blastocyst stage.
Each clinic used a different routine blastocyst embryo scoring system to assess the morphology of the Day 5 embryos. The single embryo with the highest morphological grading was chosen for transfer. Although, each clinic used a different grading system they were all based on assessment of the expansion state of the Day 5 blastocyst and a ranking of the quality of the inner cell mass (ICM) and Trophectoderm (TE). For the purpose of this analysis the morphology scores of the different clinics were separated into four morphologic grades: [A]: Expanded Blastocysts with excellent/good inner cell mass (ICM) and Trophectoderm (TE), [B]: Blastocysts with excellent/good ICM and TE, [C]: Early Blastocysts and Other Blastocysts with Poor ICM and TE and [D]: Compacted.
Following removal of the embryos in preparation for transfer, the spent Day 5 media were placed individually into labeled cryo-vials, snap frozen in liquid nitrogen, and/or stored at −80°C, and transported to Molecular Biometrics Inc. (Norwood, MA) in dry ice for analysis (maximum transport time 48 h). A control sample incubated under the same conditions without an embryo was also collected and used for normalization. Pregnancy outcomes were recorded for each patient at ~12 weeks gestational age. Positive pregnancy was defined as fetal cardiac activity (FCA) at that time.
Sample analysis by NIR spectroscopy
Analysis of the individual embryo culture media samples was performed as previously described [21, 23, 24]. Briefly, the IVF culture media samples were thawed on ice for 30 min, vortexed and then centrifuged for 5 min at 12,000 rpm, and placed in a temperature stabilizer prior to analysis for 4 min at 24.3°C. Approximately, 10–15 μl of the sample was then slowly injected using an eppendorf pipette into an individual Sample Cell designed to fit into the NIR instrument. Sample cells were then placed in the sample chamber maintained at 23.5°C and NIR transmittance spectra were measured using an indium gallium arsenide (InGaAs) array based 512 element NIR spectrometer with an operating wavelength of 920–1,675 nm. Sample analysis time was approximately 1 min. The measurement was repeated with the corresponding control medium for each sample to account for any impact of variations in culturing conditions. A final spectral profile was then generated for each embryo from the combination of the embryo culture medium and control medium analysis.
Spectral model development
We have previously reported development of spectral models for day 2 and day 3 embryos [21, 24]. As extensive data from animal models and human IVF [14–16] suggest that there are metabolic differences between cleavage stage embryos and blastocysts, a novel spectral model for day 5 embryos was developed as described in our previous publications [21, 23, 25]. Discrimination between FCA+ and FCA− embryos consisted of determining a parsimonious combination of spectral regions from the NIR metabolomic profiles that estimated pregnancy outcomes by inverse least-squares regression and genetic algorithm (GA) optimization. This method was described in detail previously [21]. Briefly, models investigated were described by the formula:
 |
1 |
where Y is the pregnancy outcome of each sample (1 = positive FCA, 0 = negative FCA), X1, X2, …, Xn are the spectral regions, and b0, b1, …, bn are their associated weighting coefficients. A GA determines the combination of X-values that best estimate Y, by employing principles of Darwinian natural selection and biological inspired operations: reproduction, crossover, and mutation. Final embryo viability indices were determined by a leave-one-out cross validation method, using the algorithm/model developed using Eq. 1 with known spectral regions and calculated weighting coefficients. All analysis was written in and evaluated using Matlab (The MathWorks Inc., MA).
Training and validation sample sets
The spent embryo culture media samples consisted of two distinct sets. The first set consisted of a total of 137 Day 5 spent embryo culture media samples collected following single embryo transfer (SET) in GSFC (n = 42) and in SGFC (n = 95). In this data set, there were 90 Day 5 embryos scoring A, 28 scoring B, 10 scoring C and 9 scoring D. This data was considered a “Training Set” as it was used to develop the spectral model predictive of pregnancy outcome (using the approach described above). The mean age (± SD) of the women in the Training Set was 34.4 ± 5.0.
