Summary
Many viruses utilize host ESCRT proteins for budding; however, influenza virus budding is thought to be ESCRT-independent. In this study we have found a role for the influenza virus M2 proton-selective ion channel protein in mediating virus budding. We observed that a highly conserved amphipathic helix located within the M2 cytoplasmic tail mediates a cholesterol-dependent alteration in membrane curvature. The 17 amino acid amphipathic helix is sufficient for budding into giant unilamellar vesicles, and mutation of this sequence inhibited budding of transfected M2 protein in vivo. We show that M2 localizes to the neck of budding virions and that mutation of the M2 amphipathic helix results in failure of the virus to undergo membrane scission and virion release. These data suggest that M2 mediates the final steps of budding for influenza viruses, bypassing the need for host ESCRT proteins.
Introduction
The budding of enveloped viruses is a complex multi-step process, the completion of which requires alterations in membrane curvature and scission at the neck of the budding virion. Many viruses such as HIV-1, Ebola virus and the paramyxovirus PIV-5 utilize the host endosomal sorting complex required for transport (ESCRT) machinery to mediate membrane scission and virion release (reviewed in Carlton and Martin-Serrano, 2009; Chen and Lamb, 2008). Matrix proteins from these and other viruses contain ‘late’ domains, sequences that bind to proteins of the ESCRT pathway, enabling the virus to utilize ESCRT proteins that are normally involved in the budding and release of endosomal vesicles into multi-vesicular bodies. However, studies indicate that influenza virus may bud independently of ESCRT proteins, utilizing an unknown mechanism of membrane scission and virion release (Bruce et al., 2009; Chen and Lamb, 2008).
Influenza virus buds from the plasma membrane of infected cells, producing both 100 nm diameter spherical and 100 nm × 2–20 μm filamentous virions. Assembly and budding is thought to begin with lipid raft-mediated clustering of the two major viral glycoproteins, the receptor binding/membrane fusion protein hemagglutinin (HA) and the enzyme neuraminidase (NA) (Chen et al., 2007; Leser and Lamb, 2005; Takeda et al., 2003). Lipid rafts are dynamic plasma membrane domains, enriched in sphingolipids and cholesterol, which are thought to serve as platforms for the concentration of proteins (Brown and Rose, 1992; Simons and Toomre, 2000). Lipid rafts are also the sites of budding for several viruses, such as influenza virus, HIV-1 and Ebola virus (reviewed in Chazal and Gerlier, 2003). Mutations in the HA transmembrane (TM) domain that eliminate association with lipid rafts, significantly attenuate viral replication and prevent the formation of filamentous virions (Chen et al., 2005).
The cytoplasmic tails of HA and NA have been inferred to bind to the matrix (M1) protein, mediating its incorporation into the budding virion (Jin et al., 1997). The M1 protein interacts with the nucleoprotein (NP) and the plasma membrane, facilitating incorporation of the genome, as a viral ribonucleoprotein complex (vRNP), into budding virions (Bui et al., 1996; Noton et al., 2007; Zhang and Lamb, 1996). Mutation of the cytoplasmic tails of HA and NA reduces the packaging of M1 and produces defective virions with greatly altered morphology (Barman et al., 2004; Jin et al., 1997). In conjunction with the HA/NA-M1 interaction, M1 binds to a third viral integral-membrane viral protein, M2. M2 is a multi-functional protein with proton-selective ion channel activity. The M1-M2 interaction may enable the recruitment of M2 to sites of virus budding and mediate the incorporation of M2 into virions (Chen et al., 2008).
