Abstract
Accurate determination of protein phosphorylation is challenging, particularly for researchers who lack access to a high-accuracy mass spectrometer. In this study, multiple protocols were used to enrich phosphopeptides, and a rigorous filtering workflow was used to analyze the resulting samples. Phosphopeptides were enriched from cultured rat renal proximal tubule cells using three commonly used protocols and a dual method that combines separate immobilized metal affinity chromatography (IMAC) and titanium dioxide (TiO2) chromatography, termed dual IMAC (DIMAC). Phosphopeptides from all four enrichment strategies were analyzed by liquid chromatography-multiple levels of mass spectrometry (LC-MSn) neutral-loss scanning using a linear ion trap mass spectrometer. Initially, the resulting MS2 and MS3 spectra were analyzed using PeptideProphet and database search engine thresholds that produced a false discovery rate (FDR) of <1.5% when searched against a reverse database. However, only 40% of the potential phosphopeptides were confirmed by manual validation. The combined analyses yielded 110 confidently identified phosphopeptides. Using less-stringent initial filtering thresholds (FDR of 7–9%), followed by rigorous manual validation, 262 unique phosphopeptides, including 111 novel phosphorylation sites, were identified confidently. Thus, traditional methods of data filtering within widely accepted FDRs were inadequate for the analysis of low-resolution phosphopeptide spectra. However, the combination of a streamlined front-end enrichment strategy and rigorous manual spectral validation allowed for confident phosphopeptide identifications from a complex sample using a low-resolution ion trap mass spectrometer.
Keywords: phosphoproteomics, DIMAC enrichment, LC-MSn, kidney proximal tubule, Wistar rat kidney proximal tubule cells
INTRODUCTION
Reversible protein phosphorylation is an essential component of numerous cellular regulatory processes.1,2 In eukaryotic organisms, protein phosphorylation occurs on serine, threonine, and tyrosine residues. Genomic sequence analysis has uncovered approximately 500 eukaryotic genes that encode protein kinases3 and more than 100 that encode protein phosphatases,4 underscoring the ubiquitous role of protein phosphorylation within the cell. Mass spectrometry (MS) has proven to be a useful method to determine the sites of protein phosphorylation.5 Recent advances in MS have resulted in significant improvements in the efficiency and accuracy of phosphopeptide analysis.6,7 For example, neutral-loss scanning can be applied with collision-induced dissociation (CID) to identify phosphopeptides based on a predominant neutral loss of phosphoric acid (H3PO4) in the MS2 spectrum. The loss of H3PO4 triggers an additional level of fragmentation, MS3, which usually produces increased backbone fragmentation for improved peptide identification.8
However, several challenges still remain. In particular, the amount and extent of protein phosphorylation within the cell are relatively low.5,9,10 Furthermore, the detection of phosphopeptides by MS is difficult as a result of low ionization efficiencies.11,12 This limitation can be overcome by the use of phosphopeptide enrichment methods. The commonly used phosphopeptide enrichment methods include immobilized metal affinity chromatography (IMAC),13–19 metal oxide affinity chromatography (MOAC),20–22 and sequential elution from IMAC (SIMAC),23 which is a recently introduced method that combines the strengths of IMAC and MOAC into a single combinatorial enrichment. Briefly, these enrichment methods use charge-charge interactions between a stationary phase consisting of positively charged metal species and a mobile phase consisting of negatively charged phosphopeptides.7
In the current study, phosphopeptides, isolated from cultured rat renal proximal tubule cells, were enriched using a streamlined phosphopeptide enrichment strategy termed dual IMAC (DIMAC). The DIMAC enrichment protocol involved parallel isolation of phosphopeptides from separate IMAC and titanium dioxide (TiO2) enrichments, resulting in two samples, which were injected separately but analyzed as a single enrichment method. The enrichment achieved by the DIMAC protocol was compared with three other commonly used protocols. The resulting phosphopeptides were analyzed using liquid chromatography-multiple levels of MS (LC-MSn) neutral-loss scanning on a low-mass accuracy linear ion trap mass spectrometer. The initial data analysis was performed using ProteinProphet and database search engine thresholds that were set to yield a false discovery rate (FDR) of <1.5%. However, only 40% of the potential phosphopeptides identified were confirmed by manual validation. Using less-stringent initial filtering criteria (7–9% FDR) followed by manual validation increased the number of confidently identified phosphopeptides from 110 to 262. The latter group included 111 novel phosphorylation sites. The overall results highlight the importance of manual validation of phosphopeptides, particularly when working with a low-mass accuracy instrument.
MATERIALS AND METHODS
Reagents
Halt protease inhibitor and Halt phosphatase inhibitor cocktails were obtained from Pierce (Rockford, IL, USA). Protein assay dye and sequencing-grade-modified trypsin were purchased from Bio-Rad (Hercules, CA, USA) and Promega (Madison, WI, USA), respectively. Sep-Pak Light C18 cartridges were obtained from Waters Corp. (Milford, MA, USA). The Gallium (Ga+3)-IMAC and TiO2 TopTips were from Glygen (Columbia, MD, USA). The PepClean C18 spin columns were from Thermo Fisher Scientific (Waltham, MA, USA). The Zorbax 300SB-C18 trap column (5 μm, 5.0×0.3 mm) and the PicoFrit BioBasic C-18 analytical column were purchased from Agilent Technologies (Santa Clara, CA, USA) and New Objective (Woburn, MA, USA), respectively.
