Abstract
While reversible histone modifications are linked to an ever-expanding range of biological functions1–5, the demethylases for histone H4 lysine 20 and their potential regulatory roles remain unknown. Here, we report that the PHD and Jumonji C (JmjC) domain-containing protein, PHF8, while utilizing multiple substrates, including H3K9me1/2 and H3K27me2, also functions as an H4K20me1 demethylase. PHF8 is recruited to promoters by its PHD domain based on interaction with H3K4me2/3 and controls G1/S transition in conjunction with E2F1, HCF-1 and Set1A, at least in part, by removing the repressive H4K20me1 mark from a subset of E2F1-regulated gene promoters. Phosphorylation-dependent PHF8 dismissal from chromatin in prophase is apparently required for the accumulation of H4K20me1 during early mitosis, which might represent a component of the Condensin II loading process. Accordingly, the HEAT repeat clusters in two non-SMC Condensin II subunits, N-CAPD3 and N-CAPG2, are capable of recognizing H4K20me1, and ChIP-seq. analysis demonstrate a significant overlap of Condensin II and H4K20me1 sites in mitotic HeLa cells. Thus, the identification and characterization of the first H4K20me1 demethylase, PHF8, has revealed an intimate link between this enzyme and two distinct events in cell cycle progression.
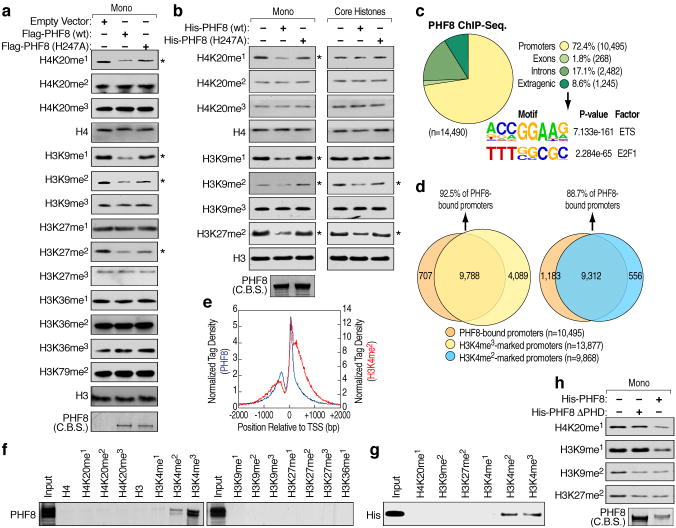
We evaluated potential substrates for the putative histone demethylase, PHF8, on mononucleosomes. Flag-tagged PHF8 immunoprecipitated from HEK293T cells was capable of demethylating H4K20me1, H3K9me1/2 and H3K27me2, but had no effects on other histone methylation marks tested. Mutation of histidine 247, predicted to be part of the Fe(II) binding site5, impaired these activities (Fig. 1a), which were similarly confirmed using bacterially-expressed PHF8 (Fig. 1b, left panels) and in a dose- and time-dependent manner (Fig. S2a). Surprisingly, when using core histones as substrates, while the activities towards H3K9me2 and H3K27me2 were preserved, only minimal activities were detected towards H4K20me1 and H3K9me1 (Fig. 1b, right panels), as similarly found for PHF8 immunoprecipitated from HEK293T cell lysates (Fig. S2b).
Figure 1. Histone demethylation mediated by PHF8.
a, Demethylase activity of Flag-tagged full-length wild type and mutant (H247A) PHF8 immunoprecipitated from HEK293T cell lysates was assessed using mononucleosomes as substrates. Expression of PHF8 proteins was visualized by Commassie blue staining (C.B.S). Asterisk denotes potential substrate. b, Demethylase activity of purified bacterially-expressed His-tagged full-length wild type and mutant (H247A) PHF8 proteins assessed using mononucleosomes (left panels) or core histones (right panels) as substrates. Expression of PHF8 proteins were visualized by Commassie blue staining (bottom left panel). Asterisk denotes potential substrate. c, Genomic distribution and top enriched motifs of PHF8 ChIP-seq. peaks (n=14,490) in HeLa cells. d, Venn diagrams showing overlap between PHF8-bound and H3K4me3 and H3K4me2-marked promoters. e, Tag density plots displaying PHF8 and H3K4me2 tags distribution relative to the transcriptional start site (TSS). f–g, Peptide pull-down assays mixing HeLa nuclear extracts (f) or purified bacterially-expressed His-tagged PHF8 PHD finger (aa1-54) (g) with biotinylated histone tails. Pull-downs were analyzed by immunoblotting. h, Demethylase activity of purified bacterially-expressed His-tagged wild type and ΔPHD finger (54–1024) PHF8 proteins assessed using mononucleosomes as substrates. Expression of PHF8 proteins were visualized by Commassie blue staining.