M2 is an essential component of the infectious virion. During virus entry by endocytosis, endosomal acidification activates the proton selective ion channel activity of the M2 protein, causing acidification of the virus interior and leading to dissociation of M1 from the vRNP (reviewed in Lamb and Pinto, 2005; Pinto and Lamb, 2006). M2 ion channel activity is inhibited by the antiviral drug amantadine, which prevents proton flux and blocks virus uncoating by preventing M1-RNP dissociation (reviewed in Pinto and Lamb, 2007). The M2 protein contains 97 amino acid residues that assemble into a homotetramer (Pinto et al., 1997). M2 contains a 24 residue N-terminal extracellular domain, a single TM domain of 19 residues that forms the pore of the ion channel, and a 54 residue cytoplasmic tail (Lamb et al., 1985; Zebedee et al., 1985; Pinto et al., 1992). The first 17 amino acid residues of the M2 protein cytoplasmic tail are predicted to form a membrane-parallel, amphipathic helix (Nguyen et al., 2008; Schnell and Chou, 2008; Tian et al., 2003). In addition to its ion channel activity, recent work has suggested that the M2 protein may also affect the morphology of budding virions and may be required for efficient vRNP packaging, assembly and budding (Chen et al., 2008; Iwatsuki-Horimoto et al., 2006; McCown and Pekosz, 2005, 2006; Rossman et al., 2010).
Whereas many of the steps required for influenza virus assembly have been determined, the molecular machinery needed to complete the budding process has not been elucidated. The available data for influenza virus indicate that the virus may bud independently of ESCRT proteins, thus the necessary components for membrane scission and virion release are not known (Bruce et al., 2009; Chen and Lamb, 2008). In this study we have identified an additional function of the M2 protein in modifying membrane curvature during virus budding. We further show that the influenza virus M2 protein mediates membrane scission, allowing for virion release independent of the host ESCRT pathway.
Results
Conservation of the M2 Amphipathic Helix
Previous work has suggested that the M2 cytoplasmic tail may play a role in virus assembly (Chen et al., 2008; Iwatsuki-Horimoto et al., 2006; McCown and Pekosz, 2005, 2006; Rossman et al., 2010). Thus, we sought to elucidate the possible functions of the M2 protein during viral assembly and budding. Analysis of the M2 protein sequence predicts that the cytoplasmic tail contains an amphipathic helix, which has been confirmed experimentally (Nguyen et al., 2008; Tian et al., 2003) (Fig. 1a). A helical wheel plot of the M2 amphipathic helix is shown in Fig. 1b with the charged and hydrophobic faces of the helix indicated. Analysis of the amino acid sequence of M2 across all influenza subtypes for 500 different strains of influenza A virus, including the recent 2009 H1N1 pandemic strain and several highly pathogenic H5N1 avian influenza virus strains, showed significant conservation along the length of the M2 amphipathic helix (Fig. S1), suggesting that the amphipathic helix has an important role for influenza virus.
Figure 1. Conservation of the M2 Amphipathic Helix.

(A) Diagram of structural motifs in the M2 protein. The TM domain/ion channel pore, CRAC motif and amphipathic helix are indicated. Alanine substituted residues in the M2(AH-Mut) protein are shown in red. (B) Helical wheel plot of the M2 amphipathic helix is shown as generated at http://heliquest.ipmc.cnrs.fr. Hydrophilic residues are shown in black and hydrophobic resides in grey. The red line separates the two faces of the helix. See also Fig. S1.
M2 Alters Membrane Curvature in a Cholesterol-dependent Manner
Many proteins, including several viral proteins, utilize an amphipathic helix for modifying membrane curvature (reviewed in Antonny, 2006). To address the role of the M2 amphipathic helix in virus budding, we first assessed the effect of the amphipathic helix on membrane curvature. As M2 has been shown to bind cholesterol (Rossman et al., 2010; Schroeder et al., 2005) and cholesterol can modify the ability of an amphipathic helix to induce membrane curvature (de Meyer and Smit, 2009; Egashira et al., 2002), we also sought to address the possibility of cholesterol-dependent alterations in membrane curvature by the M2 protein.