Sample Preparation
Wistar rat kidney proximal tubule cells24 were grown on 10 cm plates in a 50:50 mixture of DMEM and Hamm's F12 medium containing 10% FCS until ∼70% confluent. Cells were washed twice with 5 ml HEPES-buffered saline and harvested by scraping in 250 μl lysis buffer containing 150 mM NaCl, 8 M urea, 10% Halt protease and phosphatase inhibitors, and 10 mM HEPES, pH 7.4. The cells were lysed by sonicating five times, each with 10 1-s pulses (50% duty cycle, output 3). Cellular debris was removed by centrifugation at 10,000 rpm for 15 min at 4°C. The protein concentration of the supernatant was determined with a Bradford assay.25 Samples containing 6 mg protein were reduced by incubating with 5 mM DTT for 1 h at 37°C and then alkylated by incubating with 14 mM iodoacetamide for 1 h at 37°C. The urea concentration was decreased to 1.6 M by addition of 100 mM ammonium bicarbonate, and the samples were digested overnight with trypsin (1:50 w/w) at 37°C. The tryptic peptides were dried, resuspended in 3% acetonitrile/0.1% trifluroacetic acid, and desalted on a Sep-Pak Light C18 cartridge.
DIMAC Enrichment
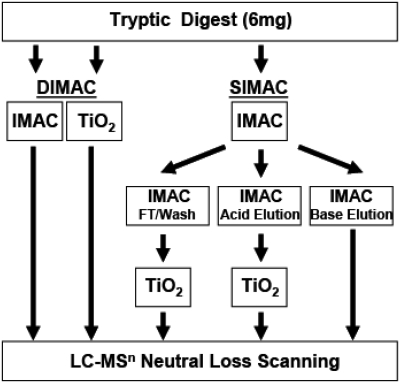
The DIMAC enrichment procedure (Fig. 1) used dual IMAC and TiO2 enrichment protocols. The dried peptides from a 6-mg sample were resuspended in 50 μl 5% acetic acid, pH 2.5, and applied to an IMAC TopTip that was pre-equilibrated with 50 μl 5% acetic acid, pH 2.5. The column was washed with 50 μl 0.1% acetic acid, pH 2.5, and then with 50 μl 0.1% acetic acid/10% acetoniltrile, pH 3.0. The IMAC TopTip was then eluted with three 50-μl aliquots of 1 M ammonium bicarbonate/20% acetonitrile, pH 9.0. The combined fractions were acidified by adding 15 μl trifluroacetic acid, dried under vacuum, and saved for mass spectrometric analysis (IMAC elution). The dried peptides from a separate 6-mg sample were resuspended in 50 μl 5% trifluroacetic acid/80% acetonitrile/1 M lactic acid. The resuspended sample was applied to a TiO2 TopTip that was pre-equilibrated with 50 μl 5% trifluroacetic acid/80% acetonitrile/1 M lactic acid. The column was then washed with 50 μl 1% trifluoroacetic acid/80% acetonitrile/1 M lactic acid and 50 μl 1% trifluroacteic acid/80% acetonitrile. The TiO2 TopTip was then eluted with three 50-μl aliquots of 0.5% ammonium hydroxide. The combined fractions were acidified by adding 15 μl trifluroacetic acid, dried, and saved for mass spectrometric analysis (TiO2 elution). Postacquisition data for IMAC and TiO2 were combined into a single analysis.
FIGURE 1.
Comparison of the DIMAC and SIMAC phosphopeptide enrichment protocols. With the DIMAC protocol, two 6-mg samples of a tryptic digest were enriched simultaneously on separate IMAC and TiO2 columns. By contrast, the SIMAC protocol involved a more complex sequential fractionation of a single sample and produced three samples for mass spectrometric analysis. FT, Flow-through.
IMAC1 and IMAC2 Enrichment
The IMAC1 enrichment procedure was adapted from Villen and Gygi.26 Briefly, 6 mg desalted peptides were dried, resuspended in 50 μl 40% acetonitrile/25 mM formic acid, and bound to an IMAC TopTip. After loading, the TopTip was washed with 50 μl binding buffer. The bound phosphopeptides were then eluted with three 50-μl aliquots of 50 mM dipotassium phosphate, adjusted to pH 10 with ammonium hydroxide (NH4OH). The combined fractions were acidified with 1 μl formic acid/10 μl sample, dried under vacuum, and saved for further analysis (IMAC1 elution). The IMAC2 enrichment procedure was adapted from Lee et al.27 The second procedure was performed in the same manner as IMAC1, except the binding buffer contained a 1:1:1 mixture of acetonitrile:methanol:H2O in 0.1% acetic acid. The column was washed initially with 50 μl of a 75:10:14:1 mixture of acetonitrile:methanol:H2O:acetic acid containing 100 mM NaCl and then with 50 μl of an 85:14:1 mixture of acetonitrile:H2O:acetic acid. The bound phosphopeptides were subsequently eluted with three 50-μl aliquots of a 45:50:5 mixture of acetonitrile:H2O:trifluroacetic acid, dried under vacuum, and saved for analysis (IMAC2 elution).
SIMAC Enrichment
The SIMAC enrichment procedure was adapted from Thingholm et al.23 SIMAC enrichment was performed as outlined in Fig. 1. Briefly, peptides from a 6-mg sample were resuspended in 50 μl 0.1% trifluroacetic acid/50% acetonitrile and bound to an IMAC TopTip. The column was washed with an additional 50-μl 0.1% trifluroacetic acid/50% acetonitrile. The IMAC flow-through and wash fractions were combined, dried under vacuum, and saved for later TiO2 enrichment. An acid elution of the IMAC column was subsequently performed using 50 μl 1.0% trifluroacetic acid/20% acetonitrile. The eluted fraction was dried under vacuum and also saved for a separate TiO2 enrichment. Finally, the IMAC column was washed with 50 μl ammonia water (10 μl NH4OH, 490 μl H2O), and the eluant was acidified with 5 μl formic acid, dried under vacuum, and saved for mass spectrometric analysis (IMAC base elution). The dried samples derived from the combined flow-through/wash and the acid elution from the IMAC column were resuspended separately in a mixture containing 4 μl 4 M urea, 6 μl 1% SDS, and 40 μl 5% trifluroacteic acid/80% acetonitrile/1 M lactic acid. The resuspended samples were applied to separate TiO2 TopTips that were pre-equilibrated with 50 μl 5% trifluroacteic acid/80% acetonitrile/1 M lactic acid. The flow-through fractions from the TiO2 TopTips were discarded, and the columns were washed with 50 μl 1% trifluoroacetic acid/80% acetonitrile/1 M lactic acid and then with 50 μl 1% trifluoroacetic acid/80% acetonitrile. The TiO2 TopTips were then eluted with three 50-μl aliquots of ammonia water (10 μl NH4OH, 490 μl H2O). The three fractions from each TiO2 column were combined, acidified by adding 15 μl trifluroacetic acid, dried under vacuum, and saved for mass spectrometric analysis (IMAC-TiO2 flow-through/wash and IMAC-TiO2 acid elution).