A second set of 61 Day 5 SET samples were considered as a “Validation Set” as Viability Scores were generated blindly using the Day 5 algorithm developed using the “Training set”. Samples in the Validation Set were collected at SGFC (n = 22), RSC (n = 12), and SYD (n = 27). In this data set there were 30 Day 5 embryos scoring A, 26 scoring B, 4 scoring C, 1 scoring D. The mean age (± SD) of the women in the Validation Set was 34.2 ± 4.3.
ROC and statistical analysis
ROC curves were generated using the ROCR library [26] of R programming language [27]. Linear models were generated using the lm function of R. The morphologic score of each embryo was re-coded to consist of four scores: A, B, C, and D from best to worst. The composite score was derived by fitting to the training data set a linear regression model of the form Composite = A + B • VS + C • morphology score.
Areas under the ROC curve (AUC) were estimated to assess the ability of the Viability Score, Morphology, and composite score to predict pregnancy outcome: the greater the AUC, the better the prediction of the model. Hosmer and Lemeshow [28] proposed thresholds to help to interpret the AUC: A perfect discrimination would have an AUC = 1, and a random prediction would result in an AUC = 0.5. The significance of the difference in performance between the three classifiers (morphology, viability and composite scores) was assessed by comparing the AUCs using the StAR server as described [29]. Alpha error of less than 0.05 was considered significant for all comparisons.
Results
ROC analysis of day 5 morphology grading and metabolomic Viability Scores on predicting implantation outcome: training sample set
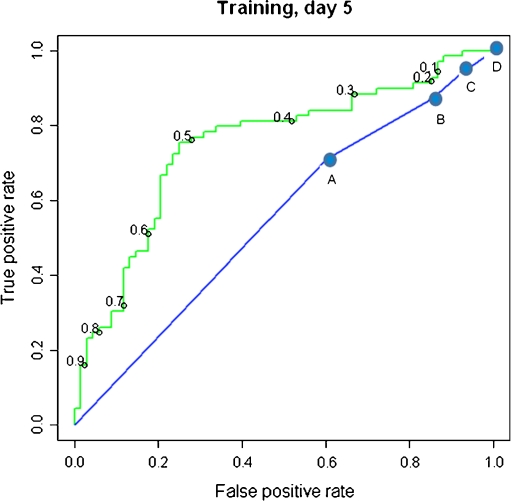
First, samples in the Training Set (n = 137) were analyzed by NIR spectroscopy and spectral regions that best discriminate between transferred embryos that did and did not result in positive FCA were identified. Using a model based on these regions, a relative Viability Score was generated for each sample, adjusting for parallel blank media controls. Then the metabolomic Viability Score and day 5 morphology were assessed and compared for their ability to predict pregnancy in women undergoing SET on day 5. The AUCROC for predicting pregnancy using the Viability Score was significantly higher compared to the AUCROC for Day 5 morphology (0.75 vs. 0.55; P = 0.0011) (Fig. 1). This indicated that, for this data set, the Viability Score is a better classifier for pregnancy outcome than morphology.
Fig. 1.
ROC curves for the Viability Score (green) and morphologic grade (blue), for day 5 SET embryo culture media samples in the Training Set. The cutoff values corresponding to the indicated true positive and false negative rates are shown on the curves. AUCROC for Viability Score = 0.75; AUCROC for morphologic grade = 0.55; P = 0.0011
ROC analysis of day 5 morphology grading and metabolomic Viability Score on predicting implantation outcome: blind assessment using the validation sample set
Next, using the model generated using the Training Sample Set, we blindly generated a Viability Score from each individual Day 5 SET from the 61 Validation Sample Set. The predictive ability of the Viability Scores was then examined in relation to FCA outcome and blastocyst grading using ROC analysis. Again, the AUCROC for predicting pregnancy using the Viability Score was significantly higher compared to the AUCROC for Day 5 morphology (0.68 vs. 0.50; P = 0.021) (Fig. 2). This verified that, for a different data set analyzed blindly using a previously established Day 5 algorithm, the Viability Score is a better classifier for pregnancy outcome than morphology.
Fig. 2.