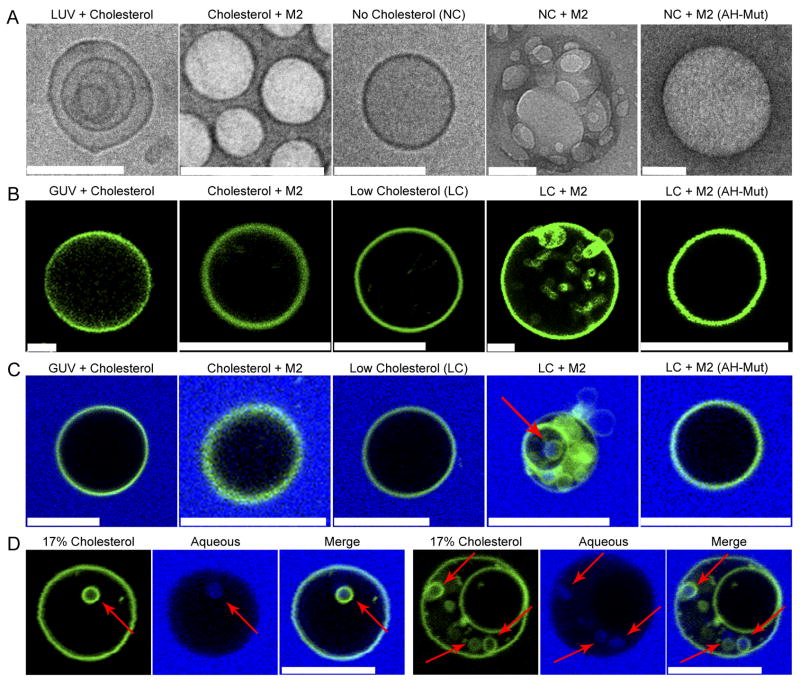
Wild-type (wt) M2 protein and an amphipathic helix mutant [M2(AH-Mut)], in which the hydrophobic face of the helix was altered by changing five bulky hydrophobic residues to alanine in an attempt to disrupt the function of the helix without disrupting its α-helicity (Rossman et al., 2010), were purified and reconstituted into large unilamellar vesicles (LUVs). Examination of LUVs reconstituted with wt M2 protein by cryo-electron microscopy, or negative staining, showed an alteration in morphology from control LUVs (Fig. 2a, S2a). Wt M2 protein induced the formation of many ‘flat’ single LUVs, suggesting that M2 may induce negative membrane curvature (Chernomordik et al., 1995; Russell et al., 2001) in the presence of cholesterol (Fig. 2a, S2a). Additionally, changes in LUV morphology were found to be independent of M2 ion channel activity, as incubation with the ion channel blocker amantadine had no effect (Fig. S2b, c).
Figure 2. M2 Alters Membrane Curvature in a Cholesterol-Dependent Manner.
(A) LUVs or cholesterol-free LUVs were reconstituted in the presence or absence of 100 μg purified M2 or M2(AH-Mut) protein and analyzed by cryo-electron microscopy. Scale bars indicate 100 nm. (B) LUVs prepared as above, with the addition of a fluorescent membrane dye (shown in green), were dehydrated, electroformed into GUVs and immediately imaged. (C) GUVs, prepared as above, with 50 μg purified M2 or 100 μg M2(AH-Mut) protein were resuspended with 0.5 mg/ml of lucifer yellow (shown in blue). NC indicates LUVs containing no cholesterol. LC indicates GUVs containing 0.5 molar % cholesterol. (D) Two examples of M2-containing GUVs, prepared as above, containing 17 molar % cholesterol. Arrows indicate lucifer yellow containing ILVs. Scale bars indicate 10 μm. See also Figs. S2, S3 and Movies S1, S2.
Control LUVs that lacked cholesterol appeared ‘flat’ and resembled cholesterol-containing LUVs reconstituted in the presence of wt M2 protein (Fig. 2a, S2a). It has been shown that cholesterol can directly modify membrane structure, providing an explanation for the different morphology seen with control LUVs in the presence and absence of cholesterol (de Meyer and Smit, 2009). When wt M2 was reconstituted into LUVs that lacked cholesterol, M2 appeared to induce positive membrane curvature, and the resulting morphology resembled control LUVs that contained cholesterol (Fig. 2a, S2a). M2-containing LUVs that lacked cholesterol showed the formation of many small, aggregated, ‘budding’ LUVs that appear morphologically similar to cholesterol-containing control LUVs (Fig. 2a, S2a). No significant change in LUV morphology was observed when the vesicles were reconstituted with M2(AH-Mut) protein in the absence of cholesterol, suggesting that the M2 amphipathic helix is necessary for the alteration of membrane curvature in the absence of cholesterol (Fig. 2a, S2a).