Peptide Desalting
The dried phosphopeptide samples were resuspended in 200 μl 0.1% formic acid/3% acetonitrile and then loaded onto a PepClean C18 spin column that was initially wet with 200 μl 50% acetonitrile and equilibrated with 0.1% formic acid/3% acetonitrile. The column was washed twice with 200 μl 0.1% formic/3% acetonitrile and then eluted with three 20-μl aliquots of 90% acetonitrile. The combined samples were dried under vacuum.
LC-MSn Neutral-Loss Scanning
The dried samples were resuspended in 10 μl 3% acetoniltrile/0.1% formic acid, and 1 μl was injected onto the Zorbax 300SB-C18 trap and PicoFrit BioBasic C-18 analytical columns. The peptides were eluted directly into a LTQ-linear ion trap mass spectrometer (Thermo Fisher Scientific) using a 42-min linear gradient of 2–100% elution buffer (80% acetonitrile/0.1% formic acid) at a flow rate of 300 nl/min. Data-dependent MS3 neutral-loss scanning was triggered when the neutral loss of H3PO4 was detected as a decrease in mass/charge ratio (m/z) of 98, 49, or 32.7 Da among the three most intense fragment ions in the MS2 scan. The normalized energy for CID was optimized at 22% for MS2 and 35% for MS3 (data not shown). The electrospray voltage was set at 2 kV, and the voltage and temperature for the ion source capillary were set at 46 V and 200°C, respectively. Compound lists of the resulting spectra were generated using Bioworks 3.0 software.
Database Searches
The compound lists were searched against the forward and reverse rat International Protein Index Database (Rat_IPI.v.3.57) containing 39,873 sequence entries using Mascot (Matrix Science, Boston, MA, USA) and SEQUEST (Bioworks 3.0, Thermo Fisher Scientific) database search engines. The peptide mass tolerances and fragment ion mass tolerances for the database searches were 2.5 Da and 1.5 Da for Mascot and 2.0 Da and 1.0 Da for SEQUEST, respectively. Additional parameters were tryptic peptides allowing for two missed cleavages, a fixed modification for cysteine carbamidomethylation (+57 Da), and variable modifications for methionine oxidation (+16 Da), phosphorylation of serine, threonine, and tyrosine residues (+80 Da) and a loss of water (−18 Da) from serine and threonine resides for MS3 scans only. Separate searches were performed for the MS2 and MS3 data.
Scaffold Analysis
MS2 and MS3 database search results were uploaded separately into Scaffold (Proteome Software, Portland, OR, USA). Data were selectively filtered for phosphopeptides using the Scaffold phospho-modification tab for MS2 data and the phospho and dehydro tabs for MS3 data. The identified phosphopeptides were validated initially using 99.9% protein and 95% peptide probability thresholds, which produced a FDR of <1.5%. A less-stringent search was subsequently performed by adjusting both thresholds to 90%. Using these criteria, the search had a FDR of 7%. The original data sets were also filtered using thresholds for the separate Mascot and SEQUEST database searches. Mascot thresholds for MS2 and MS3 data were set initially at 53 and 70, respectively. SEQUEST thresholds for MS2 data were 2.8 and 3.8 and for MS3 data were 2.5 and 3.0 for +2 and +3 charged peptides, respectively. The stringent thresholds produced FDRs of <1.5% for the Mascot and SEQUEST data when searched against a reverse database.28 A subsequent Mascot search used less-stringent filtering criteria of 30 and 55, which resulted in a FDR of 9%. In all searches, peptides with a charge state of +1 or greater than +3 were not analyzed. A number of recent phosphoproteomic studies have reported data obtained from a linear ion trap mass spectrometer using similar search criteria and FDRs.29–33 However, none of these results were confirmed by manual validation.
Manual Validation and Determination of Phosphorylation Sites
Phosphopeptides were confirmed further by manual examination. The spectra were examined initially for the presence of a predominant peak corresponding to the neutral loss of H3PO4 from a phosphorylated Ser or Thr, the continuity of b- and y-ion series (minimum of four continuous ions), and the signal-to-noise ratio or overall quality of the examined spectra. Peptides that passed these criteria were examined further for the number of H3PO4 lost from the precursor ion, the assignment of major fragment ions to b- and y-ion series and the corresponding neutral-loss ions, the presence of y- and/or b-ions corresponding to the phosphorylated sites, and high intensity of proline-directed fragment ions for proline-containing peptides. The number of manually confirmed phosphopeptides was subtracted from the number identified by database search engine analyses and used to calculate the percent of phosphopeptides rejected.
