Summary
The repair of DNA double-strand breaks by homologous recombination is essential for genomic stability. The first step in this process is resection of 5’ strands to generate 3’ single-stranded DNA intermediates. Efficient resection in budding yeast requires the Mre11–Rad50–Xrs2 (MRX) complex and the Sae2 protein, although the role of MRX has been unclear since Mre11 paradoxically exhibits 3’ to 5’ exonuclease activity in vitro. Here we reconstitute resection with purified MRX, Sae2, and Exo1 proteins and show that degradation of the 5’ strand is catalyzed by Exo1 yet completely dependent on MRX and Sae2 when Exo1 levels are limiting. This stimulation is largely the result of cooperative binding of DNA substrates by Exo1, MRX, and Sae2. This work establishes the direct role of MRX and Sae2 in promoting the resection of 5’ strands in DNA double-strand break repair.
Introduction
Accurate repair of double-strand breaks (DSBs) in chromosomal DNA is integral to the maintenance of genomic integrity in all cells, and is essential for early development in vertebrates 1. DSB repair mediated by homologous recombination utilizes an intact template for DNA replication across the break site, and in eukaryotes is preferentially used in the S and G2 phases of the cell cycle when sister chromatids are present 2,3.
Homologous recombination occurs in several distinct steps that prepare a broken DNA substrate for strand invasion into a homologous template and eventual resolution of strand invasion intermediates 4,5. The first step in this process is the resection of 5’ strands from the DSB ends, which in eukaryotes occurs typically over the course of 1 to 4 hours after a DSB is introduced, based on studies in fungi. The extent of the resection (a few hundred nucleotides to tens of kb) depends on the locus and on the availability of a homologous target for strand invasion or single-strand annealing 6−9. The 3’ single-stranded DNA (ssDNA) overhangs are bound initially by RPA, which is subsequently exchanged for the Rad51 recombinase with the help of Rad52 and other mediator proteins. The resection step in homologous recombination is a critical control point in eukaryotes, as it is regulated during the cell cycle to occur preferentially in the S and G2 phases 2,3, although partial resection of radiation-induced breaks has also been shown to occur during G1 phase 10. Extensive removal of the 5’ strand at a break also commits a cell to homologous recombination as the processed ends can no longer be joined through non-homologous end-joining pathways.
5’ strand resection of DSBs in most bacteria is catalyzed by the RecBCD helicase–nuclease complex, but RecBCD does not appear to exist beyond prokaryotes, so the mechanisms of DNA end resection in archaea and eukaryotes have remained largely unknown. Mre11–Rad50 complexes have been suggested as likely candidates for enzymes that initiate 5’ strand resection for many years, initially because of the phenotypes of null and hypomorphic mutants during meiosis in budding yeast 11−13. The efficiency of 5’ strand resection during mitotic recombination is reduced and delayed in MRX or Sae2 mutants 14,15. The fact that Mre11 shares homology with phosphodiesterases has also contributed to speculation that Mre11–Rad50 complexes directly degrade 5’ strands at DSBs. However, we and others have shown with recombinant proteins in vitro that Mre11 proteins from several different species all exhibit 3’ to 5’ exonuclease activity in vitro in the presence of manganese 16−19, which is the opposite polarity from the 5’ to 3’ excision that is required to generate 3’ overhangs. Mre11 nuclease activity is also unnecessary for the resection of endonuclease-induced DSBs in vegetatively growing yeast cells, although it is essential for the processing of meiotic DSBs which are covalently bound on the 5’ strand to the Spo11 protein 20−22.
Recent work has shown that there are two redundant pathways of further resection downstream of MRX and Sae2: the first consisting of the yeast Exo1 protein and the second consisting of a complex of proteins containing the Dna2 nuclease, the Sgs1 helicase, Rmi1, and Top3 23−25. These studies showed that the Mre11–Rad50–Xrs2 (MRX) complex, along with Sae2, are primarily responsible for the initiating stages of DSB processing.
Despite the recent identification of the components of resection in vivo in budding yeast, it is still unclear what the mechanistic role of each of these proteins is at a DSB. Why are so many nucleases present, what is the role of each, and do MRX and Sae2 actually contribute directly to the resection reaction? To address these questions, we used recombinant MRX, Sae2, and Exo1 proteins to reconstitute eukaryotic DSB resection in vitro with purified components. We show in this work that MRX and Sae2 strongly promote 5’ strand resection by Exo1 and are essential when Exo1 concentrations are functionally limiting, indicating that they play a direct role in the processing reaction. This work establishes an in vitro reaction to dissect the roles of these and other components known to regulate 5’ strand resection in cells.
Results
MRX and Sae2 stimulate Exo1 activity on DNA ends
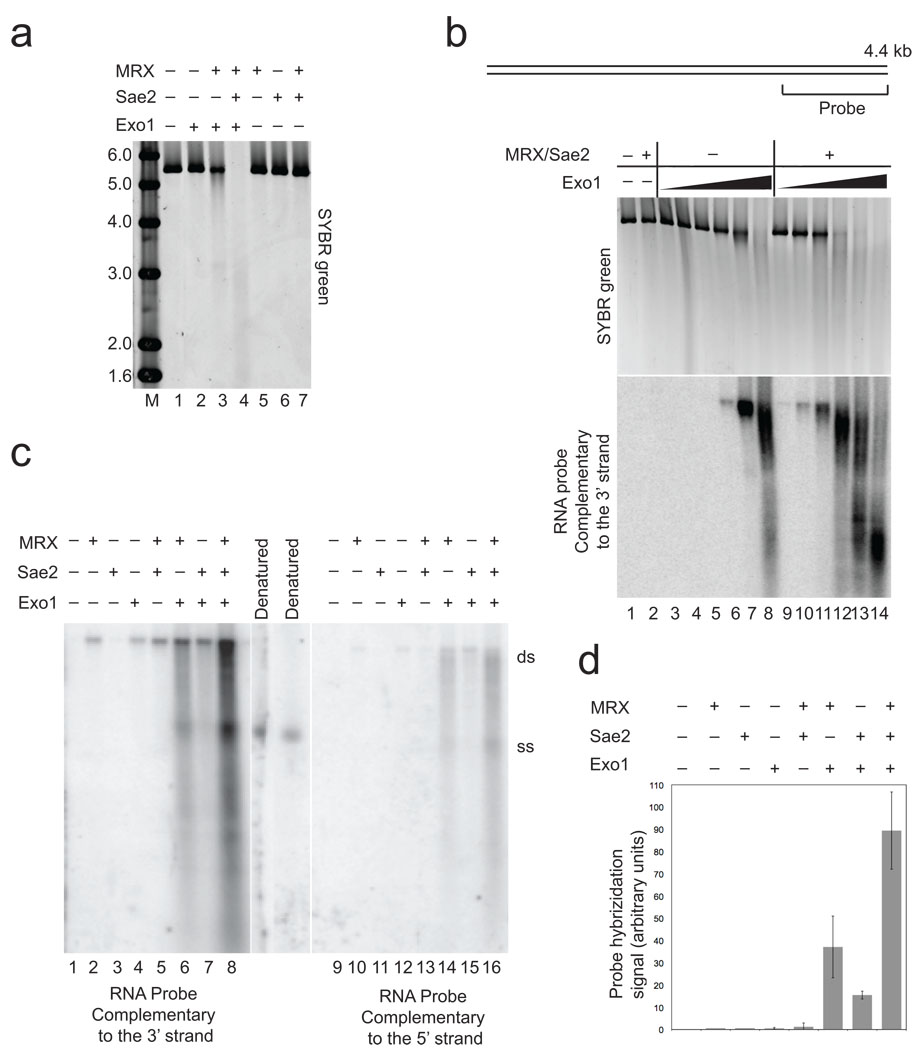
To determine if MRX and Sae2 contribute directly to the resection of 5’ strands of DNA at a double-strand break (DSB), we expressed and purified recombinant yeast Exo1 protein (Supplementary Fig. 1) and tested its activity in vitro on a linear DNA substrate in the presence or absence of recombinant MRX and Sae2. As shown in Fig. 1A, the 5.6 kb linear DNA substrate exhibits little detectable degradation in the presence of Exo1 alone, but the substrate is partially degraded with MRX present, and completely degraded in the presence of both MRX and Sae2. Yeast Exo1 is an active nuclease alone, as shown in the titration in Fig. 1B (top), although activity is seen at 4 to 8-fold lower concentrations when assayed in the presence of MRX and Sae2 (compare lanes 4–7 with lanes 10–13).
Figure 1. 5’ strand degradation at a DNA break by Exo1 is promoted by Mre11–Rad50–Xrs2 and Sae2.
