Abstract
Stroke is an age-related disease. Recovery after stroke is associated with axonal sprouting in cortex adjacent to the infarct. The molecular program that induces a mature cortical neuron to sprout a new connection after stroke is not known. We selectively isolated neurons that sprout a new connection in cortex after stroke and compared their whole-genome expression profile to that of adjacent, non-sprouting neurons. This ‘sprouting transcriptome’ identified a neuronal growth program that consists of growth factor, cell adhesion, axonal guidance and cytoskeletal modifying molecules that differed by age and time point. Gain and loss of function in three distinct functional classes showed new roles for these proteins in epigenetic regulation of axonal sprouting, growth factor–dependent survival of neurons and, in the aged mouse, paradoxical upregulation of myelin and ephrin receptors in sprouting neurons. This neuronal growth program may provide new therapeutic targets and suggest mechanisms for age-related differences in functional recovery.
Stroke is an age-related disease and the leading cause of adult disability. Reorganization of motor, sensory and language functions in cortical regions in the brain hemisphere near the stroke are the events most closely correlated with behavioral recovery1. Stroke induces axonal sprouting and the formation of new patterns of neuronal connections within these peri-infarct regions2–4. An understanding of the molecular events that underlie axonal sprouting in the adult brain after stroke, and that are affected by age, may identify new targets that promote recovery in this disease. Molecular studies of axonal sprouting in CNS injury have focused mostly on proteins that inhibit the formation of new connections, such as myelin-associated proteins and glial extracellular matrix molecules5,6. However, therapies that only block axonal growth inhibitors may not produce maximal axonal sprouting and recovery after CNS injury7. Combined therapies that both stimulate a neuronal growth state and block axonal growth inhibitors produce more robust axonal sprouting and functional recovery after stroke8.
A gene expression program that underlies a neuronal growth state after stroke has not been determined. Studies of a molecular neuronal growth program have been limited because there has been no way to selectively label and isolate the small subset of cortical neurons that undergo axonal sprouting after stroke from the larger population of neurons that do not. In contrast, the transcriptional profile of a neuronal growth state has been identified in gene expression studies of axonal sprouting in the dorsal root ganglion and retinal ganglion cells after injury9–12. Gene expression studies of neurons in cortex contralateral to stroke, some of which sprout new spinal connections, have shown activation of an injury-associated molecular program8. However, there have been no studies of the molecular mechanisms of axonal sprouting in neurons in peri-infarct cortex or of the specific gene expression profile of a sprouting neuron after adult CNS injury.
The present studies develop a method to selectively label sprouting neurons in peri-infarct cortex after stroke. This approach has the methodological advantages that the sprouting neurons are compared to non-sprouting neurons from the same rat, from the same brain region and exposed to the same stroke conditions. The results identified a sprouting or regeneration transcriptome after stroke that changed with time after injury, differed between young and aged rats and mice, and contained molecules that have different mechanistic roles in the process of brain reorganization after stroke.
RESULTS
Gene expression profile of sprouting neurons after stroke
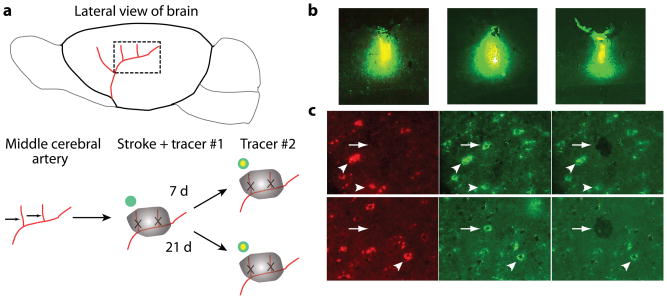
Stroke was produced in the whisker (barrel) field of the somatosensory cortex in young adult and aged rats13, which induces a region of axonal sprouting in peri-infarct sensorimotor cortex2,14. To label neurons that sprout and establish a new projection pattern in peri-infarct cortex, we injected two different fluorescent conjugates of the tracer cholera toxin B (CTB) into forelimb sensorimotor cortex. CTB back-labels neurons that project to the injection site. Alexa 488–CTB was injected at the time of the stroke; 7 or 21 d later Alexa 647–CTB was injected at the same site as Alexa 488–CTB, but at two-thirds the volume (Fig. 1a). We used for gene expression analysis only those rats with injection sites in which the second tracer injection was entirely within the first (Fig. 1b) (n = 21 in total). Neurons that were double-labeled from the two tracer injections (Fig. 1c) were those that did not change their projection to the injection site after stroke. Neurons that took up only the second tracer (Fig. 1c) were those that had not had an axonal projection to the injection site at the time of the injection of the first tracer15,16, and thus established a new projection pattern after stroke. We laser-captured neurons in peri-infarct cortex if they were double-labeled (non-sprouting) or were labeled by tracer 2 only (sprouting) (Fig. 1c, Supplementary Discussion and Supplementary Fig. 13). Whole-genome expression analysis was used to identify the distinct transcriptional profile of a sprouting neuron in peri-infarct cortex after stroke. With a P-value threshold of < 0.005 and after filtering for a minimum log2-transformed fold change of 0.2, most differential gene regulation in axonal sprouting after stroke had taken place by day 7, and more genes were differentially expressed in young adults: in young adult at 7 d, 804 genes were upregulated and 542 were downregulated; in young adult at 21 d, 572 were upregulated and 164 downregulated; in aged adults at 7 d, 384 were upregulated and 237 downregulated; in aged adults at 21 d, 157 genes were upregulated and 76 downregulated (Supplementary Tables 1, 2, 4 and 5). There were substantial differences between those genes induced in sprouting neurons in aged versus young adult at each time point. Only 79 genes were upregulated in common in both aged and young adult at day 7, and 18 genes were upregulated in common at day 21 (Supplementary Tables 4 and 5).
Figure 1.
Experimental approach and laser capture microdissection of sprouting neurons after stroke. (a) Stroke was produced by permanently occluding two anterior branches of the distal middle cerebral artery over the parietal cortex and transiently occluding bilateral jugular veins. Two different fluorescent conjugates of the tracer cholera toxin B subunit (CTB) were sequentially injected into the same site of forelimb sensorimotor cortex, the first at the time of stroke (Alexa 488–CTB) and the second at 2/3 the volume either 7 or 21 d after stroke (Alexa 647–CTB). Neurons that project to this forelimb sensorimotor site at the time of stroke are labeled with Alexa 488–CTB; neurons that establish a projection to this site only after stroke are labeled solely with Alexa 647–CTB. (b) Injection sites from three separate cases. Only mice in which the second tracer injection was entirely within the first were used for further analysis. (c) Young adult sprouting neurons are shown in the top row and sprouting neurons from an aged brain are shown in the bottom row. The left column shows neurons labeled by the first tracer injection; the middle column neurons labeled by the second tracer injection; the right column neurons seen after laser capture of a tracer 2-only neuron. Neurons with double label (non-sprouting neurons; arrowheads) or labeled by CTB-Alexa 647( tracer-2 only, sprouting neurons; arrows) were separately laser-captured in the peri-infarct cortex. Scale bars, 50 μm (c) and 10 μm (b).