To gain insight into biological substrates of PHF8, we performed ChIP-Seq. in HeLa cells, finding that ~ 72% of PHF8 peaks localized on promoters, with the most statistically-significant predicted binding sites being those for Ets and E2F1 (Fig. 1c). The great majority of PHF8-bound promoters overlapped with H3K4me3 (6) and H3K4me2 positive promoters, while very few harbored H4K20me1, H3K9me1/2, or H3K27me2 marks (Fig. 1d and Fig. S2c). The PHF8 tags distribution relative to transcription start site corresponded to that of the H3K4me2 mark (Fig. 1e), suggesting that PHF8 might be recruited to promoters by H3K4me2/3 based on interaction with its PHD finger, a known methyl lysine binding motif7. In vitro peptide pull-down revealed that PHF8 specifically bound to H3K4me2 and H3K4me3 histone tails, but not to others tested (Fig.1f). Direct interaction of PHD finger of PHF8 with H3K4me2/3 histone tails was confirmed using bacterially-expressed PHF8 PHD finger (Fig. 1g), while PHF8 ΔPHD failed to interact (Fig. S2d). Consistent with these data, PHF8ΔPHD retained its activities on mononucleosomes towards H3K9me2 and H3K27me2, but not towards H4K20me1 or H3K9me1 (Fig. 1h). These results indicate a required PHD finger-mediated targeting of PHF8 to H3K4me2/3-containing nucleosomes to efficiently demethylate H4K20me1 or H3K9me1.
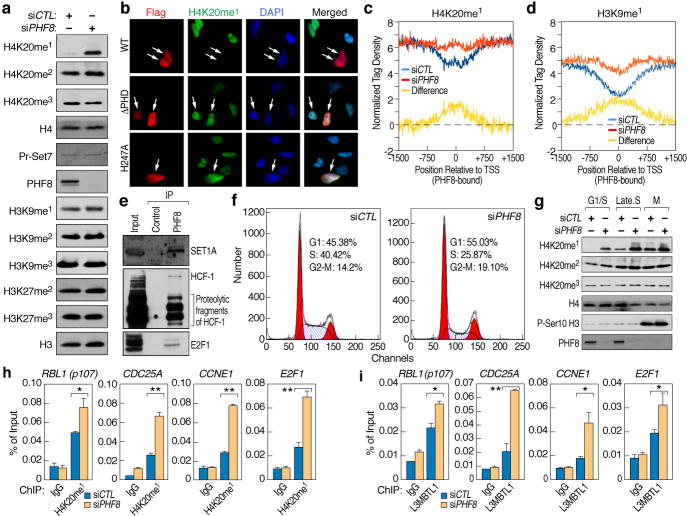
To evaluate PHF8 enzymatic activities in vivo, a PHF8-specific siRNA was transfected into HeLa cells, which led to significantly increased H4K20me1 levels and slightly increased H4K20me2 and H3K9me1, without changes in other histone marks (Fig. 2a). This H4K20me1 increase was dependent on PHF8 enzymatic activity (Fig. S2e), perhaps also partially attributed to PHF8 depletion interference with cell cycle progression (vide infra). Other known H3K9/K27 demethylases might compensate the loss of PHF8, therefore minimizing PHF8 siRNA effects. Immuno-fluorescence microscopy analysis in HeLa cells over-expressing Flag-tagged full-length PHF8 revealed that ~25% of cells exhibited significantly decreased H4K20me1 signal intensity (n=200), while PHF8 ΔPHD or PHF8 (H247A) had no effects (Fig. 2b and Fig. S2f). PHF8 over-expression caused no significant differences for other substrates (Fig. S2g–n). Similar results were obtained with PHF8 (1-447) (Fig. S3a), consistent with recent studies8,9, except that we additionally observed PHF8 activity towards H4K20me1. U2OS cells over-expressing Flag-tagged full-length PHF8 showed decrease in H4K20me1 (~20%) as well as H3K9me2 (~60%), but not H3K9me1 or H3K27me2 signal intensity (n=200) (Fig. S3b). Thus, PHF8 activity may be regulated both in a cell type-dependent manner and by specific modifications10–12. To further evaluate PHF8 enzymatic activities in HeLa cells, ChIP-Seq. analysis of H4K20me1, H3K9me1, H3K9me2 and H3K27me2 was performed in control or PHF8 siRNA-transfected HeLa cells, finding that H4K20me1 and H3K9me1, but not H3K9me2 or H3K27me2, increased significantly on PHF8-bound promoter regions upon PHF8 depletion (Fig. 2c, d and Fig. S3c, d). The difference in H4K20me1 and H3K9me1 levels between PHF8 siRNA-transfected and control cells correlated with the number of PHF8 ChIP-Seq. tags on promoters (Fig. S3e, f).