Our previous work has shown that influenza virus expressing M2(AH-Mut) protein is not impaired in ion channel activity, protein expression or protein localization during virus infection (Ma et al., 2009; Rossman et al., 2010). However, to control for possible differences in in vitro membrane incorporation, we determined the proportion of input wt and M2(AH-Mut) mutant M2 protein reconstituted into LUVs. Based on an input of 100 μg of wt M2 protein, it was found that 75% was incorporated into the LUVs. This amount was unaffected by the presence or absence of cholesterol; however, only 35% of M2(AH-Mut) protein was incorporated into LUVs, regardless of the presence or absence of cholesterol. To ensure that the differences in membrane curvature seen when comparing wt M2 and M2(AH-Mut) were due to the specific action of the protein, and not due to differences in protein incorporation, LUVs were made with 50 μg of wt M2 protein, which gave a final incorporated protein amount of 31 μg, which is comparable to the 35 μg level obtained with the M2(AH-Mut) protein. Examination of these LUVs showed that they retained their induction of positive and negative curvature and appeared indistinguishable from LUVs created with 100 μg of wt M2 protein (Fig. S2b, c).
M2 Causes Membrane Budding In Vitro
As M2 is capable of altering membrane curvature, we asked if M2 protein alone is capable of causing membrane budding. We utilized a system for imaging budding and membrane scission events within giant unilamellar vesicles (GUVs) that has been described recently for analyzing the function of proteins in the ESCRT III complex (Wollert et al., 2009). GUVs were formed in the presence of 30 or 0.5 molar % of cholesterol, incorporating purified wt M2 protein or M2(AH-Mut) protein. Low-cholesterol GUVs containing wt M2 protein exhibited a rapid budding of the GUV membrane and the formation of many intra-luminal vesicles (ILVs) (Fig. 2b LC+M2, Movie S1). Analysis of the ILVs showed free movement within the GUV, consistent with completion of membrane scission (Movie S1). Both inward and outward budding events were observed due to the random orientation of M2 protein in the lipid bilayer (Fig. 2b); however, outward budding vesicles were seen less frequently due to rapid diffusion out of the imaging plane following membrane scission. Low-cholesterol GUVs containing M2(AH-Mut) protein resembled control GUVs (cf. Fig. 2b LC and LC+M2(AH-Mut), Movie S2), suggesting that the M2 amphipathic helix is essential for M2-mediated membrane budding.
In the presence of high-levels of cholesterol, budding events were not observed for wt M2 protein (Fig. 2b Cholesterol + M2). Additionally, control GUVs showed no evidence of budding when reconstituted in the presence of high or low molar ratios of cholesterol (Fig. 2b GUV + Cholesterol and LC). It is important to note that the M2-dependent induction of negative curvature seen in high-cholesterol LUVs (Fig. 2a), was not observed in high-cholesterol GUVs (Fig. 2b). It is possible that M2-induction of negative curvature occurs in GUVs; however, the resulting changes in morphology may not be resolvable by light microscopy and changes in vesicle size may be lost in the normal background variation of GUV size. Alternatively, the cholesterol-dependent M2-induction of negative curvature may depend on the initial degree of membrane curvature, with the different LUV and GUV radii of curvature affecting the membrane association and/or curvature induction of the M2 amphipathic helix.
M2-induced budding was confirmed via the addition of the small aqueous marker lucifer yellow to the GUV resuspension buffer. Lucifer yellow fluorescence was observed only within the intraluminal vesicles of low-cholesterol GUVs reconstituted with wt M2 protein (Fig. 2c and Fig. S3a). Some lucifer yellow-negative ILVs are observed, however, the M2 protein begins budding directly after GUV formation has completed and, thus, some ILVs may complete budding before the addition of lucifer yellow to the media. Additionally, to confirm that the lack of budding observed with M2(AH-Mut) seen in Fig. 2b is not due to the lower protein incorporation levels, the GUVs used in Fig. 2c were made from LUVs incorporating 50 μg of M2 or 100 μg of M2(AH-Mut), resulting in approximately equal incorporation of these two proteins into the vesicles. No budding was observed with the M2(AH-Mut) protein, while wt M2 appeared to bud normally (Fig. 2c). As M2 is unlikely to encounter a lipid environment within the cell that is cholesterol-free, we reconstituted GUVs containing an intermediate concentration of cholesterol (17 molar %) with full-length M2 protein. In the presence of 17 molar % cholesterol, M2 caused membrane budding and the formation of lucifer yellow-containing ILVs, indicating that M2 is capable of mediating budding in physiologically-relevant levels of cholesterol (Fig. 2d, S3b).