(a) 5.6 kb double-stranded DNA (pTP407) linearized with BseRI (0.3 nM) was incubated with Exo1 (0.5 nM), MRX (5 nM), and Sae2 (5 nM) for 60 min. at 30°C. The native agarose gel containing the reactions is shown after visualization of the DNA with SYBR green. Lane marked "M" contains MW markers as indicated (kb). (b) Resection assays were performed with a 4.4 kb plasmid DNA substrate (pNO1) linearized with SphI and analyzed as in (a) with SYBR green staining (top panel), and with non-denaturing Southern hybridization with a strand-specific RNA probe for the 3' strand at one end (see diagram). Reactions contained 14 nM MRX, 3.5 nM Sae2, and 0.4, 0.8, 1.6, 3.2, 6.4, and 32 nM Exo1 and were incubated for 60 min. at 30°C. (c) Resection assays were performed as in (b) with 4 nM Exo1. The reactions were split and separated in parallel in a non-denaturing gel, followed by non-denaturing Southern hybridization and probed separately for single-stranded 3’ strand (left) or 5’ strand (right) DNA adjacent to the break site. Denatured plasmid DNA was used as a marker for the single-stranded DNA “ss”, and the position of the unresected plasmid is marked as double-stranded, “ds”. (d) Quantitation of the reactions shown in (b), in addition to reactions from 4 other independent experiments, using phosphorimager analysis to quantitate the total counts in each lane. Within each experiment, the signal from the reactions containing wild-type MRX, Exo1, and Sae2 was set to 100%, with the other values shown relative to this. Error bars indicate standard deviation.
To visualize the single-stranded DNA (ssDNA) generated during the resection reaction, we used a strand-specific RNA probe that hybridizes to a 1 kb region on one end of the substrate, following native Southern blot transfer to a nylon membrane (Fig. 1B, bottom). Using this sensitive assay, it is clear that Exo1 generates 3' ssDNA and that the extent of resection increases dramatically with MRX and Sae2 present in the reaction. Both the efficiency of resection, as measured by the intensity of the signal, as well as the extent of resection, as measured by the decrease in size of the reaction products, increased with the addition of MRX and Sae2. Comparing a 3' strand probe with a 5' strand probe in Fig. 1C, we observed that the majority of products hybridize to the 3' strand probe, thus resection by Exo1–MRX–Sae2 is in the 5' to 3' direction. A weak signal is apparent with the probe specific for the 5’ strand; we hypothesize that this is the result of resection initiating from the other DSB end, as shown previously with Pyrococcus furiosus (P. furiosus) MR in combination with the helicase–nuclease complex HerA–NurA 26. In this assay it is clear that Sae2 and MRX cooperatively increase Exo1-mediated resection (Fig. 1C, lane 8), but each also has a weak stimulatory effect on Exo1 independently (lanes 6 and 7). The stimulation of Exo1 by MRX–Sae2 combined is 60 to 300-fold over Exo1 alone, based on quantitation of the signal from 5 independent experiments (Fig. 1D). MRX by itself generates a small but detectable signal of resected 5’ strands (Fig. 1C, lane 2), as previously reported for P. furiosus Mre11–Rad50 26, that increases slightly in the presence of Sae2 (Fig. 1C, lane 5). Sae2 does not generate any resection products alone (Fig. 1C, lane 3), consistent with our previous finding that Sae2 endonuclease activity requires single-stranded DNA and is not apparent on double-stranded DNA 27.
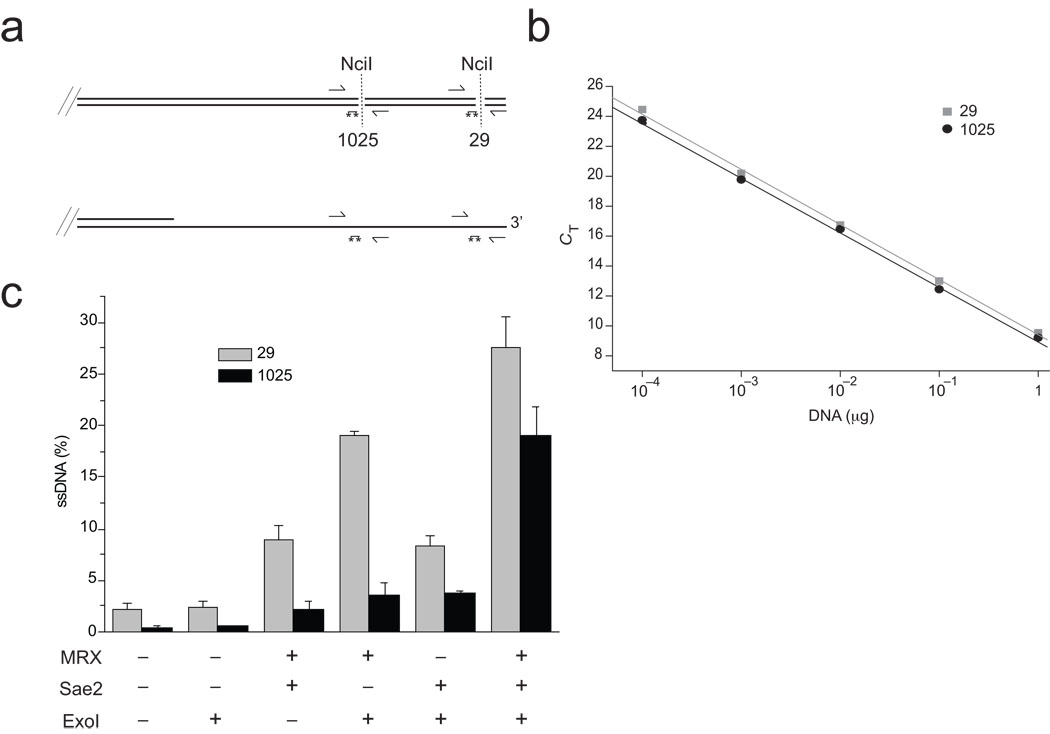
To further characterize the reaction and assess the extent of resection quantitatively, we developed a real-time PCR assay similar to a system used previously to monitor resection in vivo in yeast 8. DNA resection at restriction sites located 29 bp and 1025 bp from the DSB site was assessed by quantitative PCR using two sets of primers and probes (Fig. 2A, B). This analysis showed that, in a typical reaction with 4 nM Exo1, 25 to 30% of the substrate is single-stranded close to the DSB site in the presence of all of the factors, and that this proximal resection is mostly dependent on MRX and Exo1 (Fig. 2C, top). At the site located 1 kb from the break, all of the components are necessary for efficient resection, with ~20% of the input DNA converted to resected product (Fig. 2C, bottom).
Figure 2. Characterization of the products of cooperative DNA resection by Exo1, MRX, and Sae2.
(a) Diagram of the end of the pNO1 DNA substrate cut with SphI, with locations of NciI sites, PCR primers, and qPCR 6-FAM/TAMRA probes (asterisks). NciI digests double-stranded DNA (top) but leaves single-stranded DNA intact (bottom). (b) Standard curves using undigested DNA with the primer sets for the 29 bp (squares) and 1025 bp (circles) NciI sites. (c) qPCR results from resection assays performed as in Fig. 1B but analyzed using the qPCR technique to assess the level of DNA resection at each site: 29 nt from the end (top panel) and 1025 nt from the end (bottom panel). From each reaction, undigested aliquots were analyzed and compared with digested aliquots to obtain a ΔCT value, which was used to calculate the percentage of ssDNA in the reaction as described in the Methods. Each resection reaction was performed in triplicate and a qPCR analysis done for each; the average of these is shown in the graph with the standard error.