We focused further analysis on genes that were significantly regulated in sprouting neurons early after stroke, at day 7, as these are likely to be involved in the initiation of an axonal sprouting molecular program. Grouped by functional category, sprouting neurons in the young adult at day 7 regulate many GTPases, particularly in the RhoA and Ras pathways; axonal guidance genes; cytoskeletal modifying genes; and epigenetic or DNA-modifying genes (Table 1 and Supplementary Fig. 1). Many of the genes activated in sprouting neurons after stroke have been linked to axonal sprouting or axonal pathfinding in neurodevelopment, including MHC class I molecules, axonal guidance receptors such as neuropilin and L1 cell adhesion molecule (L1cam), complement (both C3 and C1 classes), and cytoskeletal modifying proteins such as stathmins (Stmn3 and Stmn4) (Table 1). However, stroke in young adults activated molecules that have not been linked to axonal sprouting previously, including epigenetic or DNA-modifying proteins such as histone deacetylase-4 (Hdac4), doublecortin-like kinases and several relatively uncharacterized GTPases (Table 1). Compared with the gene expression profile of sprouting neurons at day 7 in the young adult, aged sprouting neurons showed a greater activation of immune-related genes, bone morphogenic family genes, insulin-like growth factor-1 (Igf1) and osteopontin (Spp1), and a distinct set of molecules that participate in axonal growth inhibition, including the receptor tyrosine kinase ephrin type-A receptor 4 (Epha4) and leucine rich repeat and Ig domain containing 1 (Lingo1) (Table 2 and Supplementary Fig. 1). Molecular pathway analysis identified several key signaling networks, from cell surface to nuclear proteins, in aged and young adult sprouting neurons (Supplementary Fig. 2 and Supplementary Discussion).
Table I.
Genes Differentially Regulated in Sprouting Neurons at Day 7 in Young Adult
| Axonal Outgrowth/Guidance |
| Neuropilin 1 |
| Neuropilin and toll-like 2 (Neto2) |
| DEK oncogene |
| tyrosine ligase-like family, member 7 |
| Reticulon 4 receptor-like 1/NgR3 (down) |
| COUP-TFI/NR2F1 (down) |
| L1CAM (down) |
| Calcium signaling/homeostasis |
| Stromal interaction molecule 2 |
| Neurocalcin delta |
| Calmodulin 3 |
| Calreticulin |
| Calpactin |
| Intracellular phosphorylation cascades |
| Protein tyrosine phosphatase, non-receptor type 11 |
| Protein tyrosine phosphatase, receptor type, D (Ptprd) |
| Protein tyrosine phosphatase, receptor type, E (Ptpre) |
| Protein phosphatase 1A, magnesium dependent, alpha isoform (Ppm1a), |
| PLCXD3 |
| Dual specificity protein phosphatase 3 |
| Serine/threonine protein phosphatase PP1-beta |
| MAP-kinase activating death domain |
| Phosphatase and tensin homolog |
| JNK2 |
| MAPK9 |
| Protein tyrosine phosphatase, non-receptor type 1 (Ptpn1) (down) |
| Inositol 1,4,5-triphosphate receptor 1 (Itpr1) (down) |
| Dual specificity phosphatase 5 (Dusp5) (down) |
| Protein tyrosine phosphatase 2E (Ptpn21) (down) |
| Cell Surface Receptors |
| CD164 |
| integrin alpha-6 |
| UDP-glucose ceramide glucosyltransferase |
| Tetraspanin 5 |
| SNX6 sorting nexin 6 |
| CD47 |
| Cadherin 2 (N-cadherin) |
| Protocadherin 7,9,19 (19 down) |
| Extracellular matrix |
| Osteopontin |
| Laminin-5beta3 |
| Heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| Heparan sulfate (glucosamine) 3-O-sulfotransferase 2 |
| Matrilin 2 |
| Growth Factors |
| Lip1 |
| BCATc |
| Platelet derived growth factor, alpha |
| Fgf13 (down) |
| Bex1 (down) |
| LIM motif-containing protein kinase 2 (Limk2) (down) |
| Insulin-like growth factor binding protein 5 (down) |
| GTPases/G protein coupled receptors |
| Rabaptin 5 |
| Rab2 |
| RanBP1 |
| RAB6a |
| Ras homolog gene family, member E (Arhe) |
| Rab12 |
| Rab26 |
| Rab22a |
| Afadin |
| Rhoa |
| Rab11a |
| Rab7a |
| Gpr83 (down) |
| Septin 5 (down) |
| Rab9b (down) |
| Ras homolog enriched in brain like 1 Rhebl1 (down) |
| RGS4 (down) |
| Latrophilin 3 (down) |
| GTPase activating protein testicular GAP1 (down) |
| T-cell lymphoma invasion and metastasis 2 (Tiam2) (down) |
| Connector enhancer of kinase suppressor of Ras 2 (down) |
| G protein-coupled receptor 22 (down) |
| Rho family GTPase 1 (Rnd1) (down) |
| RAS, dexamethasone-induced 1 (Rasd1) (down) |
| Gpr149 G protein-coupled receptor 149 (down) |
| Guanylate cyclase 1, soluble, alpha 3 (down) |
| MCH1/immune/complement |
| C1nh/Serping 1 |
| Complement C3 |
| CD46 |
| RT1 class Ib, locus Cl (RT1-Cl) (MHCI) |
| RT1 class Ib, locus S3 (RT1-S3) (MHCI) |
| RT1 class I, CE4 (RT1-CE4) (MHCI) |
| Adipsin/complement factor D |
| Complement receptor related protein (Crry) (down) |
| Ubiquitin/proteosome |
| Usp32 |
| Praja ring finger 2 PJA2 |
| HECT type E3 ubiquitin ligase (down) |
| SNF2 histone linker PHD RING helicase (down) |
| Ubiquitin-conjugating enzyme E2 Harrier protein (down) |
| Cytoskeletal protein or trafficking/migration |
| APC |
| Adducing 3 gamma |
| Kif1a |
| Actin, gamma 2 |
| SFRS protein kinase 2 |
| Stathmin 3/SCLIP |
| Stathmin 4/RB3 |
| Protein kinase A anchoring protein 11 |
| calsyntenin 1 |
| A kinase anchor protein 13 |
| Tumor protein, translationally-controlled 1 |
| Kelch domain containing 3 (Klhdc3) |
| Olfactomedin 3/Optimedin |
| Peroxiredoxin 1 |
| Destrin |
| Doublecortin-like kinase 1 |
| Doublecortin and calcium/calmodulin-dependent protein kinase-like 1 |
| Cyclin-dependent kinase inhibitor 1B |
| Kinesin family member 5B |
| Microtubulue associated protein 4 |
| Actin related protein 2/3 complex, subunit 4 |
| Desmoglein 1 gamma |
| Actin-related protein 3 (down) |
| Retinitis pigmentosa 2 (down) |
| Kinesin-like 7 |
| Kelch-like 23 (down) |
| Transcription factors |
| Bcl11b/Ctip2 |
| Gro/TLE |
| Kruppel-like factor 7 |
| Nuclear receptor subfamily 4, group A, member 2 |
| Janus kinase 1 |
| Mesoderm development candidate 2 |
| Mesoderm induction early response 1 |
| TSC-22 |
| Recombining binding protein suppressor of hairless |
| Nuclear factor I/X (Nfix) |
| Id3 |
| Smad1 |
| Luc7l |
| Stat3 |
| Activating transcription factor 2 |
| Peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| MED10--mediator complex subunit 10 |
| LIM domain only 2 |
| Palmdelphin (down) |
| Smad 5 (down) |
| Egr 2 (down) |
| Neuron specific/related |
| Purkinje cell protein 4 |
| Snap 25a |
| mGluR3 |
| Ganglioside-induced differentiation-associated-protein 1 amyloid precursor protein |
| Synaptotagmin 12 |
| Neuronal regeneration related protein |
| Glutamate receptor, ionotropic, |
| AMPA1 (alpha 1) |
| Complexin 1, Complexin 2 |
| Receptor expression enhancing protein |
| Pantothenate kinase 1 |
| integral membrane protein 2B (Itm2b) vesicle-associated membrane protein, associated protein B and C |
| Neurexin II (down) |
| Neurobeachin (down) |
| adenylate cyclase activating polypeptide 1/PACAP (down) |
| cholinergic receptor, muscarinic 3 (down) |
| neuropeptide Y receptor Y5 (down) |
| synaptotagmin XVII (down) |
| Cytokines/Chemokines |
| CXCL10 |
| CXCL13 |
| TNFAIP2 tumor necrosis factor, alpha-induced protein 2 |
| TIGIRR-1 |
| CD40/Tnfrsf5, transforming growth factor, beta 3 TGFb3 (down) |
| TNF receptor superfamily, member 11b (osteoprotegerin) (down) |
| Epigenetic or DNA modifying |
| Hdac4 |
| Dre1 |
| Nucleolin |
| Replication factor C |
| Kin KIN17 |
| Cyclin G1 |
| REST corepressor 1 |
| NAP1L1 |
| Never in mitosis gene a-related kinase 1 |
| Nucleoporin 88 (down) |
| EP300 (down) |
All genes upregulated in sprouting neurons vs. non-sprouting neurons unless noted as (down).