To determine if the M2 amphipathic helix is sufficient to mediate GUV budding, a peptide corresponding to the M2 amphipathic helix (M2AH) was synthesized. Addition of the M2AH peptide to pre-formed GUVs containing 0.5 molar % of cholesterol lead to rapid budding from the GUV membrane (Fig. 3a LC + Pep M2AH), resulting in 80.8% (n=54) of the GUVs containing at least one ILV. Addition of M2AH peptide to pre-formed GUVs containing 30% cholesterol did not cause budding (Fig. 3a cholesterol + Pep M2AH), with only 6.2% (n=153) of the GUVs containing an ILV. As electroformed GUVs show a background level of vesicles containing ILVs [9.7% (n=281) of low cholesterol GUVs and 3.7% (n=42) of high cholesterol GUVs contain an ILV], we sought to determine if the ILVs observed in Fig. 3a were caused specifically by peptide-induced membrane budding. Peptide-induced budding was confirmed via the addition of lucifer yellow to the GUV resuspension buffer. Addition of lucifer yellow to the GUV resuspension buffer lead to fluorescence only within low-cholesterol GUVs that had been treated with the M2AH peptide [72.5% (n=54) of peptide treated low cholesterol GUVs contained a lucifer yellow positive ILV, compared to 3.5% (n=281) without peptide and 0.5% (n=153) for peptide-treated high cholesterol GUVs; Fig. S3c], thus confirming that the M2 amphipathic helix is capable of inducing budding and the uptake of surrounding aqueous media into ILVs (Fig. 3b LC + Pep M2AH). Interestingly, addition of the M2AH peptide to low-cholesterol GUVs appeared to cause membrane leakage, in addition to membrane budding, as shown by the presence of aqueous lucifer yellow marker within the bulk GUV interior, as well as within the budded intra-luminal vesicles (Fig. 3b). It is possible that the M2AH peptide possess the ability to form pores in the membrane bilayer, such as has been noted for several other amphipathic peptides (Egashira et al., 2002; Lamaziere et al., 2007; Lee et al., 2008). Pore formation leading to membrane leakage is seen with the class-A amphipathic peptide Ac-18A-NH2 (Venkatachalapathi et al., 1993) and pore formation leading to GUV lysis is seen with the class-L amphipathic peptide melittin (reviewed in Dempsey, 1990) (Fig. S4). However, in the context of the intact M2 protein, it is likely that the transmembrane domain and the full cytoplasmic tail limits the ability of the amphipathic helix to transverse the bilayer as shown by the absence of membrane leakage during M2 GUV budding (Fig. 2c, d).
Figure 3. The M2 Amphipathic Helix Causes Membrane Budding in vitro.
(A) GUVs electroformed with 30 or 0.5 molar % of cholesterol were treated with 10 μM of the indicated peptide and imaged within 1 h. (B) GUVs were prepared and treated as above except that 0.5 mg/ml of lucifer yellow (shown in blue) was added to the resuspension buffer. (C) GUVs, prepared as above, were treated with M2AH peptide at the indicated concentrations for 1 h and imaged. (D) GUVs, prepared as above with the indicated molar % of cholesterol, were treated with 10 μM of M2AH peptide for 1 h and imaged. 17% cholesterol LY-post scission indicates 17% cholesterol GUVs to which lucifer yellow was added 1 h post-treatment with 10 μM of M2AH peptide. (E) GUVs, prepared as above (with the exception of M2AH-TMR treated samples, for which lucifer yellow was omitted from the resuspension buffer), were treated with 10 μM of the indicated peptide and imaged within 1 h. The M2AH-WSN peptide corresponds to the M2 amphipathic helix of A/WSN/33, M2AH-SOIV to A/California/05/2009, M2AH-Sc to a scrambled sequence of the M2AH peptide, M2AH-TMR to rhodamine-labled M2AH peptide. LC indicates GUVs containing 0.5 molar % of cholesterol. Scale bars indicate 10 μm. See also Figs. S3, S4 and Movie S3.