Figure 2. Characterization of PHF8 protein.
a, HeLa cells transfected with control or PHF8 specific siRNAs were analyzed by immunoblotting. b, Immunohistochemical analysis of HeLa cells transfected with Flag-tagged wild type, ΔPHD or mutant (H247A) PHF8. Cells were stained with anti-flag (red), anti- H4K20me1 (green) and DAPI (blue). White arrows indicate cells transfected with PHF8. c–d, H4K20me1 (c) or H3K9me1 (d) ChIP-Seq. tags distribution over PHF8 promoter regions in control and PHF8 siRNA transfected HeLa cells. e, HeLa nuclear extracts immunoprecipitated with control IgG or PHF8 antibody and analyzed by immunoblotting. f, Flow-cytometry analysis of HeLa cells transfected with control or PHF8 siRNAs and stained with propidium iodide. g, Chromatin-bound fractions from HeLa cells transfected with control or PHF8 siRNAs and synchronized to different cell cycle phases as indicated were analyzed by immunoblotting. h–i, ChIP analysis of H4K20me1 (h) and L3MBTL1(i) on selected promoter regions in HeLa cells transfected with control or PHF8 siRNA (± s.e.m., *p<0.05, **p<0.01).
We next focused on investigating the PHF8 activity towards H4K20me1, a histone mark that has been implicated in cell cycle regulation13–17. Interestingly, gene ontology analysis of PHF8-bound promoters identified by ChIP-Seq. (Fig. 1c and Fig. S4a) revealed that one of the most statistically-significant terms was “cell cycle” (Fig. S4b), as similarly found in RNA profiling analysis for genes positively regulated by PHF8 (Fig. S4c, d), with ~72% of which proving to be PHF8 ChIP-Seq. targets. To gain further insight into PHF8 function, its associated proteins were purified, which included critical G1/S transition regulators, E2F1, HCF-1 and Set1A18,19. Interactions between PHF8 and these proteins were confirmed by immunoprecipitation (Fig. 2e). Gel filtration chromatography revealed co-fractionation of native PHF8, E2F1, HCF-1 and Set1A (Fig. S4e). Furthermore, E2F1 ChIP-Seq. analysis in HeLa cells indicated that >79% of E2F1-bound promoters corresponded to those binding PHF8 (Fig. S5a–c), suggesting a role of PHF8 in regulating E2F1 target genes.