Table 2.
Genes differentially regulated in sprouting neurons at day 7 in aged adult vs. young adult
| Chemokines/Cytokines |
| Mip1a/CCL3 |
| Mip1b/CCL4 |
| Mip2/CXCL2 |
| Interleukin 1 receptor antagonist |
| IL-27 |
| Bone morphogenic proteins |
| Growth differentiation factor 10 |
| ONECUT2 |
| Bone morphogenic protein 1 |
| Cytoskeletal proteins |
| Filamin A interacting protein |
| Supervillin |
| Actin related protein 2/3 complex, subunit 5-like |
| Actin-related protein 2 |
| Tyrosine ligase-like family member 7, transcript variant 2 |
| Cofilin 1 |
| Plastin 3 (down) |
| Calsyntenin (down) |
| Neurofilament heavy chain (down) |
| Alpha catenin (down) |
| Synaptogenesis Proteins |
| Piccolo |
| Synaptotagmin 1 |
| Synaptic vesicle glycoprotein 2b |
| Synaptophysin (down) |
| Protocadherin X |
| Adhesion molecules |
| Osteopontin |
| BEN/Alcam/CD166/Sc1 |
| Glypican 3 |
| CEA-related cell adhesion molecule 10 |
| Neurexin 1 |
| Cadherin 8 |
| Polyamine pathway |
| Spermidine/spermine N1-acetyl transferase |
| Arginase II (down) |
| Growth Factors |
| IGF-1 |
| Epidermal growth factor receptor (Egfr) |
| Glial cell line derived neurotrophic factor family receptor |
| alpha 3 |
| SORCS3 |
| Axonal Guidance |
| EphA4 |
| Lingo-1 |
| Kinases/Signaling Enzymes |
| PKCdeleta |
| Phospholipase C, gamma 2 (Plcg2) |
| Regucalcin (down) |
| Neuronal plasticity |
| Homer (down) |
| ARC (down) |
| Epigenetic or DNA modifying |
| ATRX |
| HDAC11 |
| Transcription Factors |
| Prrx1 |
| Lim Homeobox 9 |
| B cell translocation gene 1 |
| Janus kinase 1, Janus kinase 2 |
| Ring finger protein 29 (Rnf29) |
| Zinc finger protein 560 |
| E2F transcription factor 1 (E2f1) |
| General transcription factor IIa 2 (Gtf2a2) |
| Recombining binding protein suppressor of hairless |
| Egr1 (down) |
| Neuron-derived orphan receptor 1 (down) |
| Atf5 (down) |
| GTPases |
| Sec5l1 |
| Chimaerin 1 |
| Rho GDP dissociation inhibitor (GDI) alpha |
| Rho, GDP dissociation inhibitor (GDI) beta |
| Guanylate cyclase 1, soluble, alpha 3 |
| Mas1 oncogene (down) |
| Rab2b (down) |
| Immune Related |
| Toll-like receptor 2 |
| CD86 |
| Interferon gamma receptor 2 |
| MHC1RT1 class Ib_ locus S3 (RT1-S3) (down) |
mRNA and Protein Confirmation of Induced Genes
We tested the expression profiles of a subset of the genes significantly regulated in the microarray studies with quantitative, real-time reverse-transcription(qRT)-PCR. Fourteen of 21 genes that were upregulated in aged sprouting neurons versus non-sprouting neurons at day 7 in the array data set were also upregulated by qRT-PCR: Alcam (activated leukocyte cell adhesion molecule), Atrx (α-thalassemia/mental retardation syndrome X-linked), Ceacm10 (carcinoembryonic antigen-related cell adhesion molecule 10), chimaerin-1 (chimerin-1), Gdf10 (growth differentiation factor 10), Gpc3 (glypican-3), Igf1, Melk (maternal embryonic leucine zipper kinase), Nrxn1 (neurexin I), Sat2 (spermidine acetyl transferase, PRRX1 (Paired mesoderm homeobox protein 1), Spp1 (osteopontin), Svil (supervillin) and TPT1 (translationally-controlled tumor protein) (Table 2). This result is consistent with previous studies17. Igf1 and osteopontin were the most differentially regulated genes in aged sprouting neurons, with 20- and 12-fold induction compared to the control expression level of non-sprouting neurons.
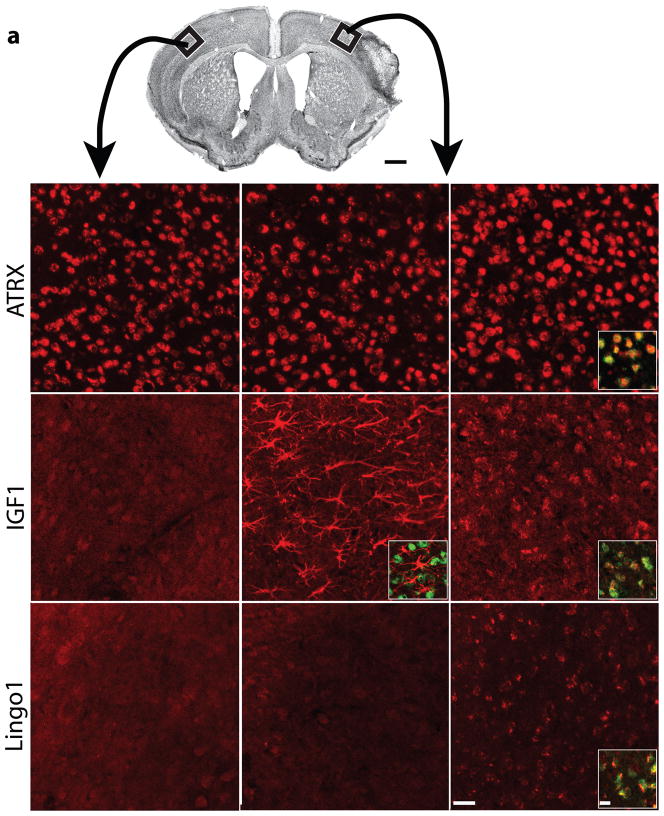
To confirm the changes in gene expression from microarray studies, and to localize these to specific cells in peri-infarct cortex, we performed multilabel immunofluorescence studies at day 7 after stroke using both aged and young adult mice and testing protein products of genes significantly induced in aged sprouting neurons: ATRX, IGF1, Lingo1, GDF10 and osteopontin. ATRX immunoreactivity was present in similar intensity in neuronal nuclei in normal and young adult peri-infarct cortex (Fig. 2b,c), but had greater immunoreactivity in aged peri-infarct neurons (Fig. 2d and Supplementary Fig. 3n,o). After stroke, IGF1 was induced in glial fibrillary acidic protein (GFAP)-positive reactive astrocytes in peri-infarct cortex of the young adult18 (Fig. 2f and Supplementary Fig. 3i,j). Unexpectedly, stroke in aged mice induced IGF1 mostly in cells positive for the neuronal marker NeuN in peri-infarct cortex (Fig. 2g and Supplementary Fig. 3d,e). Lingo1 immunoreactivity was stronger in NeuN-positive cells in aged peri-infarct cortex (Fig. 2j and Supplementary Fig. 3m,t) than in those in young adult (Fig. 2i) and control cortex (Fig. 2h). GDF10 localized to NeuN-positive neurons in peri-infarct cortex and had greater staining intensity in aged peri-infarct neurons than in young adult peri-infarct neurons (Supplementary Figs. 3s and 4i,l). Osteopontin staining was present in the apical dendrites of neurons in control and peri-infarct cortex and localized to cells positive for the microglial and macrophage marker Iba1; the intensity of immunoreactive staining was stronger in aged peri-infarct tissue than that in the young adult peri-infarct cortex (Supplementary Fig. 5). These data, together with the array and qRT-PCR studies, indicate that neurons upregulate ATRX, GDF10, Lingo1 and IGF1 in the aged brain after stroke more than they do in the young adult, and that IGF1 induction after stroke switches from astrocytes in young adults to neurons in the aged brain.