ROC curves for the Viability Score (green) and morphologic grade (blue), for day 5 SET embryo culture media samples in the Validation Set. The cutoff values corresponding to the indicated true positive and false negative rates are shown on the curves. AUCROC for Viability Score = 0.65; AUCROC for morphologic grade = 0.50; P = 0.021
ROC analysis of day 5 morphology grading, metabolomic Viability Score, and composite score on predicting implantation outcome in validation sample set
A composite score based on Viability Score and morphologic grade was then generated by fitting a linear model to the Training Sample Set for day 5. Next, this model was applied to generate a composite score for samples in the Validation Set.
The AUCROC for predicting pregnancy using the composite score was significantly higher compared to the AUCROC for Day 5 morphology in this data set (0.74 vs. 0.50; P = 0.004) (Fig. 3). While AUCROC for the composite score was also higher compared to Viability Score alone, the difference was not statistically significant (0.74 vs. 0.68; P = 0.166) (Fig. 3).
Fig. 3.
A. ROC curves for the composite score (red), Viability Score (green) and morphologic grade (blue), for day 5 SET embryo culture media samples in the Validation Set. AUCROC for composite score = 0.74, AUCROC for Viability Score = 0.68; AUCROC for morphologic grade = 0.50. Composite score vs. morphologic grade, P = 0.004; Composite score vs. Viability Score, P = nonsignificant
The effect of different morphologic grade subsets combined with Viability Score on predicting implantation outcome in a blind validation set sample
As a further method of analysis we examined the inter-relationship between the Viability Score and Day 5 morphology, and examined the ability of the Viability Score to perform across different ranges of morphology using the Validation Set (Fig. 4).
Fig. 4.
ROC curves for the Viability Score of embryos of different morphologic subsets for day 5 SET embryo culture media samples in the Validation Set. ROC curves for Viability Score of embryos of morphologic grade A (green line), grade A and B (blue line), grades A, B, and C (red line), and grades A,B,C, and D (black line) are shown. AUCROC for each group is also shown. There was no significant difference in Viability Score from between any of the subsets of morphologic grades
Comparison of the AUCROC for Viability Score in combination with the best to the worse Day 5 morphology gave no significant change regardless of the morphology. In this scenario the Viability Score within only the best morphology embryos gave an AUCROC of 0.69 while the Viability Score with all embryos regardless of morphology gave an AUCROC of 0.68.
Discussion
In this study, we found that a Viability Score generated by rapid, non-invasive metabolomic profiling of human embryo culture media using NIR spectroscopy is more accurate in predicting pregnancy outcome compared to morphologic grading in women undergoing single embryo transfer on day 5. We also found that a combination of Viability Score and morphologic grading is more accurate in predicting outcome than morphologic grading alone. These findings provide compelling evidence that metabolomic profiling of in vitro generated embryos may have clinical benefits in the IVF laboratory.
Here, we compared the ability of metabolomic profiling to predict outcome in IVF to that of morphologic grading, which currently constitutes the mainstay of embryo assessment in IVF laboratories worldwide. It is noteworthy that there are significant methodological challenges associated with the assessment of accuracy of morphologic grading as a “Diagnostic test”, and consequently, with the comparison of a novel test to morphologic grading.
“Sensitivity” and “specificity” are key parameters used to determine accuracy of diagnostic tests. In the setting of embryo viability assessment, “sensitivity” can be defined as “the proportion of embryos that resulted in a pregnancy, which were correctly identified as viable”. Similarly, “specificity” can be defined as “the proportion of embryos that failed to result in a pregnancy, which were correctly identified as non-viable”. Therefore, in order to determine “sensitivity” and “specificity” of a diagnostic test applied to embryo assessment, the outcomes of embryos identified as “viable” or “non-viable” should be known. This is not the case in morphologic embryo grading in IVF for multiple reasons. First, morphologic grading does not use a cut-off point and does not tend to identify embryos as “viable” or “non-viable” with the possible exception of embryos that are arrested and are considered very unlikely to result in a pregnancy). Rather, morphologic assessment aims to provide grades that reflect the likelihood of an embryo to result in a pregnancy. In addition, in most cases, only some of the embryos identified as most or more viable, and none of the embryos identified as less viable, are transferred.