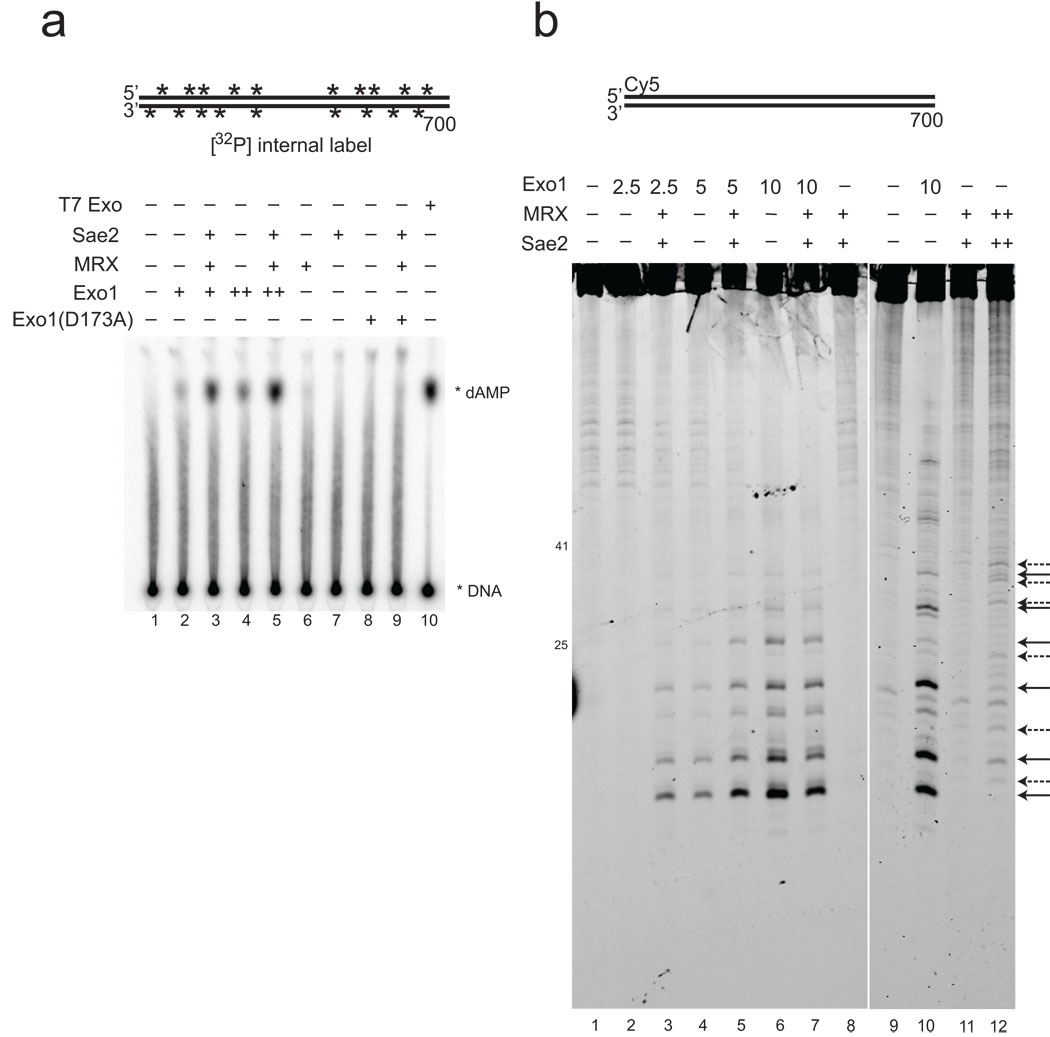
Exo1 has been shown to degrade DNA as a 5' to 3' exonuclease but also as an endonuclease on 5' flap DNA substrates 28. To determine which activity operates during resection, an internally labeled, 700 bp DNA substrate was used with Exo1, MRX, and Sae2 and the products analyzed by thin layer chromatography (Fig. 3A). This analysis shows that single nucleotides are excised by Exo1, and that MRX and Sae2 cooperatively stimulate this activity. To examine endonuclease activity by Exo1, we used 5' Cy5 fluorescently-labeled 700 bp DNA fragment, as shown in Fig. 3B. High levels of Exo1 also clearly generate oligonucleotide products from the 5' end of the 5' strand (lane 6). With lower levels of Exo1, MRX and Sae2 stimulate the appearance of these products (Fig. 3B, lanes 2 to 5), thus Exo1 acts both as an endonuclease and an exonuclease in resecting linear DNA, and MRX and Sae2 promote both activities.
Figure 3. Digestion of linear DNA by Exo1 produces both single nucleotide and oligonucleotide products.
(a) Resection assays were performed as in Fig. 1B except with 700 bp DNA substrate internally labeled with 32[P], with Exo1 (0.2, 0.4 nM), MRX (1.6 nM), and Sae2 (0.6 nM) or T7 exonuclease (1 unit per reaction). Reactions were stopped with SDS and EDTA and separated by thin layer chromatography; migration of the labeled dAMP product is indicated. T7 exonuclease was used as a positive control to generate single nucleotide products. (b) Resection assays were performed as in (a) except that the 700 bp DNA substrate was labeled on one 5' strand with Cy5. Reactions contained 2.5 (lanes 2, 3), 5 (lanes 4, 5), and 10 (lanes 6, 7, 10) nM Exo1, 7.5 nM MRX, and 1.5 nM Sae2 as indicated, with 25 nM MRX and 3 nM Sae2 in the reaction in lane 12. Arrows on the right side indicate positions of cleavage products (solid line: Exo1; dashed line: MRX–Sae2).
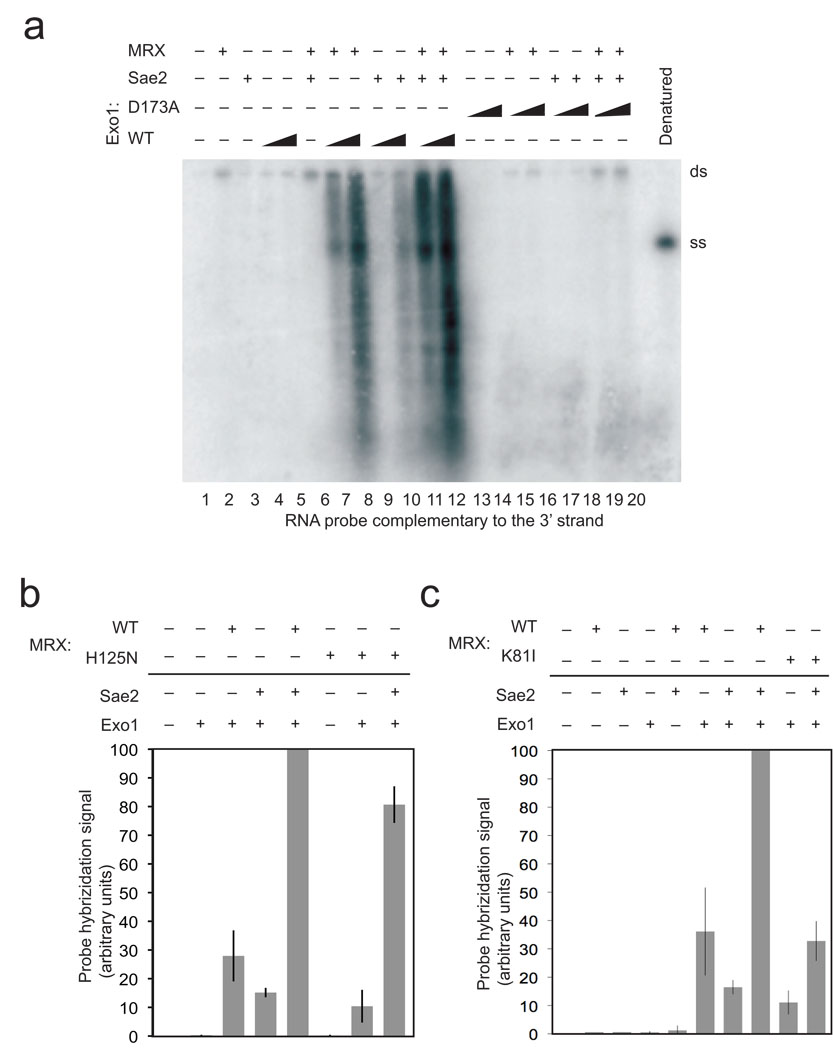
Based on this data we conclude that Exo1, MRX, and Sae2 act cooperatively to resect the 5’ strand of a DSB. Yeast Exo1, MRX, and Sae2 each exhibit nuclease activity in vitro 18,27,28, although neither Mre11 nor Sae2 would be expected to perform extensive 5’ to 3’ degradation of DNA based on the characteristics of their activities in vitro. To determine if Exo1 plays the primary catalytic role in this reaction, we expressed and purified a catalytic mutant of Exo1 which has previously been shown to lack nuclease activity 28. This mutant, D173A, exhibits no detectable activity in resection assays alone or in combination with MRX and Sae2 (Fig. 3A, lanes 8 and 9, and Fig. 4A, lanes 13, 14, 19 and 20). Thus, the catalytic activity of Exo1 is responsible for essentially all of the 5’ strand resection observed in these reactions, even though this activity is strongly dependent on MRX and Sae2.
Figure 4. Mutations in Exo1 and Rad50 prevent DNA end processing.
(a) Resection assays were performed and analyzed as in Fig. 1B using non-denaturing southern hybridication with a 3' strand probe and with a catalytic mutant of Exo1, D173A, as indicated. Concentrations of wild-type and mutant Exo1 were 4 nM and 8 nM. (b) Resection assays were performed as in Fig. 1B with the probe specific for the 3' strand but with MRX complexes containing the Mre11 mutant H125N. Results from 3 experiments were quantitated using phosphorimager analysis to quantitate the total counts in each lane and the average of these shown, with error bars indicating standard deviation. Within each experiment, the signal from the reactions containing wild-type MRX, Exo1, and Sae2 was the highest and set to 100%, with the signals in other lanes shown relative to this value. (c) Resection assays were performed, analyzed, and presented as in (b) but with the MR(K81I)X mutant complex. Reactions from three independent experiments were quantitated.