Figure 2.
Cellular pattern of ATRX, IGF1 and Lingo1 expression in the brain after stroke. (a) Nissl-stained photomicrograph of mouse barrel field stroke model. Boxes indicate region of contralateral cortex (left column) and peri-infarct cortex in young adult (middle column) and aged adult (right column). (b–j) Panels for ATRX (b–d), IGF1 (e–g) and Lingo1 (h–j) show immunoreactivity in each condition. Insets: Colocalized staining of ATRX (red) and NeuN (green) is seen as yellow (d). IGF1 staining (red) localizes to astrocytes and not neurons (green) in young adult peri-infarct cortex (f). In aged adult peri-infarct cortex, IGF1 (red) co-localizes to NeuN positive neurons (green) (g). Lingo1 staining (red) localizes to NeuN positive neurons (green) in aged peri-infarct cortex (j). Scale bars, 1 mm (a), 50 μm (b–d) and 20 μm (insets).
ATRX mediates axonal sprouting after stroke
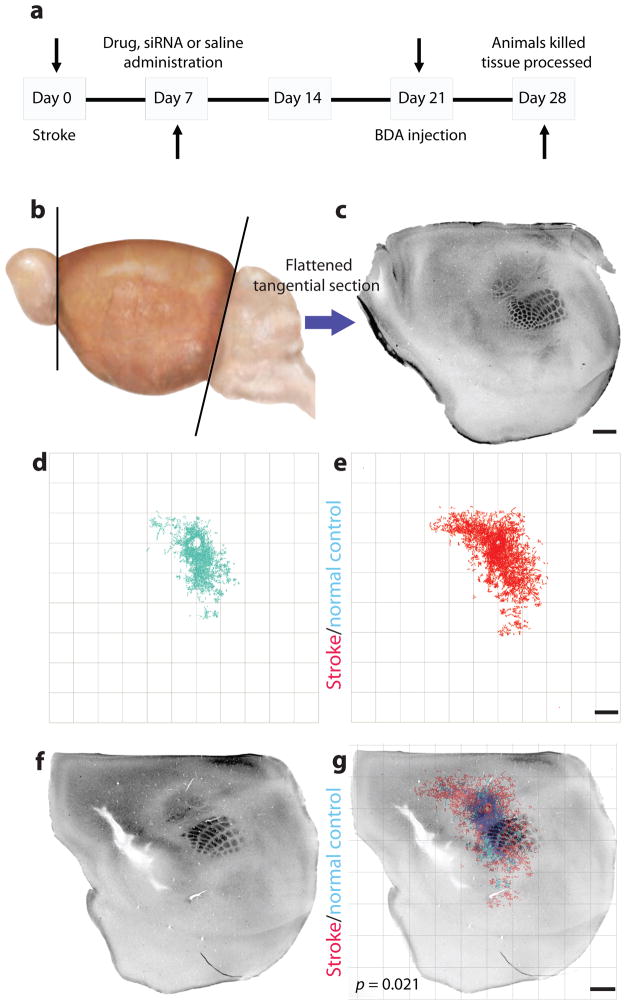
To identify the mechanistic roles of select members of this sprouting-related gene set, we tested the in vivo effect of gain or loss of function on connections in three systems that belong to distinct molecular groups: epigenetic DNA modification (ATRX), soluble growth factor (IGF1) and axonal growth inhibition (Nogo) systems. To determine the role of these three gene systems in axonal sprouting after stroke in vivo, we applied a quantitative connectional mapping technique to statistically compare the motor cortical connections of peri-infarct cortex in control, stroke and stroke plus gain- or loss-of-function conditions (Fig. 3 and Supplementary Discussion). In all experimental groups below, there was no difference in stroke size, tracer injection size or location (Supplementary Figs. 6 and 7).
Figure 3.
Quantitative connectional mapping. (a) Timeline of experimental design for all in vivo axonal tracer experiments. Mice received a sham surgery or stroke, followed 1 week later by siRNA or drug delivery. Three weeks later biotinylated dextran amine (BDA) was microinjected into forelimb motor cortex. (b,c) The cortex was then removed, flattened, tangentially cut (b) and stained for cytochrome oxidase (c) and BDA in the same sections. (d,e)The location of BDA-labeled axons was digitally mapped in x, y coordinates relative to the injection site. (d) The quantitative connectional map of forelimb sensorimotor connections in sham-operated animals (n = 5). (e) The quantitative connectional map of forelimb sensorimotor cortex connections after stroke (n = 7). (f) Cytochrome oxidase staining in layer IV identifies the mouse somatosensory body map2 after stroke. (g) Each connectional map was registered to the somatosensory body map from the same brain to produce a group connectional and functional map of cortex. Axonal sprouting was identified when a pattern of cortical connections was precisely mapped and statistically different across treatment conditions. Scale bars, 1 mm.
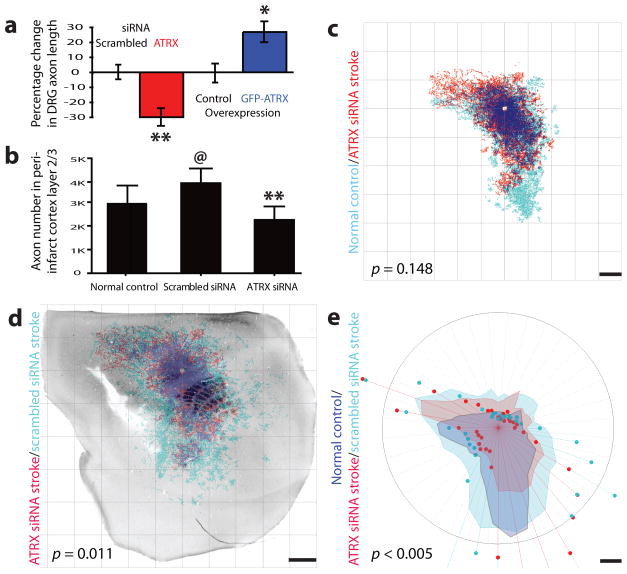
ATRX is an ATP-dependent DNA modifying enzyme that controls neuronal survival and migration during brain development19 but has not been implicated in axonal sprouting. Atrx small interfering RNA (Supplementary Fig. 9) was used to evaluate effects on axonal outgrowth in adult neurons. We used dorsal root ganglion neurons (DRG) to directly test the role of ATRX in axonal outgrowth because DRG neurons can be readily cultured from adult animals. ATRX knockdown with siRNA significantly but modestly inhibited axonal outgrowth of DRG neurons (P < 0.05 compared to scrambled control siRNA; Fig. 4a and Supplementary Fig. 9d). ATRX overexpression in adult rat DRG neurons promoted axonal outgrowth (P < 0.05 compared to the control; Fig. 4a and Supplementary Fig. 9e).