Consistent with potential roles of PHF8 in cell cycle regulation, PHF8 siRNA-treated HeLa cells exhibited a strikingly decreased cell proliferation (Fig. S6a). Flow-cytometry analysis suggested a delay in G1/S transition and revealed a slight increase in M phase population upon PHF8 knock-down (Fig. 2f). In addition, there was a decrease in cell size after PHF8 siRNA treatment (Fig. S6b). BrdU incorporation in control or PHF8 siRNA-treated HeLa cells released from mitotic arrest confirmed that PHF8 depletion led to S phase entry delay (Fig. S6c, d). To further assess PHF8 enzymatic activity at different stages during cell cycle, chromatin-bound fractions were isolated from control or PHF8 siRNA-treated HeLa cells that were synchronized following the protocols in Fig. S6e, finding that PHF8 siRNA treatment led to a significant and distinct increase in H4K20me1 at G1/S and S phases, but exhibited minimal effects at G2/M (Fig. 2g and Fig. S6f, g), which was confirmed by immuno-fluorescence microscopy analysis (Fig. S6h). Minimal effects at G2/M might be due to PHF8 dissociation from chromatin at this stage (vide infra). ChIP analysis on G1/S transition-regulated promoters bound by PHF8 and E2F1, including RBL1 (p107), CDC25A, CCNE1 and E2F1, revealed an increase in H4K20me1 levels and decrease in PHF8 binding upon PHF8 knock-down (Fig. 2h and Fig. S7a). The levels of H4K20me3, H3K9me1/2/3 or H3K27me1/2/3 were not significantly altered, except for an increase of H4K20me3 on CCNE1 promoter (Fig. S7b–h). L3MBTL1, which has been shown to associate with H4K20me1 and function in gene repression20, exhibited increased binding (Fig. 2i), while H3K4me3 decreased upon PHF8 knock-down (Fig. S7i). ChIP analysis on promoters exhibiting changes in H3K9me1 and/or H4K20me1 in ChIP-Seq experiments confirmed subsets exhibiting an increase of only H4K20me1 or H3K9me1, or both marks upon PHF8 knock-down (Fig. S8). Similarly, a change in H3K9me2 on rDNA gene promoter regions in PHF8 siRNA-treated HEK293T cells has been reported12. Therefore, PHF8 uses distinct substrates on different subsets of genes to exert its function.
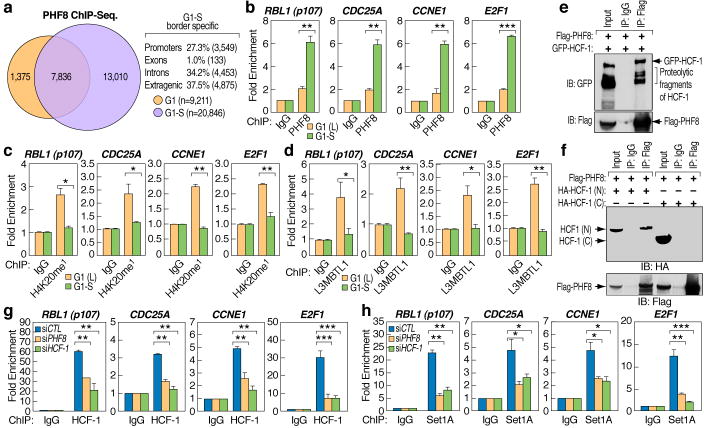
To further elucidate the role of PHF8 in G1/S transition, we performed PHF8 ChIP-Seq. in synchronized G1 and G1/S HeLa cells, revealing ~77% of binding sites at G1 localizing on promoters, as did 86% of PHF8 binding sites shared in both G1 and G1/S border. For all those shared PHF8 binding sites, ~85% showed increased PHF8 tags when cells reached G1/S border (Fig. 3a and Fig. S9), when H4K20me1 reaches its lowest levels15 (Fig. 2g). Moreover, ~13,000 new PHF8 binding sites were detected, mostly at intra- or extra-genic loci (>70%), which likely mitigate inappropriate appearance of H4K20me1 (Fig. 3a). ChIP analysis confirmed PHF8 binding increased on promoter regions of selected G1/S transition-regulated cell cycle genes (Fig. 3b). In concert with association between PHF8 and HCF-1, HCF-1 was similarly recruited (Fig. S10a). There was also a decrease in H4K20me1 levels (Fig. 3c) and L3MBTL1 binding (Fig. 3d), but an increase in H3K4me3 (Fig. S10b) during G1/S transition. Both PHF8 and HCF-1 siRNAs blocked induction of selected G1/S transition-regulated genes (Fig. S11). PHF8 and HCF-1 co-immunoprecipitated (Fig. 3e) and the interacting region was mapped to the N-terminus of HCF-1 (Fig. 3f), which has been suggested to be essential for its function as G1/S transition regulator21, further supporting their functional interactions. Furthermore, PHF8 knock-down impaired HCF-1 and Set1A recruitment (Fig. 3g, h), while HCF-1 knock-down did not impair PHF8 recruitment (Fig. S12).