Exo1 stimulation does not require Mre11 nuclease activity
The Mre11 protein exhibits endonuclease as well as 3’ to 5’ exonuclease activity in vitro, although in all of these cases it was found to be manganese-dependent 16−19,29. All of the reactions performed in this study contain only magnesium, which is the more physiologically relevant metal ion in vivo, and do not contain manganese. Nevertheless, it is still possible that a low level of Mre11 nuclease activity still occurs in magnesium, as we demonstrated previously with P. furiosus Mre11–Rad50 complexes 26. To test whether Mre11 nuclease activity contributes to the Exo1-dependent resection reaction, we expressed and purified the H125N mutant form of Mre11 in complex with Rad50 and Xrs2, M(H125N)RX, which lacks both endo- and exonuclease activity in vitro 20. We found that the nuclease-deficient complex bound to DNA in gel mobility shift assays similarly to wild-type MRX (Supplementary Fig. 2) and stimulated Exo1 activity at ~80% of the level of wild-type MRX in the presence of Sae2 (Fig. 4B). The same mutant expressed in vivo in yeast showed nearly wild-type levels of resection 20.
We also expressed and purified a “Rad50S” MRX complex, containing the Rad50 mutant K81I that was shown to block resection of Spo11-induced DSBs in meiosis and delay end processing in mitotic cells 11,15. This mutant MRX complex bound to DNA similarly to wild-type MRX (Supplementary Fig. 2) but nevertheless showed inefficient stimulation of Exo1, 3 to 4-fold lower than with wild-type MRX (Fig. 4C). Higher levels of Sae2 could reduce the defect observed with the K81I MRX mutant complex in vitro (data not shown), consistent with the observation that overexpression of Sae2 in vivo was found to partially suppress the defects in resection observed with the rad50S allele 15. However, the defect conferred by the rad50S mutation is not limited to its interaction with Sae2, because the stimulation of Exo1 observed with MRX alone is also reduced by 3.3-fold.
Sae2 also exhibits endonuclease activity in vitro 27 and is active in magnesium. In a previous study we generated N- and C-terminal truncated versions of Sae2 which lacked dimerization, DNA-binding, and catalytic activities in vitro 27. We tested both of these mutants in the resection assay in comparison with wild-type Sae2 and found that neither stimulated resection above the level of MRX and Exo1 alone (Fig. 5A). Wild-type Sae2 increased the efficiency of the resection 3-fold, while the ΔN mutant (deficient in dimerization, DNA-binding, and catalytic activity) and the ΔC mutant (deficient in catalytic activity) did not stimulate resection catalyzed by Exo1 alone or Exo1 in combination with MRX.
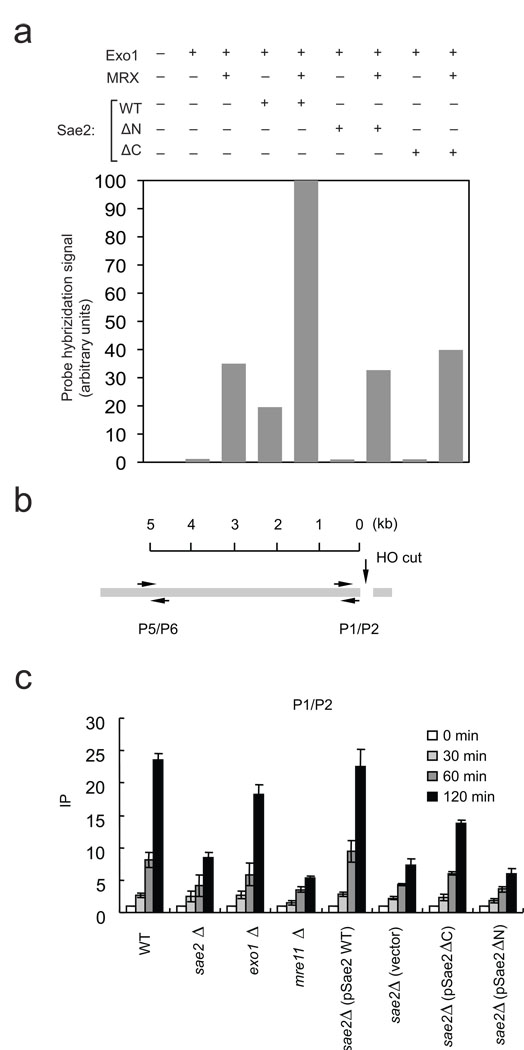
Figure 5. Mutations in Sae2 reduce the efficiency of Exo1-mediated DSB resection in vitro and in vivo.
(a) Resection assays were performed, analyzed, and presented as in Fig. 4B but with the Sae2 mutants ΔC (deletion of amino acids 251–345) and ΔN (deletion of amino acids 21 −172) as indicated. Reactions from two independent experiments were quantitated and the average value is shown. (b) Schematic of the MAT locus containing an HO endonuclease cut site, and the locations of PCR primers used to assess the levels of RPA in sae2Δ strains carrying plasmids expressing a vector control, wild type, ΔN, or ΔC truncations of Sae2. mre11Δ and exo1Δ strains are also shown for comparison. Chromatin immunoprecipitation assays were performed using an anti-RPA antibody as described previously 48. PCR signals from a primer set that anneal 0.2 kb (P1–P2) from an HO break at different durations of HO expression were quantified and plotted in (c).
Sae2 has been shown to be required for efficient resection of DSBs and repair of breaks by single-strand annealing in Saccharomyces cerevisiae in vivo 15,23. To assay for the effect of Sae2 on resection in vivo, we measured resection at an HO-induced break using chromatin immunoprecipitation (ChIP) with an antibody specific for RPA, which loads onto ssDNA after DSB processing (Fig. 5B). These results indicate that deletion of Sae2 strongly reduced RPA recruitment close to the break site as measured by the P1–P2 probe (Fig. 5C), as well as 5 kb from the cut site, as measured with the P5–P6 probe (Supplementary Fig. 3). The reduction in ChIP signal in a sae2 strain was not as severe as with an mre11 deletion but much more extreme than with an exo1 deletion, most likely because of the redundancy between Exo1 and the Sgs1–Dna2 pathway in vivo. To test whether the mutants we have analyzed are deficient in resection in cells, we expressed the ΔN and ΔC Sae2 mutants from low-copy plasmids in comparison to wild-type Sae2 in a budding yeast strain lacking endogenous Sae2. Expression of wild-type Sae2 facilitated RPA recruitment to DNA adjacent to the break site, while the ΔC showed a modest stimulatory effect over background, and the ΔN mutant did not detectably facilitate resection in vivo (Fig. 5C and Supplementary Fig. 3). Similar results were obtained when the production of ssDNA at the break site was directly measured in each strain using a PCR-based assay (Supplementary Fig. 4).
MRX and Sae2 promote Exo1 binding to DNA ends
One possible explanation for the strong stimulatory effect of MRX and Sae2 on Exo1 activity would be protein-protein interactions between the respective complexes. We have looked for such interactions but have not observed any stable interactions between MRX and Sae2 or between Exo1 and the other components in the absence of DNA (data not shown). The numerous yeast two-hybrid screens performed in budding yeast have also failed to identify such interactions. Another possibility is that the MRX, Sae2, and Exo1 proteins form complexes only in the context of a DSB. To test this hypothesis, a 32[P]-labeled DNA oligonucleotide duplex with 3' overhangs was incubated with Exo1, Sae2, and MRX and a gel mobility shift assay was performed (Fig. 6A). At protein concentrations similar to those used in the resection assay, Sae2 forms one predominant protein-DNA complex (lane 2), MRX shifts a small amount of the DNA into a large complex that is stimulated by Sae2 (lanes 3 and 7), while Exo1 by itself inefficiently forms a complex that does not enter the well (lane 6). (The D173A mutant of Exo1 is used in this experiment to prevent degradation of the DNA during the assay). In contrast we found that, in the presence of all the proteins, nearly 100% of the DNA is bound, suggesting that there is cooperative binding of the DNA when MRX, Sae2, and Exo1 are all present. This cooperative binding correlates well with the dramatic stimulation in Exo1-mediated resection observed with the plasmid DNA substrates.