Figure 4.
ATRX function in post-stroke axonal sprouting. (a) Knockdown of ATRX by siRNA reduced axonal outgrowth in vitro compared to scrambled siRNA treatment. ATRX overexpression induced axonal sprouting compared to expression of GFP alone. (b) Axon numbers in normal control or peri-infarct cortex layer 2/3 with Atrx siRNA or scrambled siRNA. (c) Sham, non stroke forelimb motor cortex connections (blue, n = 10) compared to those from Atrx siRNA siRNA group (red, n = 12). The blue label indicates the position of axons projecting from motor cortex summed from the entire sample of mice in the sham, non-stroke condition. The red label indicates that position of axons from the entire sample of mice treated with Atrx siRNA. The dark blue is the areas of dense overlap of the two projection systems. There was no significant difference in the pattern of cortical connections between normal (non-stroke, sham operated (d) Quantitative connectional map of forelimb sensorimotor connections in stroke + scrambled siRNA (blue, n = 11) and stroke + ATRX siRNA (red, n = 12), registered to the body map of underlying cortex with stroke. Dark blue shows area of dense overlap of connections in the two conditions. There is a significant increase in motor cortex projections between stroke + scrambled siRNA and stroke + ATRX siRNA. (e) Polar plots of axonal label from studies in (d). Shaded regions represent 70th percentile of the distances of labeled axons from the injection site; weighted polar vectors represent the normalized distribution of the number of points in a given segment of the graph. There was a significant difference in distribution of cortical projections between stroke + Atrx siRNA (red) and stroke + scrambled siRNA (blue) and between stroke + scrambled siRNA (blue) and non-stroke vehicle (yellow), and no significant change between stroke + Atrx siRNA (red) and non-stroke vehicle (yellow) (n = 10, P > 0.05). Scale bars, 1 mm. Error bars, s.d.; @, * and ** indicate a significant difference compared to saline normal control, GFP control and scrambled siRNA stroke, respectively.
Axonal connections increased significantly within peri-infarct cortex after stroke in the normal stroke state and after scrambled siRNA delivery in mice (P < 0.05, Fig. 4e). Stroke alone significantly induced axonal sprouting in motor, premotor and somatosensory areas in peri-infarct cortex (Fig. 3d,e,g and Supplementary Discussion). This same pattern was seen upon scrambled siRNA delivery after stroke (Supplementary Fig. 8). ATRX knockdown in vivo after stroke blocked this post-stroke axonal sprouting response. There was a significant difference between the peri-infarct connections in stroke plus Atrx siRNA versus stroke plus scrambled siRNA (Fig. 4d,e), such that knockdown of ATRX produced a pattern of cortical connections in peri-infarct cortex that did not differ from the non-stroke (control) condition (Fig. 4e). Atrx siRNA infusion reduced slightly the number of axons that were labeled from forelimb motor cortex (Fig. 4b), indicating a possible role for this protein in the normal maintenance of cortical projections. Thus, stroke normally induces axonal sprouting in peri-infarct cortex. ATRX induces axonal sprouting in vitro and is necessary for the normal process of post-stroke axonal sprouting in vivo.
IGF1 maintains cortical neurons after stroke
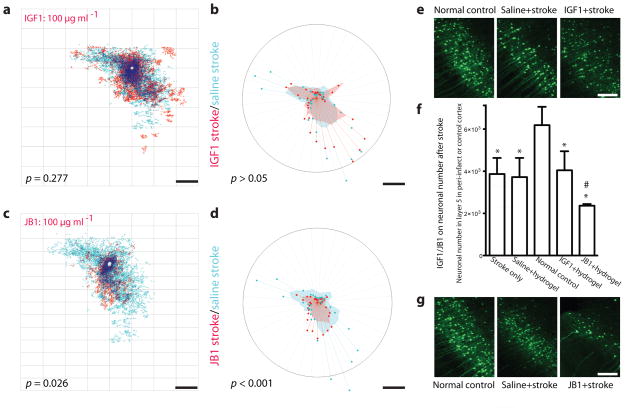
IGF1 has an established role in in vitro axonal outgrowth20 and in cortical development21. We delivered IGF1 within a biopolymer hydrogel from the infarct cavity, which produced sustained, local release of IGF1 over 4 weeks after implantation in vivo (Supplementary Fig. 10b). Quantitative connectional mapping showed that stroke plus IGF1 at either high or low doses did not induce a statistically significant change in cortical connections compared to stroke plus vehicle (Fig. 5a,b). Unexpectedly, the IGF1 antagonist JB1(ref. 22) produced a significant reduction in the distribution of cortical connections from forelimb motor cortex to below the spatial extent that existed after stroke alone (Fig. 5c,d) and below the normal pattern of forelimb motor connections (Fig. 4e).
Figure 5.
IGF1 maintains neuronal viability after stroke. (a) There was no significant difference in the pattern of cortical connections between stroke + saline (n = 5) and stroke + IGF1 (n = 7). (b) There was no significant change in the polar distribution of connections with IGF1 administration compared to stroke + saline (P > 0.05). (c) There was a significant loss of cortical connections between stroke + saline (n = 5) and stroke + JB1 (n = 8). (d) There was a significant decrease in polar distribution of connections with IGF1 blockade (P < 0.001). (e) Photomicrographs of neurons in peri-infarct cortex, showing no change in number across IGF1 treatment conditions. (f) (f) Effects of IGF1 and JB1 on neuronal number in layer 5 in normal control or peri-infarct cortex. IGF1 or JB1 were delivered beginning 7 days after stroke. Stroke (n = 6) induces neuronal cell death in peri-infarct cortex (p < 0.01). This neuronal death is not altered by delivery of IGF1 (n = 5, p > 0.05) or vehicle (n = 5, p > 0.05) into peri-infarct cortex. However, IGF1 blockade with JB1 + hydrogel significantly (n = 5, p < 0.05) increases cell death in peri-infarct cortex when compared to the saline + hydrogel (n = 5) –treated animals post-stroke. (g) Photomicrographs of neurons in peri-infarct cortex, showing decrease in number of neurons with JB1-induced IGF1 signaling blockade. Scale bars, 500 μm (c,g) and 1 mm (a,b,d,e). Note that the location of BDA tracer injection was more lateral in ATRX studies (Fig. 4) than in IGF1/JB1 and Lingo1-Fc/NgR1/NgR2 studies, the latter two of which use the same tracer coordinates; this produces slightly different patterns of intracortical connections. Error bars, s.d, *** p < 0.001 and ** p < 0.01 indicate a significant difference compared to normal control; # < 0.05 indicates a significant difference compared to stroke saline + hydrogel.
This collapse of cortical connections might occur because blockade of endogenous IGF1 signaling after stroke causes a retraction in the axonal arbors of cortical neurons or causes cell death23,24. JB1 delivery beginning 7 d after stroke produced significant neuronal cell death in peri-infarct cortex (Fig. 5). IGF1 delivery at this late stage after stroke did not affect neuronal number (Fig. 5e). IGF1 or JB1 delivery in the normal, non-stroke condition did not have any effect on neuronal cell number (Fig. 5f). Thus, stroke induces an unexpected and delayed neuronal dependency on IGF1 signaling for survival at periods of time well after the acute ischemic stimulus is over.