RESULTS
Phosphopeptide Enrichment
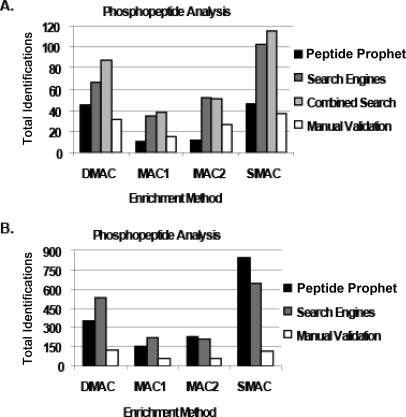
For this study, a DIMAC protocol was developed to enrich phosphopeptides for LC-MSn analysis. The DIMAC protocol involves enrichment of separate aliquots of tryptic peptides on Ga+3-IMAC and TiO2 TopTip columns. The results of this protocol were compared with those obtained using a single IMAC column (IMAC1 and IMAC2)26,27 or a more complex sequential fractionation protocol (SIMAC).23 The steps involved in the DIMAC and the more complex SIMAC enrichment protocols are illustrated in Fig. 1. For each protocol, the phosphopeptide fractions recovered from cultured rat renal proximal tubule cells were analyzed by LC-MSn using neutral-loss scanning34 without further upstream fractionation. Separate Mascot and SEQUEST database searches were performed for the individual MS2 and MS3 data obtained from triplicate injections of each enriched fraction. The database search results were combined and analyzed with Scaffold software. Phosphopeptides were initially filtered by selecting the Scaffold phospho-modification for MS2 data and phospho- and dehydro-modifications for MS3 data and by setting stringent protein and PeptideProphet probability thresholds of 99.9% and 95%, respectively. This analysis resulted in a calculated FDR of <1.5% when searched against a reverse database. In addition, the combined data were filtered using the stringent Mascot and SEQUEST thresholds described in Materials and Methods. Of the four enrichment methods tested, the SIMAC protocol produced the largest number of potential identifications of unique phosphopeptides (Fig. 2A). From the combined analyses, using Protein- and PeptideProphet and the database search engine thresholds, 115 unique phosphopeptides were potentially identified. However, only 37 of the 115 potential phosphopeptides were confirmed by the manual validation technique described below. When analyzing the DIMAC enrichment method, a total of 87 unique phosphopeptides was potentially identified from the combined searches. However, manual validation confirmed only 31 of the 87 potential phosphopeptides as confident identifications. Similarly, the IMAC1 and IMAC2 protocols produced a total of 39 and 51 unique, potential identifications, respectively. As observed with the SIMAC and DIMAC methods, a large overlap was observed in the phosphopeptides identified by the two filtering methods. However, manual validation of the IMAC1 and IMAC2 data confidently identified only 15 and 27 of the 39 and 51 potential phosphopeptides, respectively. In total, 110 phosphopeptides were confidently identified by manual validation of results obtained using the stringent search conditions that had a FDR of <1.5%. As a result, approximately 40% of the potential phosphopeptides identified from the four enrichment protocols were confirmed by manual validation. Therefore, reliance on stringent search conditions that produce a low FDR is not effective for the analysis of low-resolution phosphopeptide spectra.
FIGURE 2.
Comparison of total potential and validated phosphopeptides identified by various search procedures. (A) A greater number of potential phosphopeptides were identified for each enrichment protocol using standard search engine settings that produced a FDR of <1.5% as compared with the more rigorous manual validation. Black, dark gray, light gray, and white bars represent total identifications obtained using PeptideProphet, Mascot and SEQUEST search engines, combined PeptideProphet, Mascot, and SEQUEST search engines, and manual validation, respectively. (B) A significant increase in identifications was observed for potential and validated phosphopeptides using less-stringent filtering criteria with a combined FDR of 7–9%. Black, gray, and white bars represent total identifications obtained using PeptideProphet, Mascot and SEQUEST search engines, and manual validation, respectively.MAC1/MAC2, IMAC1/IMAC2.
The initial data sets were reanalyzed using less-stringent filtering criteria that produced FDRs between 7% and 9% to determine if additional phosphopeptides could be identified confidently. Of the four enrichment methods tested, the SIMAC protocol again produced the largest number of potential identifications of phosphopeptides (Fig. 2B). Using the newly adjusted PeptideProphet and search engine thresholds, >800 unique phosphopeptides were potentially identified. Of this set, 113 phosphopeptides were confirmed by manual validation. When analyzing the DIMAC enrichment data, >500 phosphopeptides were identified using the less-stringent PeptideProphet and database thresholds. Of these, 123 phosphopeptides were validated. Similarly, the IMAC1 and IMAC2 protocols each produced a total >200 potential identifications but only 54 and 56 phosphopeptides were validated, respectively. By manually validating the data obtained from the combined analyses of all of four enrichment protocols using the less-stringent filtering criteria, 262 unique phosphopeptides were confidently identified. Therefore, sole reliance on criteria that produce a low FDR would result in the loss of a large proportion of confident identifications that could be derived from the original data sets.
Confident Identification of Phosphopeptides
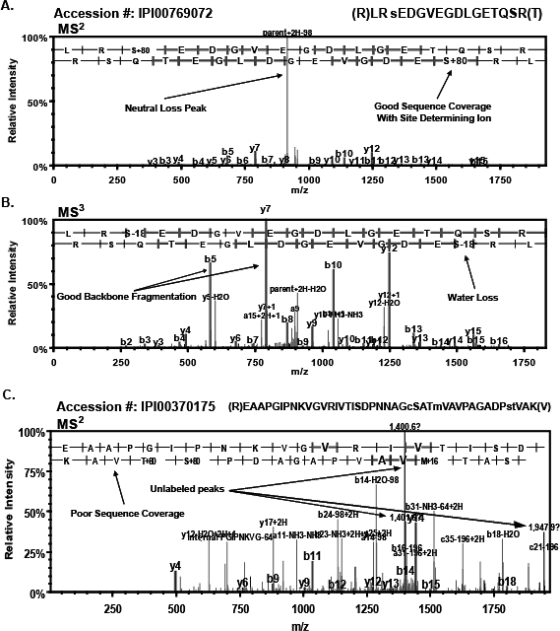
The potential phosphopeptides identified by traditional statistical filtering (PeptideProphet) and the search engine thresholds (Mascot and SEQUEST) were subjected to a rigorous manual validation protocol. The final dataset was produced by following the strict guidelines for accurate identification of phosphopeptides described in Materials and Methods. Examples of peptides that passed or failed the manual validation are illustrated in Fig. 3. The MS2 scan of the singly phosphorylated peptide (IPI00769072) contains a classic neutral-loss peak (Fig. 3A). The lower-intensity peaks were sufficient to produce excellent sequence coverage of y (upper)- and b (lower)-ions and to identify the site of phosphorylation as serine 3. In addition, the MS3 scan significantly improved the sequence coverage of y- and b-ions and identified the loss of water from serine 3 (Fig. 3B). By contrast, the MS2 scan of the potential phosphopeptide (IPI00370175) has a large proportion of unidentified peaks and produced poor sequence coverage (Fig. 3C). The scan also contained no observable neutral-loss peak. This example illustrates a potential phosphopeptide identification that passed the stringent probability-filtering and database search engine thresholds with a FDR of the <1.5% but failed to pass the manual validation criteria. This example highlights the importance of a rigorous manual spectral validation.