In other words, in IVF, outcome is only known for transferred embryos, and all transferred embryos, including those of lowest morphologic grades, are selected with the assumption that they have a potential to result in a pregnancy. Consequently, using sensitivity and specificity does not seem to be the best strategy for comparing novel embryo assessment strategies to morphological grading. Therefore, we performed a ROC analysis and assessed the accuracy of increasing morphologic grades (from D to A) in predicting pregnancy. Viability Scores were similarly evaluated.
Receiver operating characteristic (ROC) analysis is a well-established method for assessing binary classification task performance [30]. It was first used by the US Army to increase the correct detection of aircraft from radar signals during World War II. Later, ROC was applied to medicine for evaluating diagnostic testing performance and has since been extensively applied in various areas of medical research [30]. An ROC curve, is a graphical plot of the sensitivity vs. (1 − specificity) for a binary classifier system as its discrimination threshold is varied. When one ROC curve is above the other ROC curve at every point (Figs. 1, 2 and 3), it suggests that the technique producing the higher curve is superior to the technique that produced the lower curve. In particular, a system with a higher ROC curve suggests that 1) this system always has higher sensitivity for any given specificity; and 2) this system always results in a greater number of correct decisions, regardless of disease (or outcome) prevalence [30]. Thus, the higher the ROC curve, the better the system [30]. Consequently, the area under the curve (AUC) is often used as a measure of task performance [30].
In the current study, ROC curves for “Viability Score” and “Viability Score combined with morphologic grading” were higher than the ROC curve for “morphologic grading alone” at every point assessed (Figs. 1, 2 and 3). For the training set of 137 SETs the AUC was greater for the Viability Scores compared to Morphology alone [0.75 versus 0.55, P = 0.0011] (Fig. 1). Similarly, for the Day 5 Validation Set of 61 SETs the AUC was again greater for the Viability Score compared to Morphology [0.68 versus 0.50, P = 0.021] (Fig. 2). Finally, AUC for the Validation set, was also greater for the Viability Score combined with morphologic grading compared to Morphology alone [0.74 versus 0.50, P = 0.004] (Fig. 3). Therefore, our findings suggest that Viability Score alone or in combination with morphologic grading provides additional predictive value over morphologic grading alone in women undergoing SET on day 5. The use of ROC curve (rather than a cut-off point to define pregnancy as an all-or-none phenomenon) is a novel aspect of the current study, allowing direct comparison of metabolomic viability score and morphologic assessment.
Within the past two decades, an increasing number of embryo culture media have become commercially available and improvements in culture media formulations have led to an increase in the ability to maintain the human embryo in culture throughout the preimplantation period [31, 32]. A concern regarding the application of metabolomic profiling technology to different IVF centers is the substantial differences in the content of commercial culture media, which they might use. A robust and widely applicable methodology for the evaluation of spent culture media should be able to detect the changes associated with embryo viability independent of type of culture media used in a particular setting. In this study, the “training set” consisted of 137 samples that were collected following day 5 SET in two centers using two different types of culture media. More importantly, using the algorithm developed with the training set, Viability Scores obtained for the “Validation set” achieved a high accuracy in 61 samples collected in 3 centers using different types of culture media. Our findings, combined with previous reports [21, 22], suggest that an algorithm developed using spectroscopy-based metabolomic profiling can aid in the accurate prediction of viability across a multitude of culture media types and culture volumes.