Nogo receptor complex-1 signaling restricts axonal sprouting after stroke
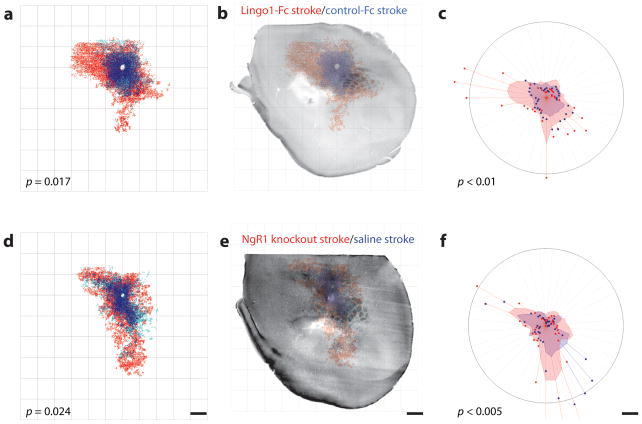
We mapped cortical connections following delivery of a Lingo1 function-blocking protein (Lingo1-Fc)25 and in Nogo receptor complex-1 (NgR1, Rtn4r, reticulon 4 receptor) knockout mice. In addition to the upregulation of Lingo1 after stroke in aged adults (Fig. 2d and Table 2), myelin-associated glycoprotein (MAG), a ligand for NgR1, NgR2, and PirB26,27, was also uniquely induced in the aged brain after stroke13. Because NgR2 (Rtn4rl2, reticulon 4 receptor-like 2) is a receptor selective for MAG, we also mapped the cortical connections of NgR2 knockout mice (Supplementary Fig. 12e).
Blockade of Lingo1 produced a significant induction in post-stroke axonal sprouting (P < 0.05, compared to control immunoglobulin G Fc, Fig. 6a–c), inducing new patterns of connections in premotor and motor cortex and in primary and secondary somatosensory areas (P < 0.001, Fig. 6b). There was also a significant increase in axonal connections in the NgR1 knockout after stroke, with a distribution of new axonal connections that is similar to that seen with Lingo1 blocakde(P < 0.05 compared to saline stroke, Fig. 6d–f). There was no significant difference in axonal sprouting between NgR2 knockout stroke and saline stroke groups (P > 0.05, Supplementary Fig. 12d). There was no difference in the pattern of connections from forelimb motor cortex between NgR1 and NgR2 knockout mice and C57BL/6 mice in non-stroke conditions (Supplementary Fig. 12a–c).
Figure 6.
Lingo1/NgR1 restricts cortical axonal sprouting after stroke. Quantitative connectional maps of forelimb motor connections with NgR1 signaling blockade after hydrogel delivery of Lingo1-Fc + stroke (n = 6) versus hydrogel control-Fc + stroke (n = 7). (a) Lingo1-Fc release into peri-infarct cortex induced a significantly different pattern of forelimb motor cortex connections. (b) Registration of motor cortical connections with the underlying cytochrome oxidase body map shows that Lingo1-Fc blocked the connections that form lateral and rostral to mouse somatosensory cortex, in secondary somatosensory cortex and motor cortex. (c) Polar analysis of forelimb sensorimotor connections from same maps. Lingo1-Fc induced a significant increase in motor cortex connections in the sites seen in the body map registration motor cortex, SI and SII (P < 0.001). (d). Quantitative connectional map of forelimb sensorimotor connections in NgR1 knockout mice with stroke (n = 5) versus control mouse stroke (n = 5). There was a significant increase in the pattern of cortical connections in NgR1 knockout. (e) Registering the underlying mouse body map to the motor cortical projections indicates that new projections could be seen in the NgR1 knockout in lateral SI, SII and to a lesser extent in motor cortex. (f) Polar analysis of forelimb motor connections in stroke + NgR1 knockout versus control. Significantly different connections were seen in motor and lateral somatosensory regions (P < 0.005). Scale bars, 1 mm.
DISCUSSION
Cortical plasticity after stroke is associated with recovery. Neurons in the peri-infarct region sprout new connections2,3. Cortical connections are normally show minimal overall change over the course of months in dendritic spines or axonal terminals even in the presence of sensory deprivation28,29. Axonal sprouting after stroke thus represents a phenotypic response to central injury: an adult neuron forms new connections within an environment that normally restricts this process. Here we have used a sequential tracer approach to selectively identify the gene expression profile of sprouting neurons after stroke in young and aged adults. Cortical neurons that sprouted after stroke activated a distinct gene expression profile, or ‘sprouting transcriptome’, which differed markedly in aged versus young adult sprouting neurons. Mechanistic studies of molecules in three distinct functional classes of this cortical sprouting transcriptome identified a new epigenetic regulatory protein in axonal sprouting after stroke, an unexpected growth factor dependency in the adult cortex after stroke and a paradoxical upregulation of inhibitory myelin receptors after stroke in aged neurons that limits axonal sprouting.
Sprouting transcriptome in cortical neurons after stroke
Sprouting neurons activated a distinct sprouting transcriptome that included both newly identified and established growth-associated genes. Sprouting neurons regulated axonal guidance molecules, including downregulating L1CAM and upregulating neuropilin-1, the structurally related molecule NETO2 and several MHC1 class I and complement genes. Neuropilin-1 transduces the growth cone–collapsing signals of semaphorin 3a, but also the axonal outgrowth–promoting signals of vascular endothelial growth factor (VEGF)30. Stroke induces both semaphorin 3a14 and VEGF31 within the region of sprouting neurons after stroke, placing both ligands near sprouting neurons with this induced neuropilin-1 receptor. MHC class I molecules have a role in activity-dependent axonal remodeling in the developing and adult brain, where they control topographic mapping in the retino-geniculate system and ocular dominance plasticity in visual cortex8. Other immune molecules also function in synaptic remodeling during development, including the complement proteins C3 and C1q32. C1q is activated in retinal ganglion8 and facial motor neurons33 during axonal sprouting, but not in sprouting neurons in cortex after stroke, pointing up differences in these cell populations during axonal regeneration. MHC class I and complement molecules provide a signal for synaptic elimination in development but may have a permissive role for axonal sprouting in adult CNS injury34.
Sprouting neurons activate many genes with a role in cytoskeletal modification, cell-cell adhesion and extracellular matrix interactions. In general, these gene products promote actin or microtubule cytoskeletal changes associated with cell migration or axonal outgrowth in development, and are likely to act similarly in the subcellular remodeling that occurs in sprouting neurons in peri-infarct cortex after stroke. Actin-related protein 2/3, destrin, cortactin, kelch domain containing-3, α-dystrobrevin and supervillin all interact with actin to influence actin cytoskeletal turnover, actin interactions with cell adhesion signals, or actin-associated motility. Integrin and integrin signaling molecules, induced in sprouting neurons after stroke, link extracellular matrix binding events to actin cytoskeleton remodeling and axonal outgrowth35,36. Sprouting neurons induce microtubule-modifying genes (Table 1), notably microtubule-associated protein 4, the neuronal polarity molecule tyrosine ligase-like family member 7 (TTLL7), stathmins and doublecortin-motif proteins. Stathmin-family proteins promote a more dynamic cytoskeletal structure that provides outgrowth10,37. TTL7 polyglutamylates tubulin and promotes neurite outgrowth38. In sprouting neurons many of these genes are associated with larger molecular systems, in which functionally related cytoskeleton-modifying or cell adhesion molecules are co-regulated in systems of molecular pathways with molecules such as small GTPases, amyloid-β precursor protein (APP), kinesins, growth factors and transcription factorsvfrom the cell surface to the nucleus (Supplementary Figs. 1 and 2 and Supplementary Discussion). Some of the components of this sprouting neuron cytoskeletal network resemble those seen in growth cone proteomic screens36 and in peripheral nerve regeneration after injury10. These data suggest that a sprouting or regeneration transcriptome after stroke involves a coordinated regulation in gene expression that underlies a switch in cellular phenotype toward a state of active neuronal outgrowth within the adult brain environment.