FIGURE 3.
Confident identification of phosphopeptides. The importance of manual validation is illustrated by comparing the spectra of phosphopeptides that were confirmed or rejected. (A) The MS2 scan for a singly phosphorylated peptide (IPI00769072) contains a classic neutral-loss peak with lower intensity sequence coverage of y (lower)- and b (upper)-ions containing the phosphorylation site, determining ion on serine 3. (B) The MS3 scan of the same peptide (IPI00769072) showing improved sequence coverage of y- and b-ions as well as a loss of water from serine 3. (C) The MS2 scan of potential phosphopeptide (IPI00370175) lacks an observable neutral-loss peak, has poor sequence coverage, and has several unidentified, prominent peaks. This spectra illustrates a potential phosphopeptide that passed the probability-filtering and the traditional-score thresholds for the more-stringent filtering criteria but did not satisfy the criteria for manual validation.
Overall, the manual validation technique resulted in the rejection of a large proportion of potential phosphopeptides identified by PeptideProphet probability thresholds or the combined thresholds for the Mascot and SEQUEST database searches (Fig. 2). Therefore, exclusive use of PeptideProphet or Mascot and SEQUEST database search engine thresholds resulted in a large number of “false positive” phosphopeptide identifications when compared with the more accurate manual validation, even when using the more-stringent filtering criteria. For example, with the DIMAC method, the percent of the combined phosphopeptides identified by the stringent PeptideProphet and Mascot and SEQUEST thresholds but rejected by manual validation was 64%. This rejection rate is much greater than anticipated for a false discovery rate of <1.5%. These analyses again highlight the importance of manual validation for confident identification of phosphopeptides.
It is important to note that with the less-stringent search conditions, the DIMAC method produced the greatest number of confident phosphopeptide identifications (Fig. 2). In addition, this method is also significantly more streamlined and cost-efficient to perform than the SIMAC method. The DIMAC method used dual phosphopeptide enrichments that can be performed simultaneously and produced two fractions for subsequent analysis. By contrast, the SIMAC method required an elaborate sequential fractionation scheme that necessitated more time to complete and produced three separate fractions for subsequent analysis (Fig. 1). Therefore, the DIMAC protocol is faster and more cost-efficient and produces a greater number of confident phosphopeptide identifications.
Comparison of Phosphopeptide Enrichment Protocols
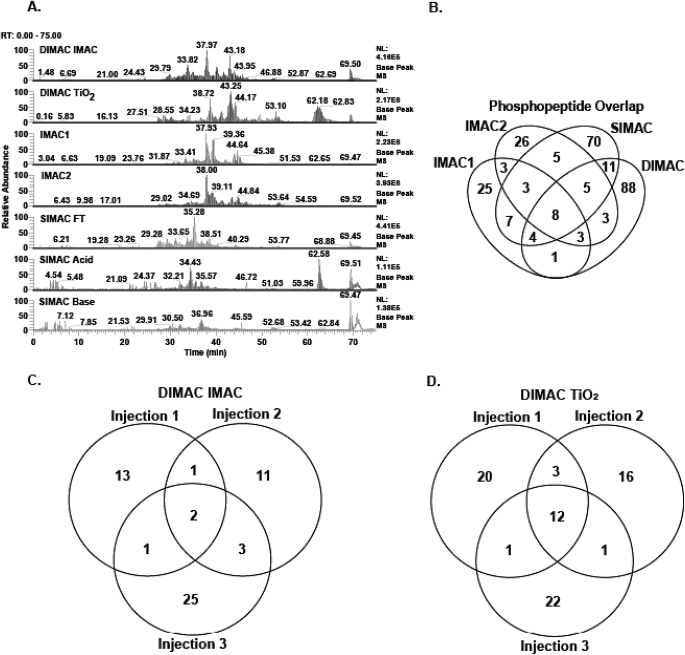
The HPLC base peak chromatograms of each fraction obtained from the four enrichment protocols produced a unique elution profile (Fig. 4A). By contrast, the triplicate injections of the individual samples were highly reproducible (Supplemental Fig. 1). Therefore, each protocol enriches a unique set of phosphopeptides. Manual validation of the data obtained from triplicate injections of all fractions from the four enrichment protocols resulted in the confident identification of 262 phosphopeptides. However, only 53 phosphopeptides (20%) were identified in fractions obtained from more than one enrichment protocol (Fig. 4B). By contrast, 88 (34%) and 70 (27%) of the total phosphopeptides were unique identifications from the DIMAC and SIMAC enrichment methods, respectively.
FIGURE 4.
Comparison of phosphopeptide enrichment protocols. Each phosphopeptide enrichment strategy led to the identification of a unique set of phosphopeptides, but the DIMAC protocol generated the largest number of confident identifications. (A) Base peak chromatograms for the individual phosphopeptide enrichment samples illustrate the uniqueness of each fraction. RT, retention time; NL, natural log. (B) Venn diagram of the phosphopeptides confidently identified from each of the four enrichment protocols. The minimal overlap verifies that the four protocols are highly complementary and illustrates that the DIMAC and SIMAC strategies identified the majority of the phosphopeptides. (C and D) Venn diagrams demonstrating the uniqueness of multiple injections of the DIMAC–IMAC enrichment (C) and the DIMAC–TiO2 enrichment (D) fractions.