In this study we evaluated spent culture media samples collected after SET on day 5. We had previously published that metabolomic profiling is predictive of embryo viability in blinded analyses of spent culture media collected after embryo transfer on day 2 or 3 [21, 24]. The current study is the first report on utilization of NIR spectroscopy-based metabolomics for model development and blinded analysis to determine viability of day 5 embryos. Extended culture to blastocyst stage, made possible by the advent of more physiological culture media and improvements in the IVF laboratory, resulted in high implantation and pregnancy rates [33–36]. A meta-analysis by Blake et al. [37] including 18 randomized controlled trials, reported that blastocyst stage embryo transfer is associated with increased pregnancy rates compared to embryo transfer at cleavage stage. This effect of extended culture is believed to be largely due to an improvement in the assessment of embryo viability. However, while exclusion of embryos that do not reach the compaction stage on day 5 may have had beneficial effect on outcome, we found the accuracy of morphologic grading to be poor (Figs. 1, 2 and 3).
It is noteworthy that our initial studies on the application of metabolomic profiling to embryo assessment used both Raman and NIR-spectroscopies [21, 22], both vibrational spectroscopies (see [16] for comparison) that can be performed on-site using small size instruments compatible with the environment of an IVF laboratory. In our more recent studies [23, 24], and in the current study, we utilized NIR spectroscopy as this technology has a sensitivity and specificity comparable to Raman, and is less costly, therefore has more potential for clinical application.
Our findings extend previously published data [21–24] by demonstrating that metabolomic models alone or in combination with morphology allow an improved prediction of embryo viability compared to morphology. Therefore, our study introduces metabolomic profiling as a rapid, and non-invasive technology that can provide an objective assessment aiding in the decision of which embryo(s) to transfer in the IVF laboratory. It is noteworthy that an improved understanding of embryo viability will aid in identifying the embryo(s) that are most likely to result in a pregnancy. Consequently, increased pregnancy rates may be achieved in women undergoing SET. In addition, in women undergoing multiple embryo transfer, more accurate decisions can be made about the number of embryos to be transferred, potentially reducing the likelihood of multiple gestation while maintaining or even increasing pregnancy rates. Further studies using randomized prospective study design will be necessary to determine the true value and limitations of the use of metabolomics in IVF.
Footnotes
Capsule
In women undergoing single embryo transfer on day 5, receiver operating characteristic (ROC) analysis suggests that metabolomic Viability Score alone or in combination with morphologic grading has the potential to be a better classifier for pregnancy outcome than morphology alone.
References
- 1.SART. Assisted reproductive technology success rates. National summary and fertility clinic reports. Centers for disease control, USA; 2007. [DOI] [PubMed]
- 2.Ata B, Seli E. Economics of assisted reproductive technologies. Curr Opin Obstet Gynecol. 2010; epub Feb 1. [DOI] [PubMed]
- 3.Steptoe PC, Edwards RG. Birth after the reimplantation of a human embryo. Lancet. 1978;2:366. doi: 10.1016/S0140-6736(78)92957-4. [DOI] [PubMed] [Google Scholar]
- 4.Trounson A, Leeton J, Wood C, Webb J, Wood J. Pregnancies in humans by fertilization in vitro and embryo transfer in the controlled ovulatory cycle. Science. 1981;216:681–682. doi: 10.1126/science.7221557. [DOI] [PubMed] [Google Scholar]