Sprouting neurons from the peri-infarct cortex of aged animals had a transcriptional profile that differed in key molecular classes from that of young adult sprouting neurons, particularly in cytokine and chemokine, axonal guidance, bone morphogenic protein, cell adhesion and growth factor molecules (Table 2 and Supplementary Discussion). Aged sprouting neurons paradoxically upregulate, EphA4 and Lingo1, receptors for axonal growth inhibitory proteins. In addition to the cell surface receptor EphA4, a downstream mediator of EphA4 signaling, the Rho-GAP chimerin-1 (ref. 39), is also induced in aged sprouting neurons. Lingo1 is part of the Nogo receptor-1 complex and is associated with inhibition of axonal sprouting in spinal cord injury and in saxonal sprouting after that occurs into the spinal cord5,40. These data suggest that a reduced recovery in aged stroke may relate to increased axonal growth inhibition directly in sprouting neurons in the aged peri-infarct brain. Not only did the absolute levels of specific molecules differ between aged and young adult sprouting neurons, but the cellular expression profile also was distinct, with IGF1 induction in peri-infarct astrocytes in the young adult (Fig. 2f) but in peri-infarct neurons in the aged adult (Fig. 2g). Gene products that are involved in DNA epigenetic or structural regulation were upregulated in both aged and young adult, and differed by age (Table 2). Epigenetic control of DNA structure and gene expression has a prominent role in brain development and neural stem cell differentiation41 and may influence the phenotypic change characterized by post-stroke axonal sprouting. ATRX was one of the most highly induced genes in aged sprouting neurons after stroke, and it was also upregulated in young adult sprouting neurons, but this upregulation, at P = 0.036, fell short of the statistical cutoff (P ≤ 0.005) established in this data set.
Growth factor dependence, epigenetic modifiers and growth inhibitors after stroke
Stroke itself induces axonal sprouting within motor and somatosensory areas of peri-infarct cortex (Supplementary Discussion) both locally and up to several millimeters away from the infarct. Knockdown of ATRX expression blocked post-stroke axonal sprouting. This establishes a direct role for stroke-induced neuronal ATRX expression in the process of post-stroke axonal sprouting. IGF1 delivery did not induce any significant change in motor cortical connections above the normal degree of post-stroke axonal sprouting. This result was unexpected, as IGF1 mediates axonal sprouting from cortical neurons in development21 and acts in axonal and dendritic remodeling in many systems20. One possible explanation might be that IGF1 is at a biological maximum after stroke, with increases already at 3 d after stroke (Supplementary Fig. 10a). However blockade of IGF1, induced 1 week after stroke, produced widespread neuronal cell death in peri-infarct cortex and a contraction in the pattern of intracortical connections. One week after stroke is a time period in which the initial injury from stroke is long past; most cell death ceases within 3 d in this and other experimental models of stroke42. The death of neurons when IGF1 signaling was locally blocked late after stroke suggests a delayed neuronal growth factor dependency in peri-infarct cortex long after the stroke stimulus. Because the amount of neuronal cell death does not differ between aged and young adults in stroke13, it is possible that the upregulation of IGF1 in the aged brain functions to support peri-infarct neurons and limit a progressive neuronal cell death. Blockade of Lingo1 and knockout of its receptor partner NgR1 enhanced post-stroke axonal sprouting in motor cortex. These data indicate that the NgR1–Lingo1–Troy–p75 signaling system is critical for normally limiting post-stroke axonal sprouting in peri-infarct cortex, as it does in contralateral cortex after stroke8,42, but extends this concept to indicate that sprouting neurons in the aged brain upregulate a myelin protein receptor.
These results indicate a widespread cellular and molecular alteration in peri-infarct cortex after stroke. This tissue survives the stroke and appears morphologically intact to gross histological examination in both humans and experimental animals. However, neurons in this region are not static survivors of cerebral ischemia: they are induced to express an age-related growth-associated genetic program that controls axonal sprouting and mediates the formation of new patterns of connections within the sensorimotor system. Axonal sprouting or regeneration after stroke thus resembles a neurodevelopmental cellular program: a specific molecular program mediates formation of new axonal connections in neurons which require a growth factor for their survival.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/natureneuroscience/.
Supplementary Material
Acknowledgments
We thank S. Raike (University of Michigan) for help with obtaining NgR2 mutant mice. This research was funded by US National Institutes of Health NS045729, NS04733, NS049041, AHA 09SDG2310180 and 0525144Y; the Unitesd States Department of Veterans Affairs; the Larry L Hillblom Foundation; the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation and the American Federation of Aging Research.
Footnotes
Accession codes. Gene Expression Omnibus: Microarray data have been deposited with accession code GSE24442.
Note: Supplementary information is available on the Nature Neuroscience website.
AUTHOR CONTRIBUTIONS
S.L. designed and performed experiments, analyzed data and wrote the paper; J.J.O. developed cortical connectional mapping methodology and analyzed data; D.K. designed and performed experiments and analyzed data; S.V.K. developed a transgenic mouse first reported in this manuscript; C.J.D. and J.L.T. designed and performed the DRG in vitro studies; R.J.G. provided transgenic mice and reagents. G.C. and D.H.G. analyzed the microarray data; S.T.C. designed experiments, analyzed data and wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Carmichael ST. Cellular and molecular mechanisms of neural repair after stroke: making waves. Ann Neurol. 2006;59:735–742. doi: 10.1002/ana.20845. [DOI] [PubMed] [Google Scholar]
- 2.Carmichael ST, Wei L, Rovainen CM, Woolsey TA. New patterns of intracortical projections after focal cortical stroke. Neurobiol Dis. 2001;8:910–922. doi: 10.1006/nbdi.2001.0425. [DOI] [PubMed] [Google Scholar]
- 3.Dancause N, et al. Extensive cortical rewiring after brain injury. J Neurosci. 2005;25:10167–10179. doi: 10.1523/JNEUROSCI.3256-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown CE, Aminoltejari K, Erb H, Winship IR, Murphy TH. In vivo voltage-sensitive dye imaging in adult mice reveals that somatosensory maps lost to stroke are replaced over weeks by new structural and functional circuits with prolonged modes of activation within both the peri-infarct zone and distant sites. J Neurosci. 2009;29:1719–1734. doi: 10.1523/JNEUROSCI.4249-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu BP, Cafferty WBJ, Budel SO, Strittmatter SM. Extracellular regulators of axonal growth in the adult central nervous system. Phil Trans R Soc Lond B. 2006;361:1593–1610. doi: 10.1098/rstb.2006.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fitch MT, Silver J. CNS injury, glial scars, and inflammation: inhibitory extracellular matrices and regeneration failure. Exp Neurol. 2008;209:294–301. doi: 10.1016/j.expneurol.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Case LC, Tessier-Lavigne M. Regeneration of the adult central nervous system. Curr Biol. 2005;15:R749–R753. doi: 10.1016/j.cub.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 8.Zai L, et al. Inosine alters gene expression and axonal projections in neurons contralateral to a cortical infarct and improves skilled use of the impaired limb. J Neurosci. 