Figure 3. PHF8 regulates E2F target genes in conjunction with HCF-1 and Set1A during G1/S transition.
a, Venn diagram showing overlapping between PHF8 ChIP-Seq. peaks in G1 and G1/S phases (n= 9,211 and 20,846, respectively). b–d, ChIP analysis of PHF8 (b), H4K20me1 (c) and L3MBTL1(d) on selected promoter regions in HeLa cells synchronized to G1 or G1/S phases. e, Cell extracts from Flag-PHF8 and GFP-HCF-1 co-transfected HEK293T cells were immunoprecipitated and analyzed by immunoblotting as indicated. f, Cell extracts from Flag-PHF8 and HA-HCF-1 (N1011) (amino-terminus of HCF-1) or HA-HCF-1 (C600) (Carboxyl-terminus of HCF-1) co-transfected HEK293T cells were immunoprecipitated and analyzed by immunoblotting as indicated. g–h, ChIP analysis of HCF-1 (g) or Set1A (h) on selected promoter regions in HeLa cells transfected with control, PHF8 or HCF-1 siRNAs. (± s.e.m., *p<0.05, **p<0.01, ***p<0.001)
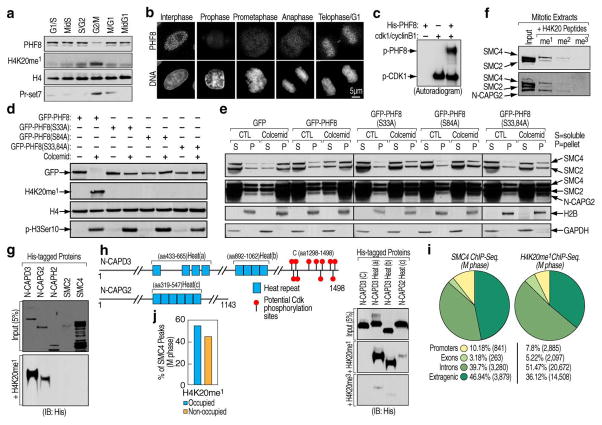
We examined chromatin fractions isolated from different phases in cell cycle, confirming PHF8 disappearance from chromatin when cells enter mitosis, which correlates with an increase in H4K20me1 and Pr-Set7 (Fig. 2g and Fig. 4a), while minimal or no increase in other marks was observed (Fig. 13a). Immunostaining of PHF8 without pre-extraction revealed no significant signal intensity change between interphase and prophase cells (Fig. S13b), while with pre-extraction revealed its dissociation from chromatin in prophase and re-association in telophase (Fig. 4b), as similarly found for GFP-tagged PHF8 (Fig. S13c). In contrast, H4K20me1 levels increased dramatically in prophase (Fig. S13c, d). PHF8 dissociation became more evident as chromosomes condensed (Fig. S13e). These results suggest that PHF8 dissociation in prophase might be involved in H4K2ome1 increase at this stage, as similarly proposed due to the increase of methyltransferase, Pr-Set715. Interestingly, the five-subunit Condensin II complex begins loading onto chromosomes starting in prophase22–25. Cellular fractionation revealed that PHF8 dissociation from chromatin paralleled chromosomal loading of Condensin II during M-phase (Fig. 13f). We therefore wanted to test whether PHF8 dissociation from chromatin permits accumulation of H4K20me1 and, potentially, Condensin II loading.
Figure 4. Phosphorylation-dependent PHF8 dissociation from chromatin in prophase links H4K20me1 with Condensin II.