- 5.Edwards R, Fishel S, Cohen J. Factors influencing the success of in vitro fertilization for alleviating human infertility. J In Vitro Fertil Embryo Transf 1984:3–23. [DOI] [PubMed]
- 6.Gardner DK, Schoolcraft WB. In vitro culture of human blastocysts. In: MD JR, editor. Towards reproductive certainty: fertility and genetics beyond. Carnforth: Parthenon; 1999. pp. 378–388. [Google Scholar]
- 7.Gerris J, Neubourg D, Mangelschots K, et al. Prevention of twin pregnancy after in-vitro fertilization or intracytoplasmic sperm injection based on strict embryo criteria: a prospective randomized clinical trial. Hum Reprod. 1999;14:2581–2587. doi: 10.1093/humrep/14.10.2581. [DOI] [PubMed] [Google Scholar]
- 8.Scott LA, Smith S. The successful use of pronuclear embryo transfer the day following oocyte retrieval. Hum Reprod. 1998;13:1003–1013. doi: 10.1093/humrep/13.4.1003. [DOI] [PubMed] [Google Scholar]
- 9.Tesarik J, Greco E. The probability of abnormal preimplantation development can be predicted by a single static observation on pronuclear state morphology. Hum Reprod. 1999;14:1318–1323. doi: 10.1093/humrep/14.5.1318. [DOI] [PubMed] [Google Scholar]
- 10.VanRoyan E, Mangelschots K, Neubourg D, Valkenburg M, Meerssche M, Ryckaert G, et al. Characterization of a top quality embryo, a step towards single-embryo transfer. Hum Reprod. 1999;14:2345–2349. doi: 10.1093/humrep/14.9.2345. [DOI] [PubMed] [Google Scholar]
- 11.Veeck L. An atlas of human gametes and conceptuses: an illustrated reference for assisted reproductive technology. New York: Parthenon; 1999. [Google Scholar]
- 12.Sakkas D, Percival G, D’Arcy Y, Sharif K, Afnan M. Assessment of early cleaving in vitro fertilized human embryos at the 2-cell stage before transfer improves embryo selection. Fertil Steril. 2001;76:1150–1156. doi: 10.1016/S0015-0282(01)02901-6. [DOI] [PubMed] [Google Scholar]
- 13.Toner JP. Progress we can be proud of: U.S. trends in assisted reproduction over the first 20 years. Fertil Steril. 2002;78:943–950. doi: 10.1016/S0015-0282(02)04197-3. [DOI] [PubMed] [Google Scholar]
- 14.Bromer JG, Seli E. Assessment of embryo viability in assisted reproductive technologies: shortcomings of current approaches and the emerging role of metabolomics. Curr Opin Obstet Gynecol. 2008;20:234–241. doi: 10.1097/GCO.0b013e3282fe723d. [DOI] [PubMed] [Google Scholar]
- 15.Sakkas D, Gardner DK. Noninvasive methods to assess embryo quality. Curr Opin Obstet Gynecol. 2005;17:283–288. doi: 10.1097/01.gco.0000169106.69881.3e. [DOI] [PubMed] [Google Scholar]
- 16.Botros L, Sakkas D, Seli E. Metabolomics and its application for non-invasive embryo assessment in IVF. Mol Hum Reprod. 2008;14:679–690. doi: 10.1093/molehr/gan066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katz-Jaffe MG, Schoolcraft WB, Gardner DK. Analysis of protein expression (secretome) by human and mouse preimplantation embryos. Fertil Steril. 2006;86:678–685. doi: 10.1016/j.fertnstert.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 18.Fuzzi B, Rizzo R, Criscuoli L, Noci I, Melchiorri L, Scarselli B, et al. HLA-G expression in early embryos is a fundamental prerequisite for the obtainment of pregnancy. Eur J Immunol. 2002;32:311–315. doi: 10.1002/1521-4141(200202)32:2<311::AID-IMMU311>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 19.Vercammen MJ, Verloes A, Velde H, Haentjens P. Accuracy of soluble human leukocyte antigen-G for predicting pregnancy among women undergoing infertility treatment: meta-analysis. Hum Reprod Update. 2008;14:209–218. doi: 10.1093/humupd/dmn007. [DOI] [PubMed] [Google Scholar]
- 20.Oliver SG, Winson MK, Kell DB, Baganz F. Systematic functional analysis of the yeast genome. Trends Biotechnol. 1998;16:373–378. doi: 10.1016/S0167-7799(98)01214-1. [DOI] [PubMed] [Google Scholar]