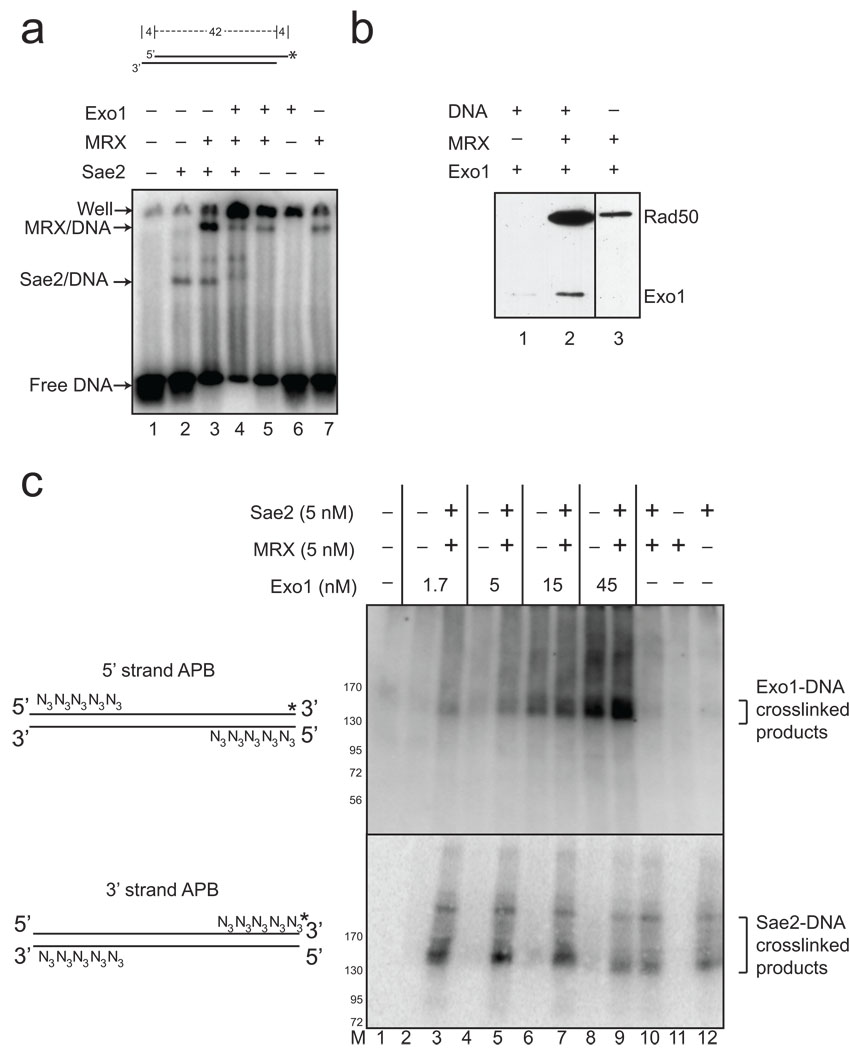
Figure 6. MRX and Sae2 promote Exo1 DNA binding.
(a) Gel mobility shift assays were performed with wild-type MRX (2.5 nM), Sae2 (2.5 nM), and Exo1 D173A (4 nM) proteins as indicated and a 32[P]-labeled, double-stranded oligonucleotide substrate containing 4 nt 3’ overhangs on both ends. Reactions were incubated for 15 min on ice before separation on a native acrylamide gel. (b) MRX and Exo1 D173A proteins were incubated with biotinylated, blunt 100 bp duplex DNA as indicated, crosslinked with formaldehyde, and proteins bound to the DNA were isolated using streptavidin-coated magnetic beads. Bound protein were visualized by SDS-PAGE and western blotting with anti-Flag antibody for Exo1 and Rad50. (c) Protein-DNA binding assays were performed with a 90 bp blunt DNA substrate, containing 5 azide groups (N3) on the 5' ends of the 5' strands or the 3' ends of the 3' strands as shown in the diagram. Both DNA substrates were labeled with 32[P] ("*"). Proteins were incubated with the DNA substrates on ice, UV irradiated, separated by SDS-PAGE and transferred to a PVDF membrane to remove all uncrosslinked DNA before phosphorimager analysis. Migration of molecular weight markers in the gels are shown in the lane marked “M”.
To specifically examine the binding of Exo1 to DNA and its recruitment by MRX, we also performed a pull-down assay in which MRX and Exo1 were incubated together with biotinylated DNA, the DNA molecules were isolated with streptavidin beads, and then bound proteins were visualized by SDS-PAGE and western blotting with anti-Flag antibody to visualize both Rad50 and Exo1. As shown in Fig. 6B, considerably more Exo1 was bound to the DNA when MRX was also present in the reaction. Binding of MRX, Sae2, and Exo1 to DNA was also analyzed using Surface Plasmon Resonance (SPR) (Supplementary Fig. 5). In this assay, MRX, Exo1, and Sae2 at 2 nM concentrations bound with nearly equivalent affinity to the DNA on the surface, but Exo1 and MRX together showed faster association and slower dissociation, consistent with the formation of a higher affinity complex.
As a final test of protein-DNA binding, we used strand-specific crosslinking to determine where on the DNA end each component is associating. In this assay, azidophenacylbromide (APB) is used to modify DNA containing phosphorothioates on the ends of either the 5' strand or the 3' strand with azido groups, which can be crosslinked to proteins using UV radiation 30. The 32[P]-labeled DNA substrate is incubated with the proteins on ice, crosslinked with UV, separated by denaturing SDS-PAGE, and transferred to a PVDF membrane. During this process the free labeled DNA is removed, while DNA that is covalently attached to protein appears on the membrane in a protein-DNA complex. The results show that, at high levels, Exo1 crosslinks to the 5' strand (Fig. 6C, lane 8, top). However, crosslinking of Exo1 to the 5' strand is strongly enhanced by the inclusion of MRX and Sae2 in the reaction (lanes 2 to 9, top), while neither MRX nor Sae2 form distinct crosslinking products with the 5' azido-conjugated DNA under these conditions (lanes 10 to 12, top). In contrast, the DNA substrate with azido groups on the 3' strand is specifically crosslinked to Sae2 independently of MRX or Exo1 in the reaction (lane 12, bottom). Based on these results, we postulate that MRX partially opens a DSB into a branched structure, which could be bound by both Sae2 on the 3' strand and Exo1 on the 5' strand. In support of this model, we found that Exo1 shows a higher affinity for a branched end structure relative to double-stranded linear DNA ends in gel mobility shift assays (Supplementary Fig. 6).
5' strand processing by MRX and Sae2 promotes Exo1 activity
Here we have demonstrated a recruitment role for MRX and Sae2 in stabilizing Exo1 on DNA ends. However, it is also apparent in the resection analysis (Fig. 1C, 2C, and Fig. 3B) and in our previous characterization of Sae2 in vitro 27 that MRX and Sae2 together catalyze a weak but detectable processing of DNA ends that removes a short region of the 5' strand at a break. To determine the extent of the contribution of this processing event, we performed the resection assay in two stages, with MRX–Sae2 in the first reaction only, followed by precipitation of the DNA substrate and incubation with Exo1 in the second reaction. As shown in Fig. 7A, the resection of the plasmid DNA by Exo1 is more efficient in the second stage when the DNA was incubated with MRX and Sae2 in the first stage (compare lanes 2 and 6). This is likely due to the fact that MRX and Sae2 catalyze inefficient endonucleolytic cleavage of the 5' strand of DNA to produce oligonucleotide products (seen with the 5' end-labeled substrate in Fig. 3B, lane 12). The limited 5' strand removal occurring in the first stage reaction is likely promoting Exo1 activity during the second stage reaction. To test this idea, a unrelated, distributive 5' to 3' exonuclease (T7 exonuclease)31 was used in the first stage reaction instead of MRX and Sae2, using levels of enzyme that generate approximately the same amount of digestion as MRX–Sae2 based on the non-denaturing southern assay (data not shown). When this pre-resected DNA was subsequently used as a substrate for Exo1, we also observed increased Exo1 activity in the second stage reaction (Fig. 7B, compare lanes 2 and 6), confirming that limited 5' strand resection can promote further 5' strand resection by Exo1. However, even with the pre-resected substrate, there is still a dramatic effect of MRX–Sae2 on Exo1 (Fig. 7B, lane 7), thus the recruitment of Exo1 by MRX–Sae2 still plays an important role even when the DNA is already partially processed. This result is also consistent with our observation that Exo1-mediated resection is more efficient on linear substrates containing 3' overhangs in comparison to substrates containing 5' overhangs (Supplementary Fig. 7).