a, Chromatin-bound fractions from HeLa cells synchronized to different cell cycle phases were analyzed by immunoblotting. b, Asynchronously growing HeLa cells were pre-extracted with 0.1% Triton X-100, fixed and stained with PHF8 and Hoechst dye (DNA). Representative images for different cell cycle phases were shown as indicated. Scale bar, 5 μm. c, His-tagged PHF8 phosphorylation by CDK1/cyclinB1 in vitro. d, HeLa cells transfected with GFP-tagged wild type or phosphorylation mutant PHF8 were treated with or without colcemid and chromatin bound fractions were analyzed by immunoblotting. e, HeLa cells transfected with GFP empty vector, wild type or phosphorylation mutant PHF8 in the presence or absence of colcemid were sorted. Chromatin-free (S) and –bound (P) fractions were analyzed by immunoblotting. f, Peptide pull-down assays performed mixing HeLa mitotic extracts with biotinylated histone tails. Pull-downs were analyzed by immunoblotting. g, Peptide pull-down assays performed mixing bacterially-expressed full length proteins with biotinylated histone tail as indicated. Input (upper panel) and pull-downs (bottom panel) were analyzed by immunoblotting. h, Schematic representation of N-CAPD3 and N-CAPG2 proteins27. Peptide pull-down assays were performed by mixing His-tagged C-terminus of N-CAPD3 (lane 1) or HEAT repeat clusters from N-CAPD3 or N-CAPG2 (lane 2–4) with biotinylated histone tails as indicated. Input (upper panel) and pull-downs (bottom two panels) were analyzed by immunoblotting. i, The genomic distribution of SMC4 and H4K20me1 ChIP-Seq peaks in M phase HeLa cells (n=8,263 and 40,162, respectively). j, Bar graph displaying the association of SMC4 and H4K20me1 peaks in M phase HeLa cells.
To test these hypotheses, we first investigated the mechanisms underlying PHF8 dissociation from chromatin. CDK1/cyclin B1 activity is known to fluctuate during cell cycle26, with its activity being inverse to PHF8 association with chromatin. In vitro kinase assays revealed CDK1 phosphorylates PHF8 (Fig. 4c). Two putative phosphorylation sites at serine 33 and 84 were identified by mass spectrometry (Fig. S14a, b), and mutation at either site proved to be a poor substrate, with double mutation further diminishing the phosphorylation (Fig. S14c). Inhibition of CDK activity impaired PHF8 dissociation from chromatin in mitosis (Fig. S14d). Similarly, mutation of the phosphorylation sites disrupted PHF8 dissociation from chromatin and increase in H4K20me1 levels that occurred in colcemid-treated cells (Fig. 4d). Thus, PHF8 phosphorylation at both serine 33 and 84 appears to be required for triggering its dissociation from chromatin and accumulation of H4K20me1 levels in prophase. In accord with the possibility that PHF8 dissociation from chromatin could potentially participate in Condensin II loading, M phase-synchronized cells transfected with an empty vector or wild type PHF8, but not mutant PHF8, exhibited significant increase of chromatin-associated Condensin II components compared with asynchronized cells (Fig. 4e).
The Condensin II complex from HeLa mitotic extracts was selectively pulled down by H4K20me1 and weakly by H4K20me2, but not by other histone tails tested (Fig. 4f and Fig. S15a). No interaction was detected between H4K20me1 and the non-SMC subunits in the Condensin I complex or one component in the Cohesin complex, Rad21, as similarly found for Condensin II from asynchronized cells (Fig. S15b). Furthermore, purified Condensin II bound preferentially to mononucleosomes mono-methylated at H4K20 (Fig. S15c). We then asked which subunit in Condensin II could mediate these interactions. As shown in Fig. 4g, N-CAPD3 strongly and N-CAPG2, to a less extent, interacted with H4K20me1 histone tails, but not the other three subunits. Little, if any, interaction was detected with H4K20me3 (Fig. S16a). Since none of the subunit in Condensin II contains known motifs associating with H4K20me1, we hypothesized that a distinct motif harbored in N-CAPD3 and N-CAPG2, the HEAT repeat27–30, might be responsible for these events (Fig. S16b). Indeed, the three predicted HEAT repeat clusters in N-CAPD3 and N-CAPG2 exhibited interactions with H4K20me1 histone tails, with the ones from N-CAPD3 showing stronger affinity than that from N-CAPG2. In contrast, minimal, if any, interaction with H4K20me3 was detected (Fig. 4h, lane 2, 3 and 4). As control, H4K20me1 histone tails failed to pull down N-CAPD3 C-terminus (Fig. 4h, lane 1). Furthermore, ChIP-Seq. analysis found SMC4 and H4K2ome1 co-localized predominately at intra- and extra-genic regions (Fig. 4i), with ~55% of SMC4-bound regions occupied by H4K20me1 (Fig. 4j). The specificity of a number of SMC4 binding sites was validated by ChIP-qPCR (Fig. S17b, c). Given that SMC4 is present in both Condensin I and II complexes, which have different distributions along chromosomes27, the ~55% overlap of SMC4 and H4K20me1 loci supports the specific association between H4K20me1 and Condensin II.