2009;29:8187–8197. doi: 10.1523/JNEUROSCI.0414-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer D, Petkova V, Thanos S, Benowitz LI. Switching mature retinal ganglion cells to a robust growth state in vivo: gene expression and synergy with RhoA inactivation. J Neurosci. 2004;24:8726–8740. doi: 10.1523/JNEUROSCI.2774-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao HS, et al. Identification of gene expression profile of dorsal root ganglion in the rat peripheral axotomy model of neuropathic pain. Proc Natl Acad Sci USA. 2002;99:8360–8365. doi: 10.1073/pnas.122231899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanabe K, Bonilla I, Winkles JA, Strittmatter SM. Fibroblast growth factor-inducible-14 is induced in axotomized neurons and promotes neurite outgrowth. J Neurosci. 2003;23:9675–9686. doi: 10.1523/JNEUROSCI.23-29-09675.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang F, et al. Nogo-A is involved in secondary axonal degeneration of thalamus in hypertensive rats with focal cortical infarction. Neurosci Lett. 2007;417:255–260. doi: 10.1016/j.neulet.2007.02.080. [DOI] [PubMed] [Google Scholar]
- 13.Li S, Carmichael ST. Growth-associated gene and protein expression in the region of axonal sprouting in the aged brain after stroke. Neurobiol Dis. 2006;23:362–373. doi: 10.1016/j.nbd.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 14.Carmichael ST, et al. Growth-associated gene expression after stroke: evidence for a growth-promoting region in peri-infarct cortex. Exp Neurol. 2005;193:291–311. doi: 10.1016/j.expneurol.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 15.Callaway EM, Katz LC. Emergence and refinement of clustered horizontal connections in cat striate cortex. J Neurosci. 1990;10:1134–1153. doi: 10.1523/JNEUROSCI.10-04-01134.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Callaway EM, Katz LC. Development of axonal arbors of layer 4 spiny neurons in cat striate cortex. J Neurosci. 1992;12:570–582. doi: 10.1523/JNEUROSCI.12-02-00570.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costigan M, et al. Replicate high-density rat genome oligonucleotide microarrays reveal hundreds of regulated genes in the dorsal root ganglion after peripheral nerve injury. BMC Neurosci. 2002;3:16. doi: 10.1186/1471-2202-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao Y, et al. Insulin-like growth factor (IGF)-1 suppresses oligodendrocyte caspase-3 activation and increases glial proliferation after ischemia in near-term fetal sheep. J Cereb Blood Flow Metab. 2003;23:739–747. doi: 10.1097/01.WCB.0000067720.12805.6F. [DOI] [PubMed] [Google Scholar]
- 19.Bérubé NG, et al. The chromatin-remodeling protein ATRX is critical for neuronal survival during corticogenesis. J Clin Invest. 2005;115:258–267. doi: 10.1172/JCI22329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salie R, Steeves JD. IGF-1 and BDNF promote chick bulbospinal neurite outgrowth in vitro. Int J Dev Neurosci. 2005;23:587–598. doi: 10.1016/j.ijdevneu.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 21.Özdinler PH, Macklis JD. IGF-I specifically enhances axon outgrowth of corticospinal motor neurons. Nat Neurosci. 2006;9:1371–1381. doi: 10.1038/nn1789. [DOI] [PubMed] [Google Scholar]
- 22.Topalli I, Etgen AM. Insulin-like growth factor-I receptor and estrogen receptor crosstalk mediates hormone-induced neurite outgrowth in PC12 cells. Brain Res. 2004;1030:116–124. doi: 10.1016/j.brainres.2004.09.057. [DOI] [PubMed] [Google Scholar]
- 23.Kooijman R, Sarre S, Michotte Y, De Keyser J. Insulin-like growth factor I: a potential neuroprotective compound for the treatment of acute ischemic stroke? Stroke. 2009;40:e83–e88. doi: 10.1161/STROKEAHA.108.528356. [DOI] [PubMed] [Google Scholar]
- 24.Zhu ML, Kyprianou N. Androgen receptor and growth factor signaling crosstalk in prostate cancer cells. Endocr Relat Cancer. 2008;15:841–849. doi: 10.1677/ERC-08-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji B, et al. LINGO-1 antagonist promotes functional recovery and axonal sprouting after spinal cord injury. Mol Cell Neurosci. 2006;33:311–320. doi: 10.1016/j.mcn.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 26.Venkatesh K, et al. The Nogo-66 receptor homolog NgR2 is a sialic acid-dependent receptor selective for myelin-associated glycoprotein. J Neurosci. 2005;25:808–822. doi: 10.1523/JNEUROSCI.4464-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Atwal JK, et al. PirB is a functional receptor for myelin inhibitors of axonal regeneration. Science. 2008;322:967–970. doi: 10.1126/science.1161151. [DOI] [PubMed] [Google Scholar]
- 28.Chklovskii DB, Mel BW, Svoboda K. Cortical rewiring and information storage. Nature. 2004;431:782–788. doi: 10.1038/nature03012. [DOI] [PubMed] [Google Scholar]
- 29.Petreanu L, Mao T, Sternson SM, Svoboda K. The subcellular organization of neocortical excitatory connections. Nature. 2009;457:1142–1145. doi: 10.1038/nature07709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenstein JM, Mani N, Khaibullina A, Krum JM. Neurotrophic effects of vascular endothelial growth factor on organotypic cortical explants and primary cortical neurons. J Neurosci. 2003;23:11036–11044. doi: 10.1523/JNEUROSCI.23-35-11036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stowe AM, et al. VEGF protein associates to neurons in remote regions following cortical infarct. J Cereb Blood Flow Metab. 2007;27:76–85. doi: 10.1038/sj.jcbfm.9600320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stevens B, et al. The classical complement cascade mediates CNS synapse elimination. Cell Mol Neurobiol. 2007;131:1164–1178. doi: 10.1016/j.cell.2007.10.036. [DOI] [PubMed] [Google Scholar]
- 33.Zujovic V, et al. The facial motor nucleus transcriptional program in response to peripheral nerve injury identifies Hn1 as a regeneration-associated gene. J Neurosci Res. 2005;82:581–591. doi: 10.1002/jnr.20676. [DOI] [PubMed] [Google Scholar]
- 34.Boulanger LM. Immune proteins in brain development and synaptic plasticity. Neuron. 2009;64:93–109. doi: 10.1016/j.neuron.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 35.Lemons ML, Condic ML. Integrin signaling is integral to regeneration. Exp Neurol. 2008;209:343–352. doi: 10.1016/j.expneurol.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 36.Pertz OC, et al. Spatial mapping of the neurite and soma proteomes reveals a functional Cdc42/Rac regulatory network. Proc Natl Acad Sci USA. 2008;105:1931–1936. doi: 10.1073/pnas.0706545105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du J, Fu C, Sretavan DW. Eph/ephrin signaling as a potential therapeutic target after central nervous system injury. Curr Pharm Des. 2007;13:2507–2518. doi: 10.2174/138161207781368594. [DOI] [PubMed] [Google Scholar]
- 38.Ikegami K, et al. TTLL7 is a mammalian beta-tubulin polyglutamylase required for growth of MAP2-positive neurites. J Biol Chem. 2006;281:30707–30716. doi: 10.1074/jbc.M603984200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dalva MB. There’s more than one way to skin a chimaerin. Neuron. 2007;55:681–684. doi: 10.1016/j.neuron.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee JK, Kim JE, Sivula M, Strittmatter SM. Nogo receptor antagonism promotes stroke recovery by enhancing axonal plasticity. J Neurosci. 2004;24:6209–6217. doi: 10.1523/JNEUROSCI.1643-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacDonald JL, Roskams AJ. Epigenetic regulation of nervous system development by DNA methylation and histone deacetylation. Prog Neurobiol. 2009;88:170–183. doi: 10.1016/j.pneurobio.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 42.Lipton P. Ischemic cell death in brain neurons. Physiol Rev. 1999;79:1431–1568. doi: 10.1152/physrev.1999.79.4.1431. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.