As suggested previously,35,36 triplicate injections of each sample were performed to increase the total number of unique phosphopeptides identified. Venn diagrams were constructed to compare the results obtained from multiple injections of the two DIMAC samples (Fig. 4C and D). Only 16% and 23% of the total phosphopeptides were confidently identified in more than one injection of the DIMAC–IMAC and DIMAC–TiO2 enrichment samples, respectively. This is not surprising given the complexity of the injected sample. To obtain greater coverage of the phosphoproteome, it would be necessary to incorporate an additional separation technique, such as strong cation exchange chromatography, prior to or post-phosphopeptide enrichment. However, development of an optimal pipeline to maximize coverage was not the focus of this study.
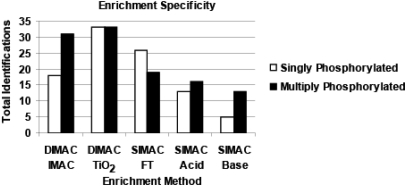
Previous studies have concluded that IMAC protocols preferentially enrich for multiply phosphorylated peptides.20,37 Consistent with this conclusion, the DIMAC–IMAC protocol also selectively enriches for multiply phosphorylated peptides (63%) compared with singly phosphorylated peptides (37%; Fig. 5). However, the DIMAC–TiO2 method had an equal preference to enrich for singly and multiply phosphorylated peptides. In addition, the observed preference of the individual SIMAC fractions to enrich for singly and multiply phosphorylated peptides was consistent with previous observations.7,23
FIGURE 5.
Method-dependent enrichment of phosphopeptides with different states of phosphorylation. The bar graph illustrates the number of phosphopeptides indentified in the individual DIMAC and SIMAC fractions that were singly (white bars) or multiply phosphorylated (black bars).
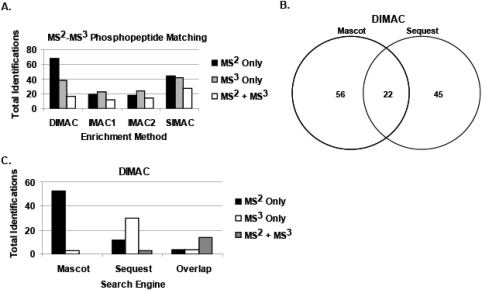
The data produced in this study also highlight the advantage of using neutral-loss scanning for the identification of phosphopeptides. The final set of manually validated phosphopeptides was subdivided into those identified by MS2 only, MS3 only, or a combination of MS2 + MS3 data (Fig. 6A). This comparison illustrates the importance of analyzing the spectra obtained from both fragmentation schemes to identify the greatest number of phosphopeptides. For example, for the DIMAC enrichment strategy, 68 (55%), 38 (31%), and 17 (14%) of the total phosphopeptides were identified from MS2 only, MS3 only, and MS2 + MS3 data, respectively. In addition, there was a clear advantage to using both database search engines. For example, from the DIMAC enrichment strategy, 56 (46%), 45 (36%), and 22 (18%) of the phosphopeptides were identified from Mascot only, SEQUEST only, or a combination of both search engines, respectively (Fig. 6B). As illustrated for the DIMAC data, further analysis indicated that the Mascot database search engine analysis identified phosphopeptides almost exclusively from the MS2 scans, whereas the majority of identified phosphopeptides from the SEQUEST database search engine was identified from MS3 scans (Fig. 6C). Overall, these results indicate the observed advantage of performing neutral-loss scanning as well as using multiple search engines to increase the confident identification of phosphopeptides.
FIGURE 6.
Advantages of multiple scans and database search engines. A combination of neutral-loss scanning and multiple database search engines resulted in a greater number of confident identifications of phosphopeptides. (A) The bar graph compares the number of phosphopeptides identified from each enrichment protocol using data derived from the MS2 scan only (black bars), MS3 scan only (gray bars), and MS2 and MS3 scans (white bars). (B) Venn diagram showing the total phosphopeptides identified from the DIMAC enrichment protocol using the Mascot or SEQUEST database search engines. (C) The bar graph compares the total phosphopeptides identified by each search engine from the MS2 scan only (black bars), MS3 scan only (white bars), and MS2 and MS3 scans (gray bars).
Identification of Novel Phosphorylation Sites
The sites of phosphorylation were determined by analyzing the series of b- and y-ions as well as the presence of key phosphosite-determining ions. Of the 262 phosphopeptides identified by manual validation of the data from the combined enrichment protocols, 111 were novel phosphorylation sites, as revealed by comparison with the PhosphoSitePlus database (www.phosphosite.org). The novel phosphopeptides identified by the DIMAC protocol are listed in Table 1 along with the protein names, accession numbers, and putative kinases, which were determined using the predictive software of the NetPhosK 1.0 website (www.cbs.dtu.dk/services/NetPhosK/). In addition, a list of all of the novel sites of phosphorylation, identified from the combined enrichment protocols, is shown in Supplemental Table 1, as well as a complete profile (e.g., observed m/z, Mascot, and SEQUEST scores, etc.) of all phosphopeptides obtained from MS2 and MS3 data. The complete list of phosphopeptides identified from MS2 spectra, including peptide sequence, site of phosphorylation, and PeptideProphet probability, SEQUEST XCorr, and Mascot ion scores, is provided in Supplemental Table 2. The complete profile list of MS3 phosphopeptides, with phosphorylation and dehydro-modifications, is provided in Supplemental Tables 3 and 4. Overall, the combined analyses led to the identification of a large number of novel phosphorylation sites.
TABLE 1.