In conclusion, our study has revealed that PHF8 acts as a cell cycle regulator, at least partially based on its H4K20me1 demethylase activity (Supplementary Fig. 1). The absence of PHF8 leads to a delay in G1/S transition and its dissociation from chromatin in early mitosis, which in conjunction with increased expression of Pr-Set7, leads to a surge of the H4K20me1 mark capable of interacting with the Condensin II complex through the HEAT repeat clusters harbored in two non-SMC subunits, N-CAPD3 and N-CAPG2.
Methods
PHF8 protein purification and Demethylation assay
Flag-PHF8 proteins were expressed in HEK293T cells and cells were lysed in lysis buffer containing 50mM Tris HCl, pH 7.4, 450mM NaCl, 1mM EDTA, 1% TRITON X-100 followed by sonication. High salt concentration and sonication were applied here due to PHF8 is mainly and tightly associated with chromatin. Flag-PHF8 proteins were then affinity purified by using Anti-flag M2-agarose as described in the technical bulletin (Sigma A2220) and washed extensively. Before elution with 3X Flag peptides (Sigma), the affinity resins were washed with demethylation buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 50 mM [NH4]2Fe[SO4]2, 1 mM α-ketoglutarate, and 2 mM ascorbic acid) twice. His-tagged PHF8 proteins were expressed in BL21 (DE3) bacterial cells (Stratagene) and purified by using Ni-NTA Agarose (Qiagen). For demethylation reactions, PHF8 proteins were incubated with 5μg of bulk histones (Sigma H9250) or mononucleosomes prepared from HeLa cells in demethylation buffer at 37 °C.
Constructs, antibodies and other methods are described in the Supplementary Methods.
Supplementary Material
Acknowledgments
We thank Dr. Winship Herr for generously providing HCF-1 and Set1A antibody; Dr. Angus, C. Wilson for generously providing pCGN-GAL4-HA-HCF-1(N1011) and pCGN-GAL4-HA-HCF-1(C600) expression vectors; Dr. Angus I. Lamond for generously providing YFP-HCF-1 expression vector; Kaki Wang for experimental assistance; Justin Nand for assistance with the ChIP-Seq. data analysis; C. Nelson for cell culture assistance; J. Hightower and D. Benson for assistance with figure and manuscript preparation and the UCSD BIOGEM laboratory for RNA profiling. M.G.R. is an investigator with the Howard Hughes Medical Institute. This work was supported by grants from NIH and NCI to A.K.A., P.D., C.K.G, A.D., and M.G.R. and from DOD and PCF to M.G.R. We apologize that we were not able to cite all the studies in the primary references characterizing PHF8 family enzymatic activities while our work was under review, due to Reference limitation.
Footnotes
Supplementary information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions W.L. and M.G.R. designed the experiments and W.L. performed most of the experiments. W.L. and M.G.R. prepared the manuscript with contributions of I.G.B., A.K.A., A.D., P.C.D. and C.K.G. B.T. and C.B. analyzed the ChIP-Seq. and microarray data. T.Z and K.A.O generated Flag-PHF8 constructs. R. G. performed confocal microscopy in mitosis. I.G.B performed gel filtration chromatograph.
Author information Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests. Correspondence and requests for materials should be addressed to M.G.R (mrosenfeld@ucsd.edu).