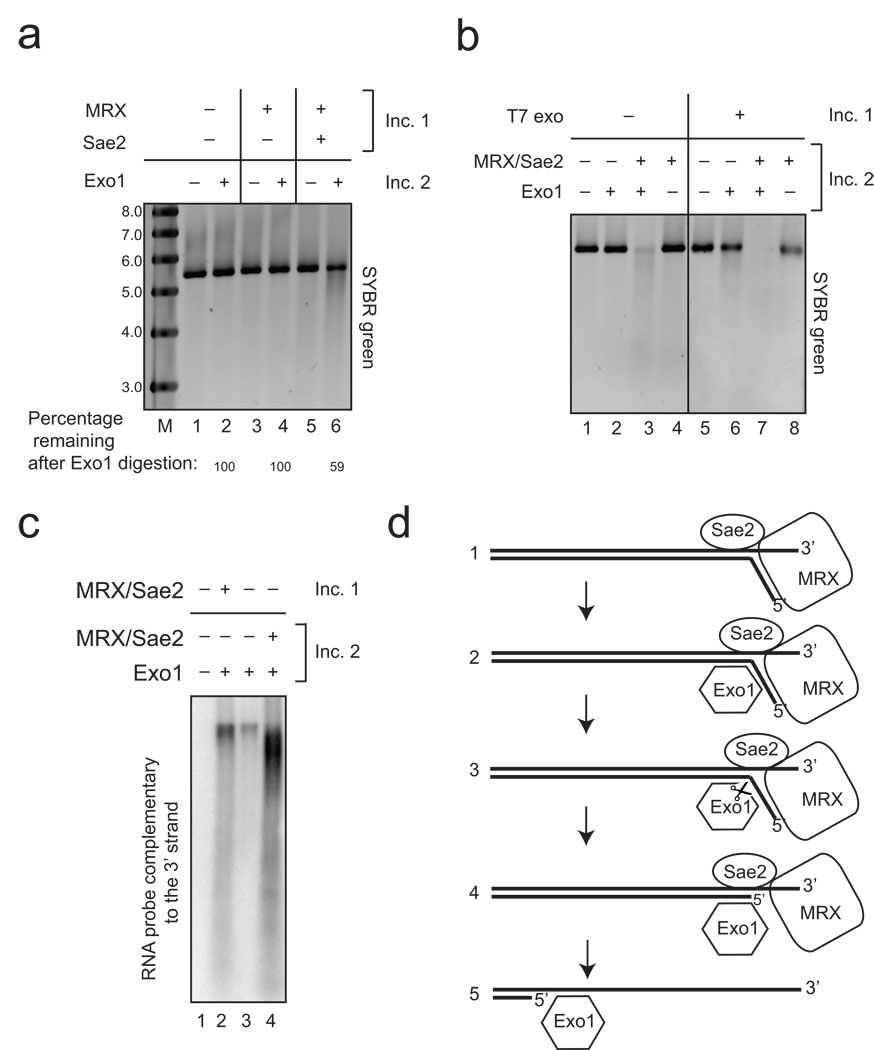
Figure 7. MRX and Sae2 facilitate Exo1 activity through distinct "processing" and “recruitment” pathways.
The resection reaction was performed as in Fig. 1A using pTP407 plasmid DNA linearized with BseRI except that the reaction was done in two stages. The first incubation (“Inc. 1”), included MRX (50 nM) and Sae2 (26 nM) as indicated but the DNA was deproteinized with SDS and proteinase K, followed by ethanol precipitation. The DNA from each reaction was incubated in a second reaction (“Inc. 2”) that included Exo1 (1 nM) as indicated. Reactions were separated by native agarose gel electrophoresis and DNA stained with SYBR green. Lane marked “M” contains molecular weight markers with sizes in kb as indicated. (b). 2-stage reactions were performed as in (a) except that T7 exonuclease (1 unit) was used in the first incubation and MRX (5 nM) and Sae2 (5 nM) were also added in the second incubation as indicated. (c) 2-stage reactions were performed as in (b) except with pNO1 plasmid DNA linearized with Sph1 and analyzed by non-denaturing southern hybridization using a probe specific for the 3' strand as in Fig. 1B. Reactions included MRX (14 nM), Sae2 (3.5 nM), and Exo1 (4 nM) as indicated. Lane marked “M” shows migration of molecular weight markers with sizes in kb as indicated. (d) Working model for association and cleavage of DNA ends by MRX, Sae2, and Exo1. We hypothesize a DNA-unwinding step followed by inefficient MRX–Sae2 cleavage, Exo1 recruitment, and further excision catalyzed by Exo1 (see text for details).
Another version of the 2-stage experiment is shown in Fig. 7C, in which the extent of resection was monitored using the southern assay and compared to the normal coupled reaction and a reaction in which Exo1 was present alone. The results indicate that resection by Exo1 alone is ~3-fold more efficient when MRX–Sae2 were present in the first incubation (Fig. 7C, compare lanes 2 and 3). However, this split reaction is still several-fold less efficient than the combined reaction (lane 2 vs lane 4). Most likely the combined reaction benefits from the recruitment of Exo1 by MRX that we have demonstrated, yet there is still a discernible effect of MRX–Sae2 that is distinct from recruitment which is likely the effect of 5' end processing by MRX and Sae2.
Recent studies with yeast Dna2 and the Sgs1–RmiI–Top3 complex also showed a stimulatory effect of MRX on 5' strand resection 32,33. With the Dna2–Sgs1–RmiI–Top3 reaction, the presence of yeast RPA was critical for maximal efficiency of resection as RPA increased the rate of DNA unwinding by Sgs1 and specifically protected the 3' single-strands from degradation by Dna2. With yeast Exo1 we found that RPA is inhibitory to its activity in vitro, although we observed that MRX and Sae2 can partially restore its activity in the presence of RPA (Supplementary Fig. 8).
Discussion
In this study we have used purified, eukaryotic proteins to reconstitute 5’ strand resection of DNA DSBs in vitro. Our results show that removal of the 5’ strand of DNA is catalyzed by Exo1, yet its activity is strongly dependent on MRX and Sae2. MRX–Sae2 dramatically stimulate Exo1 activity (60 to 300-fold) when Exo1 is present in limiting concentrations, less than ~6 nM. Above this level, Exo1 exhibits MRX–Sae2-independent activity; however, the extent of resection is still markedly increased by MRX and Sae2. The ability of Exo1 to function in the absence of MRX is consistent with the observation that overexpression of Exo1 in yeast partially suppresses the DNA damage sensitivity of strains lacking MRX 13,34−37.
The concentration dependence of Exo1 stimulation suggests that MRX and Sae2 affect the recruitment of Exo1. Consistent with this idea, we find that Exo1, MRX, and Sae2 each bind a short DNA duplex minimally in a gel mobility shift assay but the presence of all three complexes results in nearly 100% binding of the labeled DNA (Fig. 6). Similar cooperativity between Exo1 and MRX was observed using SPR (Supplementary Fig. 5), and Exo1 binding to DNA was stimulated by MRX in pull-down assays. Lastly we showed MRX–Sae2 stimulation of Exo1-DNA binding, specifically at the 5' strand at a break, using UV crosslinking (Fig. 6). Taken together, these results strongly suggest that the major effect of MRX on Exo1 activity is through an increase in the affinity of Exo1 for DNA.
In the absence of any evidence for protein-protein interactions, we postulate that MRX may create a specific DNA structure that results in a higher-affinity binding site for Exo1 (Fig. 7D). Based on previous work with human MRN, we think that a likely possibility for such a structure is an unwound end. We previously showed that MRN can dissociate a short (15 nt) oligonucleotide from a complementary DNA strand in an ATP-stimulated manner 38, and that unwinding of a DNA end is important in MRN-dependent ATM activation 39. DNA unwinding is also an attractive idea because Sae2 and Exo1 both can cleave 5’ flaps in branched DNA 27,28. We also show in this work that Exo1 preferentially binds to a branched DNA structure (Supplementary Fig. 6), and that Exo1 generates oligonucleotide products from the 5' strands of a DSB (Fig. 3). There is no indication from genetic experiments in budding yeast that Exo1 acts with Sgs1 or any other helicase in vivo 23,24, although human Exo1 was shown to interact physically and functionally with BLM helicase, a relative of Sgs1 in the RecQ family 40.
We also uncovered evidence for a pathway of Exo1 stimulation involving DNA end processing. Endonucleolytic cleavage of the 5' strand occurs in the presence of MRX and Sae2 alone, and this limited resectin of the DNA stimulates Exo1 by ~3-fold in comparison to Exo1 activity on unprocessed DNA. An unrelated 5' exonuclease can be used in place of MRX–Sae2 for this processing (Fig. 7). However, in the context of the coupled reaction where recruitment of Exo1 is taking place, the processing pathway does not play a major role since we find that resection stimulated by MRX complexes containing nuclease-deficient Mre11 (mre11-H125N, 20) is ~80% of that seen with wild-type MRX. This result is very similar to our observations with nuclease-deficient P. furiosus Mre11 in a reaction with archaeal components 26. We postulate that the 5' end processing by MRX–Sae2 is likely to play a much more important role in situations where the 5' strand is blocked, as in the case of Spo11 conjugates or other polypeptide or chemical adducts 20,21. Mre11 nuclease activity is similarly essential for Spo11 removal in fission yeast 41−43, but also plays an important role in the repair of covalent topoisomerase I and II complexes from DNA 44 and is essential for embryonic development in the mouse 45.