Novel Phosphorylation Sites Identified Using the DIMAC Enrichment Protocol and Manual Validation
| Protein name | Accession number | Phosphosites | Putative kinase |
|---|---|---|---|
| 32 kDa Protein | IPI00566235 | Y133 | EGFR |
| 77 kDa Protein | IPI00392830 | S404 | CKII |
| 97 kDa Protein | IPI00764372 | T622 | GSK3 |
| Actin, cytoplasmic 1 | IPI00189819 | S365 | GSK3 |
| Calpain-6 | IPI00210533 | Y532, T536 | INSR, PKC |
| Heat shock protein 90-α | IPI00210566 | T426 | CKII |
| Hypothetical protein LOC364073 | IPI00196210 | S618, S620, S627 | CDC2, CKI, PKC |
| Isoform 1 of synaptopodin | IPI00417225 | S114, S132, T133 | PKC, GSK3, CAM-II |
| Isoform 3 of Ras GTPase-activating protein SynGAP | IPI00212566 | S320, S475 | GSK3, CAM-II |
| Isoform B23.1 of nucleophosmin | IPI00197553 | S112 | PKA |
| Nodal homolog | IPI00361228 | S104, T111 | CDK5, GSK3 |
| Nuclear pore complex protein Nup88 | IPI00194687* | S158 | GSK3 |
| RAB2, member RAS oncogene family-like | IPI00417839 | S129, S130 | GSK3, CDK5 |
| Similar to PI3K-related kinase SMG-1 | IPI00763254 | T3166 | PKC |
| Similar to titin isoform N2-B | IPI00210193 | Y28357, T28358, T28360 | INSR, PKA, PKC |
The listed sites were not reported previously in the PhosphoSitePlus database (www.phosphosite.org). The putative kinases were obtained from NetPhosK 1.0 (www.cbs.dtu.dk/services/NetPhosK/). CKII/I, Casein kinase II/I; INSR, insulin receptor; CAM-II, Ca2+/calmodulin-dependent protein kinase II, CDK5, cyclin-dependent kinase 5, SMG-1, serine/threonine-protein kinase.
DISCUSSION
Many studies have shown the importance of prior phosphopeptide enrichment to identify phosphopeptides by mass spectrometric analysis. In the current study, a comparison of four phosphopeptide enrichment techniques was performed, and a rigorous phosphoproteomic filtering workflow was used to analyze data from a linear ion trap mass spectrometer. The combined results demonstrate that it is beneficial to perform IMAC and TiO2 enrichments to enhance the total number of confident identification of phosphopeptides. The DIMAC enrichment method identified more phosphopeptides and is significantly more time- and cost-efficient than the SIMAC method. However, the two protocols were found to be complementary. Approximately 60% of the potential phosphopeptides that were identified using database search engine thresholds and statistical filtering with a FDR of <1.5% were rejected by the more-rigorous manual analysis. Thus, sole reliance on stringent filtering criteria was not sufficient to yield confident phosphoprotein identifications. Starting with less-stringent filtering criteria, the combined enrichment protocols and rigorous manual spectral analysis led to the confident identification of 262 unique phosphopeptides, including 111 novel phosphorylation sites. Therefore, a large proportion of the confident identifications would have been lost if the filtering were performed using only a stringent cutoff of <1.5% FDR. The combined findings highlight the requirement for a rigorous manual validation approach for the analysis of low-mass accuracy spectra.
ACKNOWLEDGMENT
This research was supported in part by National Institutes of Diabetes and Digestive and Kidney Diseases grants DK-037124 and DK-075517 awarded to N.P.C.
Footnotes
Disclosures: None of the authors derive financial support or have associations that may constitute a conflict of interest with the submitted manuscript.
REFERENCES
- 1. Graves JD, Krebs EG. Protein phosphorylation and signal transduction. Pharmacol Ther 1999;82:111–121 [DOI] [PubMed] [Google Scholar]
- 2. Hunter T. Signaling—2000 and beyond. Cell 2000;100:113–127 [DOI] [PubMed] [Google Scholar]
- 3. Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science 2002;298:1912–1934 [DOI] [PubMed] [Google Scholar]
- 4. Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science 2001;291:1304–1351 [DOI] [PubMed] [Google Scholar]
- 5. Mann M, Ong SE, Gronborg M, Steen H, Jensen ON, Pandey A. Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol 2002;20:261–268 [DOI] [PubMed] [Google Scholar]
- 6. Dunn JD, Reid GE, Bruening ML. Techniques for phosphopeptide enrichment prior to analysis by mass spectrometry. Mass Spectrom Rev 2009;29:29–54 [DOI] [PubMed] [Google Scholar]
- 7. Thingholm TE, Jensen ON, Larsen MR. Analytical strategies for phosphoproteomics. Proteomics 2009;9:1451–1468 [DOI] [PubMed] [Google Scholar]
- 8. Beausoleil SA, Jedrychowski M, Schwartz D, et al. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci USA 2004;101:2130–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aebersold R, Goodlett DR. Mass spectrometry in proteomics. Chem Rev 2001;101:269–295 [DOI] [PubMed] [Google Scholar]
- 10. Simpson RJ. Proteins and Proteomics: A Laboratory Manual, Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory, 2003 [Google Scholar]
- 11. Craig AG, Hoeger CA, Miller CL, Goedken T, Rivier JE, Fischer WH. Monitoring protein kinase and phosphatase reactions with matrix-assisted laser desorption/ionization mass spectrometry and capillary zone electrophoresis: comparison of the detection efficiency of peptide-phosphopeptide mixtures. Biol Mass Spectrom 1994;23:519–528 [DOI] [PubMed] [Google Scholar]
- 12. Liao PC, Leykam J, Andrews PC, Gage DA, Allison J. An approach to locate phosphorylation sites in a phosphoprotein: mass mapping by combining specific enzymatic degradation with matrix-assisted laser desorption/ionization mass spectrometry. Anal Biochem 1994;219:9–20 [DOI] [PubMed] [Google Scholar]