References
- 1.Kouzarides T. Histone methylation in transcriptional control. Curr Opin Genet Dev. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- 2.Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol. 2007;14:1008–16. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- 3.Lachner M, O’Sullivan RJ, Jenuwein T. An epigenetic road map for histone lysine methylation. J Cell Sci. 2003;116:2117–24. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- 4.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–94. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7:715–27. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 6.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–12. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mellor J. It takes a PHD to read the histone code. Cell. 2006;126:22–4. doi: 10.1016/j.cell.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 8.Loenarz C, et al. PHF8, a gene associated with cleft lip/palate and mental retardation, encodes for an Nepsilon-dimethyl lysine demethylase. Hum Mol Genet. 19:217–22. doi: 10.1093/hmg/ddp480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fortschegger K, et al. PHF8 targets histone methylation and RNA polymerase II to activate transcription. Mol Cell Biol. doi: 10.1128/MCB.01520-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horton JR, et al. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat Struct Mol Biol. 17:38–43. doi: 10.1038/nsmb.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kleine-Kohlbrecher D, et al. A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol Cell. 38:165–78. doi: 10.1016/j.molcel.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng W, Yonezawa M, Ye J, Jenuwein T, Grummt I. PHF8 activates transcription of rRNA genes through H3K4me3 binding and H3K9me1/2 demethylation. Nat Struct Mol Biol. 17:445–50. doi: 10.1038/nsmb.1778. [DOI] [PubMed] [Google Scholar]
- 13.Jorgensen S, et al. The histone methyltransferase SET8 is required for S-phase progression. J Cell Biol. 2007;179:1337–45. doi: 10.1083/jcb.200706150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tardat M, Murr R, Herceg Z, Sardet C, Julien E. PR-Set7-dependent lysine methylation ensures genome replication and stability through S phase. Cell Biol. 2007;179:1413–26. doi: 10.1083/jcb.200706179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houston SI, et al. Catalytic function of the PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for mitotic entry and genomic stability. J Biol Chem. 2008;283:19478–88. doi: 10.1074/jbc.M710579200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin Y, Yu VC, Zhu G, Chang DC. SET8 plays a role in controlling G1/S transition by blocking lysine acetylation in histone through binding to H4 N-terminal tail. Cell Cycle. 2008;7:1423–32. doi: 10.4161/cc.7.10.5867. [DOI] [PubMed] [Google Scholar]
- 17.Oda H, et al. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol. 2009;29:2278–95. doi: 10.1128/MCB.01768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tyagi S, Chabes AL, Wysocka J, Herr W. E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol Cell. 2007;27:107–19. doi: 10.1016/j.molcel.2007.05.030. [DOI] [PubMed] [Google Scholar]
- 19.Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 2003;17:896–911. doi: 10.1101/gad.252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trojer P, et al. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell. 2007;129:915–28. doi: 10.1016/j.cell.2007.03.048. [DOI] [PubMed] [Google Scholar]
- 21.Julien E, Herr W. Proteolytic processing is necessary to separate and ensure proper cell growth and cytokinesis functions of HCF-1. Embo J. 2003;22:2360–9. doi: 10.1093/emboj/cdg242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Losada A, Hirano T. Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev. 2005;19:1269–87. doi: 10.1101/gad.1320505. [DOI] [PubMed] [Google Scholar]
- 23.Hudson DF, Marshall KM, Earnshaw WC. Condensin: Architect of mitotic chromosomes. Chromosome Res. 2009;17:131–44. doi: 10.1007/s10577-008-9009-7. [DOI] [PubMed] [Google Scholar]
- 24.Belmont AS. Mitotic chromosome structure and condensation. Curr Opin Cell Biol. 2006;18:632–8. doi: 10.1016/j.ceb.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 25.Haering CH. Foreword: the many fascinating functions of SMC protein complexes. Chromosome Res. 2009;17:127–9. doi: 10.1007/s10577-008-9008-8. [DOI] [PubMed] [Google Scholar]
- 26.Wolf F, Sigl R, Geley S. ‘... The end of the beginning’: cdk1 thresholds and exit from mitosis. Cell Cycle. 2007;6:1408–11. [PubMed] [Google Scholar]
- 27.Ono T, et al. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell. 2003;115:109–21. doi: 10.1016/s0092-8674(03)00724-4. [DOI] [PubMed] [Google Scholar]
- 28.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 29.Neuwald AF, Hirano T. HEAT repeats associated with condensins, cohesins, and other complexes involved in chromosome-related functions. Genome Res. 2000;10:1445–52. doi: 10.1101/gr.147400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andrade MA, Bork P. HEAT repeats in the Huntington’s disease protein. Nat Genet. 1995;11:115–6. doi: 10.1038/ng1095-115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.