In contrast to the Mre11 nuclease domain mutations, the K81I (Rad50S) mutation caused a more severe reduction in resection, with only 25 to 30% of products observed compared to that seen with wild-type MRX. This result agrees well with published data showing that the kinetics of DSB resection and single-strand annealing are delayed in vivo in a strain expressing the rad50S mutant 15. rad50S strains share the same phenotype as Δsae2 strains in meiosis, where Spo11-induced DSBs are made but not resected 11. Our results with the K81I Rad50S complex suggest that the mutation may cause a defect in the protein-DNA structure formed by MRX on the DNA end, since the ability of the mutant to stimulate Exo1 is reduced in comparison to wild-type MRX, even in the absence of Sae2.
Based on this evidence we propose a working model for the initiation of resection (Fig. 7D). MRX and Sae2 recognize and bind stably to the DSB site, concomitant with MRX-dependent unwinding of the DNA strand, which facilitates higher-affinity binding of Exo1 to the DSB end. We have observed Exo1 endonucleolytic activity on the 5' ends of DNA substrates that is stimulated by MRX–Sae2 (Fig. 3B), but there is also extensive degradation of the duplex via exonucleolytic activity, generating single nucleotide products (Fig. 3A). We envision that Exo1 may bind initially at a branched structure, make an endonucleolytic cut, followed by exonucleolytic cleavage events. We have also observed Exo1-generated oligonucleotide products internal to the DNA ends (data not shown), suggesting that endonucleolytic cuts may occur away from the initial 5' end. MRX–Sae2 can also inefficiently cleave the 5' strand to generate oligonucleotide products, an activity that also promotes Exo1 activity. Based on immunofluorescence and ChIP studies we postulate that MRX–Sae2 dissociates after stable binding of Exo1 46.
The resection reactions we have reconstituted with archaeal and budding yeast components show that Mre11–Rad50 complexes are important for efficient removal of the 5' strand at a DSB, even though the resection is carried out by other enzymes. In eukaryotes, one reason for this role of MRN–X is clearly to incorporate activation of ATM(Tel1) signaling during the earliest stages of DNA double-strand break repair. MRN(X) recruits and activates ATM(Tel1) at DSB sites, and the branched DNA structure we are proposing here may be the same structure that stimulates ATM activity. The successful establishment of this in vitro system suggests that it may be possible to dissect these important functions of prokaryotic and eukaryotic Mre11–Rad50 complexes using model DNA substrates and purified protein factors to clarify the molecular roles of this multi-functional enzyme and other factors involved in DSB resection in eukaryotic cells.
Methods
Expression Constructs
Plasmids and bacmids expressing MBP-Sae2 and components of MRX were described previously 27. Baculovirus transfer vectors containing the yeast Exo1 gene with a C-terminal Flag epitope tag, wild-type and D173A, were gifts from Michael Liskay 28.
Protein Expression and Purification
Recombinant yeast Mre11–Rad50–Xrs2 complexes were co-expressed in Sf21 insect cells (Supplementary Fig. 1) and recombinant MBP-Sae2 was expressed in E. coli. as previously described 27. Monomeric Sae2 was used for all of the experiments shown here. Exo1 was expressed using the Bac-to-Bac system (Invitrogen) in Sf21 cells. For details of purification, see Supplemental Methods, Purification of yeast Exo1b.
Resection assays
Resection assays with plasmid DNA substrates were performed with either pTP407 (a 5.6 kb derivative of pRS314) linearized with BseRI, which generates 2 nt 3' overhangs (Fig. 1A; 7A and B) or with pNO1 (a 4.4 kb derivative of pBR322) linearized with SphI, which generates 4 nt 3' overhangs (Fig. 1B, C, D; Fig. 2; Fig. 4; Fig. 5A; Fig. 7C). Resection reactions contained 10 ng (0.30 nM pTP407 or 0.35 nM pNO1) plasmid DNA, 25 mM MOPS pH 7.0, 50 mM NaCl, and 1 mM DTT. Some reactions contained 10 mM MgCl2, and 1 mM ATP (Fig. 1B, C, D, Fig. 2, Fig. 4, Fig. 5A, and Fig. 7C). Others contained 5 mM MgCl2, and 0.5 mM ATP (Fig. 1A, Fig. 3, and Fig. 7A, B). There were no differences in activity between the 10 mM MgCl2 conditions and the 5 mM MgCl2 conditions. All reactions were performed in a volume of 10 µl and were incubated at 30°C for 1 hr unless indicated otherwise. Reactions were stopped by the addition of 0.2% SDS and 10 mM EDTA, incubated with 1 µg proteinase K at 37°C for 15 to 30 min., analyzed on 0.8% native agarose gels run in 1X Tris-acetate-EDTA (TAE; 40 mM Tris-acetate, 1 mM EDTA) buffer, and visualized with SYBR green (Invitrogen). Resection assays with pNO1 were further analyzed by non-denaturing Southern hybridization, in which agarose gels were washed into 20× SSC (3 M NaCl, 0.3 M sodium citrate) and DNA was transferred by capillary action onto nylon membranes (NEN) overnight in 20× SSC. Membranes were probed with RNA complementary to either the 3' or 5' strand of the DNA substrate in a 1 kb region on one end, adjacent to the SphI site. The probes were internally labeled with [α-32P]CTP (NEN) and were made using Riboprobe System T7 (Promega) and purified with RNAeasy extraction (Qiagen) according to the manufacturer's instructions. Denatured DNA controls consisted of substrate DNA denatured with NaOH.
Resection reactions with the 700 bp internally labeled substrate (Fig. 3A) were identical to the standard reactions with 5 mM MgCl2 and 0.5 mM ATP (above) except that they utilized a [32P]-labeled DNA substrate made by PCR. Resection reactions were stopped by the addition of 0.2% SDS and 10 mM EDTA and analyzed by TLC as described previously 47. The Cy5-labeled 700 bp DNA substrate (Fig. 3B) was also made by PCR; reactions contained 10.8 nM DNA with 25 mM MOPS pH 7.0, 2 mM DTT, 5 mM MgCl2, 0.5 mM ATP, 60 mM NaCl, with protein concentrations as described in the figure legends, in a total volume of 100 µL. Reactions were performed at 30°C for 30 min., and were stopped with 0.2% SDS and 10 mM EDTA before ethanol precipitation. Precipitated reactions were resuspended in formamide and separated on 20% acrylamide sequencing gels before analysis using a Typhoon Imager (GE). For PCR reaction conditions, see Supplemental Methods.
Quantitative PCR
See Supplemental Methods.
Chromatin Immunoprecipitation
See Supplemental Methods.
Gel Mobility Shift Assay
See Supplemental Methods.
Pull-down Assay
Recombinant Exo1 D173A (7.5 nM) and MRX (15.4 nM) were incubated on ice with 4 nM double-stranded DNA consisting of biotinylated TP2399 annealed to TP2400 (see Supplementary Materials for sequences). TP2399 was labeled on the 3' end with α-[32P]-cordycepin using terminal deoxytransferase (Roche). The reaction volume was 10 µl and included 25 mM MOPS pH 7.0, 60 mM NaCl, 1 mM DTT, 5 mM MgCl2, and 0.5 mM ATP. Reactions were incubated for 15 min. before the addition of 0.74% formaldehyde. After 10 min. crosslinking on ice, 100 mM Tris pH 8.0 was added, followed by 5 µl Dynal streptavidin-coated magnetic beads (Invitrogen) which had previously been diluted 20-fold and washed into PBS. The reactions were rotated at 4°C for 1 hr, the beads were separated using a magnetic stand and washed with 50 µl wash buffer (25 mM MOPS pH 7.0, 50 mM NaCl, 0.1% CHAPS) 3 times, and eluted with 1X SDS-PAGE loading buffer. After separation in a 6% acrylamide gel, the proteins were transferred to a PVDF membrane and probed with anti-Flag antibody (which recognizes both Exo1 and Rad50), and visualized using a HRP-conjugated anti-mouse secondary antibody by chemiluminescence (Pierce).
Strand-specific Crosslinking
Recombinant Exo1 D173A (1.7, 5, 15, or 45 nM), MRX (5 nM), or Sae2 (5 nM) were incubated on ice with 10 nM 90 bp double-stranded DNA that was conjugated with 5 azido groups on either the 5' end of the 5' strands (TP2722/TP2723) or the 3' end of the 3' strands (TP771/TP828) at phosphorothioate linkages. Modification of the oligonucleotides with azidophenacylbromide was performed as described previously 30. TP771 and TP2722 were labeled on the 3' end with α-[32P]-cordycepin using terminal deoxytransferase (Roche). The reaction volume was 10 µl and included 25 mM MOPS pH 7.0, 60 mM NaCl, 1 mM DTT, 5 mM MgCl2, and 0.5 mM ATP. Reactions were incubated for 15 min. on ice before UV crosslinking on ice for 5 min. using a 254 nm handheld UV lamp. Reactions were stopped with the addition of 1X SDS-PAGE loading buffer; the reactions were separated with a 6% acrylamide gel, transferred to a PVDF membrane, and analyzed by phosphorimager (GE).