- 13. Nuhse TS, Bottrill AR, Jones AM, Peck SC. Quantitative phosphoproteomic analysis of plasma membrane proteins reveals regulatory mechanisms of plant innate immune responses. Plant J 2007;51:931–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Michel HP. Identification of the phosphorylation site of an 8.3 kDa protein from photosystem II of spinach. FEBS Lett 1987;212:103–108 [Google Scholar]
- 15. Gruhler A, Olsen JV, Mohammed S, et al. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol Cell Proteomics 2005;4:310–327 [DOI] [PubMed] [Google Scholar]
- 16. Figeys D, Gygi SP, McKinnon G, Aebersold R. An integrated microfluidics-tandem mass spectrometry system for automated protein analysis. Anal Chem 1998;70:3728–3734 [DOI] [PubMed] [Google Scholar]
- 17. Li S, Dass C. Iron(III)-immobilized metal ion affinity chromatography and mass spectrometry for the purification and characterization of synthetic phosphopeptides. Anal Biochem 1999;270:9–14 [DOI] [PubMed] [Google Scholar]
- 18. Nuhse TS, Stensballe A, Jensen ON, Peck SC. Large-scale analysis of in vivo phosphorylated membrane proteins by immobilized metal ion affinity chromatography and mass spectrometry. Mol Cell Proteomics 2003;2:1234–1243 [DOI] [PubMed] [Google Scholar]
- 19. Posewitz MC, Tempst P. Immobilized gallium(III) affinity chromatography of phosphopeptides. Anal Chem 1999;71:2883–2892 [DOI] [PubMed] [Google Scholar]
- 20. Jensen SS, Larsen MR. Evaluation of the impact of some experimental procedures on different phosphopeptide enrichment techniques. Rapid Commun Mass Spectrom 2007;21:3635–3645 [DOI] [PubMed] [Google Scholar]
- 21. Sugiyama N, Masuda T, Shinoda K, Nakamura A, Tomita M, Ishihama Y. Phosphopeptide enrichment by aliphatic hydroxy acid-modified metal oxide chromatography for nano-LC-MS/MS in proteomics applications. Mol Cell Proteomics 2007;6:1103–1109 [DOI] [PubMed] [Google Scholar]
- 22. Yu L-R, Issaq HJ, Veenstra TD. Phosphoproteomics for the discovery of kinases as cancer biomarkers and drug targets. Proteomics Clin Appl 2007;1:1042–1057 [DOI] [PubMed] [Google Scholar]
- 23. Thingholm TE, Jensen ON, Robinson PJ, Larsen MR. SIMAC (sequential elution from IMAC), a phosphoproteomics strategy for the rapid separation of monophosphorylated from multiply phosphorylated peptides. Mol Cell Proteomics 2008;7:661–671 [DOI] [PubMed] [Google Scholar]
- 24. Woost PG, Orosz DE, Jin W, et al. Immortalization and characterization of proximal tubule cells derived from kidneys of spontaneously hypertensive and normotensive rats. Kidney Int 1996;50:125–134 [DOI] [PubMed] [Google Scholar]
- 25. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976;72:248–254 [DOI] [PubMed] [Google Scholar]
- 26. Villen J, Gygi SP. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat Protoc 2008;3:1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee J, Xu Y, Chen Y, et al. Mitochondrial phosphoproteome revealed by an improved IMAC method and MS/MS/MS. Mol Cell Proteomics 2007;6:669–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 2007;4:207–214 [DOI] [PubMed] [Google Scholar]
- 29. Jorge I, Casas EM, Villar M, et al. High-sensitivity analysis of specific peptides in complex samples by selected MS/MS ion monitoring and linear ion trap mass spectrometry: application to biological studies. J Mass Spectrom 2007;42:1391–1403 [DOI] [PubMed] [Google Scholar]
- 30. Black TM, Andrews CL, Kilili G, Ivan M, Tsichlis PN, Vouros P. Characterization of phosphorylation sites on Tpl2 using IMAC enrichment and a linear ion trap mass spectrometer. J Proteome Res 2007;6:2269–2276 [DOI] [PubMed] [Google Scholar]
- 31. Domon B, Bodenmiller B, Carapito C, Hao Z, Huehmer A, Aebersold R. Electron transfer dissociation in conjunction with collision activation to investigate the Drosophila melanogaster phosphoproteome. J Proteome Res 2009;8:2633–2639 [DOI] [PubMed] [Google Scholar]
- 32. Han G, Ye M, Zhou H, et al. Large-scale phosphoproteome analysis of human liver tissue by enrichment and fractionation of phosphopeptides with strong anion exchange chromatography. Proteomics 2008;8:1346–1361 [DOI] [PubMed] [Google Scholar]
- 33. Carrascal M, Gay M, Ovelleiro D, Casas V, Gelpi E, Abian J. Characterization of the human plasma phosphoproteome using linear ion trap mass spectrometry and multiple search engines. J Proteome Res 2010;9:876–884 [DOI] [PubMed] [Google Scholar]
- 34. Hoffert JD, Pisitkun T, Wang G, Shen RF, Knepper MA. Quantitative phosphoproteomics of vasopressin-sensitive renal cells: regulation of aquaporin-2 phosphorylation at two sites. Proc Natl Acad Sci USA 2006;103:7159–7164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhai B, Villen J, Beausoleil SA, Mintseris J, Gygi SP. Phosphoproteome analysis of Drosophila melanogaster embryos. J Proteome Res 2008;7:1675–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bakalarski CE, Haas W, Dephoure NE, Gygi SP. The effects of mass accuracy, data acquisition speed, and search algorithm choice on peptide identification rates in phosphoproteomics. Anal Bioanal Chem 2007;389:1409–1419 [DOI] [PubMed] [Google Scholar]
- 37. Ficarro SB, McCleland ML, Stukenberg PT, et al. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol 2002;20:301–305 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.