Titration of the peptide concentration required to induce budding showed no effect at 5 μM, robust inward budding at 10 μM, a switch to outward budding causing GUV deformation at 50 μM and robust outward budding leading to lysis of the GUV at 100 μM (Fig. 3c and Movie S3), indicating a dose-dependent alteration in membrane curvature. The ability of amphipathic helices to modify membrane curvature and cause budding is well established (reviewed in Drin and Antonny, 2010) and is shown for the amphipathic peptide RW16 (Lamaziere et al., 2007) (Fig. S4). Additionally, several amphipathic helices have been shown to induce both inward and outward budding depending on the peptide concentration (Lamaziere et al., 2007), possibly due to concentration-dependent shifts in peptide orientation in the membrane. A similar activity may occur with the M2 amphipathic helix peptide, causing the different forms of budding and membrane leakage observed in Fig. 3c.
Because full-length M2 protein is capable of causing budding in the presence of intermediate levels of cholesterol, we titrated the amount of cholesterol that was required to enable GUV budding by the M2AH peptide. The M2AH peptide induced GUV budding at cholesterol levels at and below 17 molar % (Fig. 3d), a level that is likely comparable to many regions of the bulk plasma membrane and is consistent with the cholesterol-dependence of the full-length M2 protein. Interestingly, treatment of GUVs, containing 17 molar % cholesterol, with the M2AH peptide caused budding without membrane leakage (Fig. 3d); further suggesting that the membrane leakage observed in Fig. 3b is not relevant to the budding activity of the M2 protein. To further confirm the completion of M2-mediated scission, GUVs containing 17 molar % of cholesterol were treated with the M2AH peptide for 1 hr and then lucifer yellow was added to the imaging buffer. Lucifer yellow was not found in the GUV or the ILVs, indicating that the majority of ILVs had completed scission within the 1 h treatment and were no longer assessable to the outside buffer (Fig. 3d 17% Ch LY-Post Scission).
A synthetic peptide corresponding to the amino acid sequence of the M2(AH-Mut) amphipathic helix did not cause GUV budding in the presence of low-levels of cholesterol (Fig. 3a, b LC + Pep M2AH-Mut), nor did a peptide containing a scrambled version of the M2AH peptide sequence (Fig. 3e LC + Pep M2AH-Sc). M2AH peptide-induced budding was further confirmed by the observation that a fluorescently-labeled version of the M2AH peptide colocalized with budded ILVs in low-cholesterol GUVs (Fig. 3e LC + Pep M2AH-TMR) and with the GUV membrane in the presence of high-cholesterol levels (Fig. 3e Chol + Pep M2AH-TMR). Additionally, treatment of low-cholesterol GUVs with peptides corresponding to the M2 amphipathic helix of the influenza virus strain A/WSN/33 or of the consensus sequence for the 2009 H1N1 pandemic swine-origin influenza virus (SOIV) strain lead to a rapid budding of the GUV membrane (Fig. 3e LC + Pep M2AH-WSN and LC + Pep M2AH-SOIV). The M2AH peptides exhibited a slight variation in their effects, with the M2AH-WSN peptide showing reduced membrane leakage and the M2AH-TMR peptide showing enhanced GUV lysis at the 10 μM concentration used. However, the consistent induction of membrane budding in a low-cholesterol environment suggests that M2-mediated membrane budding is a highly conserved function of the M2 protein.