Supplementary Material
Acknowledgments
We are grateful to Michael Liskay (Oregon Health & Science University) for yeast Exo1 reagents, to Steve Brill (Rutgers University) for RPA antibodies, and Marc Wold (University of Iowa) for RPA constructs, and to members of the Paull lab for critical comments about this project. This work was supported by NIH grant R01 CA094008 to T.T.P. Studies performed in the Lee laboratory were supported by R01 GM083010, and S.E.L. is a scholar of the Leukemia and Lymphoma Society.
References
- 1.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 2.Aylon Y, Liefshitz B, Kupiec M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 2004;23:4868–4875. doi: 10.1038/sj.emboj.7600469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ira G, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wyman C, Kanaar R. DNA double-strand break repair: all's well that ends well. Annu Rev Genet. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- 5.Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 6.Vaze MB, et al. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol Cell. 2002;10:373–385. doi: 10.1016/s1097-2765(02)00593-2. [DOI] [PubMed] [Google Scholar]
- 7.Fishman-Lobell J, Rudin N, Haber JE. Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol Cell Biol. 1992;12:1292–1303. doi: 10.1128/mcb.12.3.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zierhut C, Diffley JF. Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. EMBO J. 2008;27:1875–1885. doi: 10.1038/emboj.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung WH, Zhu Z, Papusha A, Malkova A, Ira G. Defective resection at DNA double-strand breaks leads to de novo telomere formation and enhances gene targeting. PLoS Genet. 6:e1000948. doi: 10.1371/journal.pgen.1000948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barlow JH, Lisby M, Rothstein R. Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol Cell. 2008;30:73–85. doi: 10.1016/j.molcel.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- 12.Nairz K, Klein F. mre11S--a yeast mutation that blocks double-strand-break processing and permits nonhomologous synapsis in meiosis. Genes & Dev. 1997;11:2272–2290. doi: 10.1101/gad.11.17.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsubouchi H, Ogawa H. A novel mre11 mutation impairs processing of double-strand breaks of DNA during both mitosis and meiosis. Mol. Cell. Biol. 1998;18:260–268. doi: 10.1128/mcb.18.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivanov EL, Sugawara N, White CI, Fabre F, Haber JE. Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol. Cell. Biol. 1994;14:3414–3425. doi: 10.1128/mcb.14.5.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clerici M, Mantiero D, Lucchini G, Longhese MP. The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J Biol Chem. 2005;280:38631–38638. doi: 10.1074/jbc.M508339200. [DOI] [PubMed] [Google Scholar]
- 16.Paull TT, Gellert M. The 3' to 5' exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 17.Connelly JC, de Leau ES, Leach DRF. DNA cleavage and degradation by the SbcCD protein complex from escherichia coli. Nucleic Acids Res. 1999;27:1039–1046. doi: 10.1093/nar/27.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trujillo KM, Sung P. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50/Mre11 complex. J Biol Chem. 2001;13:13. doi: 10.1074/jbc.M105482200. [DOI] [PubMed] [Google Scholar]
- 19.Hopfner KP, et al. Mre11 and rad50 from pyrococcus furiosus: cloning and biochemical characterization reveal an evolutionarily conserved multiprotein machine. J Bacteriol. 2000;182:6036–6041. doi: 10.1128/jb.182.21.6036-6041.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moreau S, Ferguson JR, Symington LS. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol. 1999;19:556–566. doi: 10.1128/mcb.19.1.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Llorente B, Symington LS. The Mre11 nuclease is not required for 5' to 3' resection at multiple HO-induced double-strand breaks. Mol Cell Biol. 2004;24:9682–9694. doi: 10.1128/MCB.24.21.9682-9694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Usui T, et al. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell. 1998;95:705–716. doi: 10.1016/s0092-8674(00)81640-2. [DOI] [PubMed] [Google Scholar]
- 23.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Budd ME, Campbell JL. Interplay of Mre11 nuclease with Dna2 plus Sgs1 in Rad51-dependent recombinational repair. PLoS ONE. 2009;4:e4267. doi: 10.1371/journal.pone.0004267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hopkins B, Paull TT, The P. furiosus Mre11/Rad50 complex promotes 5' strand resection at a DNA double-strand break. Cell. 2008;135:250–260. doi: 10.1016/j.cell.2008.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT. Sae2 Is an Endonuclease that Processes Hairpin DNA Cooperatively with the Mre11/Rad50/Xrs2 Complex. Mol Cell. 2007;28:638–651. doi: 10.1016/j.molcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tran PT, Erdeniz N, Dudley S, Liskay RM. Characterization of nuclease-dependent functions of Exo1p in Saccharomyces cerevisiae. DNA Repair (Amst) 2002;1:895–912. doi: 10.1016/s1568-7864(02)00114-3. [DOI] [PubMed] [Google Scholar]
- 29.Trujillo KM, Yuan SS, Lee EY, Sung P. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J. Biol. Chem. 1998;273:21447–21450. doi: 10.1074/jbc.273.34.21447. [DOI] [PubMed] [Google Scholar]
- 30.Yang SW, Nash HA. Specific photocrosslinking of DNA-protein complexes: identification of contacts between integration host factor and its target DNA. Proc Natl Acad Sci U S A. 1994;91:12183–12187. doi: 10.1073/pnas.91.25.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas KR, Olivera BM. Processivity of DNA exonucleases. J Biol Chem. 1978;253:424–429. [PubMed] [Google Scholar]
- 32.Niu H, et al. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 467:108–111. doi: 10.1038/nature09318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cejka P, et al. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 467:112–116. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chamankhah M, Fontanie T, Xiao W. The Saccharomyces cerevisiae mre11(ts) allele confers a separation of DNA repair and telomere maintenance functions. Genetics. 2000;155:569–576. doi: 10.1093/genetics/155.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moreau S, Morgan EA, Symington LS. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics. 2001;159:1423–1433. doi: 10.1093/genetics/159.4.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewis LK, Karthikeyan G, Westmoreland JW, Resnick MA. Differential suppression of DNA repair deficiencies of Yeast rad50, mre11 and xrs2 mutants by EXO1 and TLC1 (the RNA component of telomerase) Genetics. 2002;160:49–62. doi: 10.1093/genetics/160.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee SE, Bressan DA, Petrini JH, Haber JE. Complementation between N-terminal Saccharomyces cerevisiae mre11 alleles in DNA repair and telomere length maintenance. DNA Repair (Amst) 2002;1:27–40. doi: 10.1016/s1568-7864(01)00003-9. [DOI] [PubMed] [Google Scholar]
- 38.Paull TT, Gellert M. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes & Dev. 1999;13:1276–1288. doi: 10.1101/gad.13.10.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 40.Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc Natl Acad Sci U S A. 2008;105:16906–16911. doi: 10.1073/pnas.0809380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Milman N, Higuchi E, Smith GR. Meiotic DNA double-strand break repair requires two nucleases, MRN and Ctp1, to produce a single size class of Rec12 (Spo11)-oligonucleotide complexes. Mol Cell Biol. 2009;29:5998–6005. doi: 10.1128/MCB.01127-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hartsuiker E, et al. Ctp1CtIP and Rad32Mre11 nuclease activity are required for Rec12Spo11 removal, but Rec12Spo11 removal is dispensable for other MRN-dependent meiotic functions. Mol Cell Biol. 2009;29:1671–1681. doi: 10.1128/MCB.01182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rothenberg M, Kohli J, Ludin K. Ctp1 and the MRN-complex are required for endonucleolytic Rec12 removal with release of a single class of oligonucleotides in fission yeast. PLoS Genet. 2009;5:e1000722. doi: 10.1371/journal.pgen.1000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hartsuiker E, Neale MJ, Carr AM. Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol Cell. 2009;33:117–123. doi: 10.1016/j.molcel.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buis J, et al. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 47.Bhaskara V, et al. Rad50 Adenylate Kinase Activity Regulates DNA Tethering by Mre11/Rad50 complexes. Molecular Cell. 2007;25:647–661. doi: 10.1016/j.molcel.2007.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee K, Zhang Y, Lee SE. Saccharomyces cerevisiae ATM orthologue suppresses break-induced chromosome translocations. Nature. 2008;454:543–546. doi: 10.1038/nature07054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.