- 21.Seli E, Sakkas D, Scott R, Kwok JS, Rosendahl S, Burns DH. Non-invasive metabolomic profiling of human embryo culture media using Raman and near infrared spectroscopy correlates with reproductive potential of embryos in women undergoing in vitro fertilization. Fertil Steril. 2007;88:1350–1357. doi: 10.1016/j.fertnstert.2007.07.1390. [DOI] [PubMed] [Google Scholar]
- 22.Scott RT, Seli E, Miller K, Sakkas D, Scott K, Burns DH. Non-invasive metabolomic profiling of human embryo culture media using Raman spectroscopy predicts embryonic reproductive potential: a prospective blinded pilot study. Fertil Steril. 2008;90:77–83. doi: 10.1016/j.fertnstert.2007.11.058. [DOI] [PubMed] [Google Scholar]
- 23.Vergouw CG, Botros LL, Roos P, Lens JW, Schats R, Hompes PGA, et al. Metabolomic profiling by near-infrared spectroscopy as a tool to assess embryo viability: a novel, non-invasive method for embryo selection. Hum Reprod. 2008;23:1499–1504. doi: 10.1093/humrep/den111. [DOI] [PubMed] [Google Scholar]
- 24.Seli E, Vergouw CG, Morita H, Botros L, Roos P, Lambalk CB, et al. Non-invasive metabolomic profiling as an adjunct to morphology for non-invasive embryo assessment in women undergoing single embryo transfer. Fertil Steril. 2010;94:535–542. doi: 10.1016/j.fertnstert.2009.03.078. [DOI] [PubMed] [Google Scholar]
- 25.Seli E, Botros L, Henson M, Roos P, Sakkas D, group. Ms. Viability scores determined by metabolomic assessment of embryo culture media correlate with IVF outcome in women undergoing single embryo transfer on day 2: a prospective multi-center trial. In: Annual Meeting of American Society of Reproductive Medicine; 2009; Atlanta, GA.; 2009.
- 26.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21:3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 27.R Development Core Team. R: A language and environment for statistical computing. In: R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL: http://www.R-project.org; 2005.
- 28.Hosmer DW, Lemeshow S. Applied logistic regression. 2nd ed. Wiley; 2000.
- 29.Vergara IA, Norambuena T, Ferrada E, Slater AW, Melo F. StAR: a simple tool for the statistical comparison of ROC curves. BMC Bioinform. 2008;9:265. doi: 10.1186/1471-2105-9-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He X, Frey E, ROC, LROC, FROC, AFROC An alphabet soup. J Am Coll Radiol. 2009;6:652–655. doi: 10.1016/j.jacr.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 31.Gardner DK. Dissection of culture media for embryos: the most important and less important components and characteristics. Reprod Fertil Dev. 2008;20:9–18. doi: 10.1071/RD07160. [DOI] [PubMed] [Google Scholar]
- 32.Biggers JD, Summers MC. Choosing a culture medium: making informed choices. Fertil Steril. 2008;90:473–483. doi: 10.1016/j.fertnstert.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 33.Gardner DK, Schoolcraft WB, Wagley L, Schlenker T, Stevens J, Hesla J. A prospective randomized trial of blastocyst culture and transfer in in-vitro fertilization. Hum Reprod. 1998;13:3434–3440. doi: 10.1093/humrep/13.12.3434. [DOI] [PubMed] [Google Scholar]
- 34.Behr B, Pool TB, Milki AA, Moore D, Gebhardt J, Dasig D. Preliminary clinical experience with human blastocyst development in vitro without co-culture. Hum Reprod. 1999;14:454–457. doi: 10.1093/humrep/14.2.454. [DOI] [PubMed] [Google Scholar]
- 35.Auwera I, Debrock S, Spiessens C, Afschrift H, Bakelants E, Meuleman C, et al. A prospective randomized study: day 2 versus day 5 embryo transfer. Hum Reprod. 2002;17:1507–1512. doi: 10.1093/humrep/17.6.1507. [DOI] [PubMed] [Google Scholar]
- 36.Gardner DK, Surrey E, Minjarez D, Leitz A, Stevens J, Schoolcraft WB. Single blastocyst transfer: a prospective randomized trial. Fertil Steril. 2004;81:551–555. doi: 10.1016/j.fertnstert.2003.07.023. [DOI] [PubMed] [Google Scholar]
- 37.Blake DA, Farquhar CM, Johnson N, Proctor M. Cleavage stage versus blastocyst stage embryo transfer in assisted reproduction. Cochrane Database Syst Rev. 2007;4:CD002118. doi: 10.1002/14651858.CD002118.pub3. [DOI] [PubMed] [Google Scholar]