M2 Causes Budding at Lipid Phase Boundaries
As the M2 protein is capable of inducing budding from single-phase GUVs, we asked if M2 could mediate budding from a phase-separated GUV that may better mimic domains within the plasma membrane (Kaiser et al., 2009). GUVs containing sphingomyelin, poly-unsaturated phosphocholine and cholesterol segregate into lipid ordered (Lo) and lipid disordered (Ld) phases. Incorporation of fluorescent markers allows visualization of the two phases (Fig. 4a). Addition of a fluorescently-tagged M2AH peptide to phase-separated GUVs showed that the amphipathic helix binds to the Ld phase and clusters at the phase boundary (Fig. 4b). Treatment of high-cholesterol GUVs with the M2 amphipathic helix peptide did not cause budding and had no observable effect on lipid phase separation (Fig. 4c). However, M2AH peptide-treatment of low-cholesterol phase-separated GUVs lead to rapid outward budding beginning with the excision of the Lo phase from the GUV, in a manner that may be similar to the scission and release of the lipid-raft rich budding virus from the bulk-phase plasma membrane, and resulting in numerous, smaller, single-phase vesicles (Fig. 4d, 4e, S5a, Movie S4). This suggests that the M2 amphipathic helix may modify the line tension between lipid phases, subsequently driving budding as has been previously observed for other proteins that mediate budding and scission (Allain and Ben Amar, 2006; Liu et al., 2006; Romer et al., 2010).
Figure 4. The M2 Amphipathic Helix Causes Membrane Budding at Phase Boundaries.
Phase-separated GUVs, electroformed with 20 or 5 molar % of cholesterol and fluorescent markers for the Lo and Ld phase, were treated as indicated and imaged within 1 h. (A) Representative example of phase-separated GUV containing 20 molar % of cholesterol. (B) Phase-separated GUVs were prepared as above with the Lo marker omitted. GUVs were treated with 10 μM of the M2AH-TMR peptide for 1 h and imaged. Arrows indicate clustered M2AH peptide at the phase boundary. (C) Phase-separated GUVs prepared as in Fig. 4a containing 20 molar % cholesterol or (D) 5 molar % were resuspended with lucifer yellow, treated with 10 μM of the M2AH peptide for 1 h and imaged. Ld phase is shown in red, the Lo phase in green and the aqueous media in blue. (E) Time lapse images of a GUV from Fig. 4d. Note: loss of the Ld signal in later time points is due to the rapid bleaching of the red signal. (F) Plasma membrane spheres were prepared from wt M2 expressing cells or (G) M2(AH-Mut) expressing cells, GM1 was crosslinked to mark the Lo phase (green) and spheres were stained for M2 (red). Scale bars indicate 10 μm. Arrows indicate sites of budding. See also Fig. S5 and Movie S4.
To confirm that the full-length M2 protein similarly localizes to the Ld phase we created plasma membrane spheres (Lingwood et al., 2008) from M2 and M2 (AH-Mut) expressing cells. By crosslinking GM1 and using immunofluorescence staining to detect M2, we observed segregation of the Lo phase and M2, suggesting that M2 is localized to the Ld phase in living cells (Fig. 4f). Additionally, budding Lo vesicles were seen at the phase boundary between Lo and M2 with cells expressing wt M2 but not the mutant M2 (AH-Mut) (Fig. 4g, S5b, S5c), further suggesting that while the M2 amphipathic helix does not appear to be necessary for the localization of M2 to the Ld phase, the modulation of line tension causing Lo phase budding requires the wt M2 amphipathic helix.
M2 Causes Membrane Budding In Vivo
As the M2 amphipathic helix is sufficient to induce budding in an in vitro liposome assay, we assessed the ability of the M2 protein to induce budding in vivo. Transfection of an M2 expression plasmid into 293T cells led to the budding and release of M2-containing vesicles that could be detected by immunoblotting (Fig. 5a) and by electron microscopy (Fig. 5b). Wt M2 vesicles appeared aggregated; however, it is unlikely that this reflects any biologically-relevant activity, as aggregation may have occurred during concentration of the vesicle-containing supernatant prior to imaging. The release of M2-containing vesicles into the culture supernatant was dependent on an intact amphipathic helix, as expression of M2(AH-Mut) in 293T cells led to a significant reduction in M2 vesicle release, even when accounting for the reduced expression level as compared to wt M2 (Fig. 5). We have shown previously that M2 exhibits only minimal cholesterol binding when expressed in the absence of other viral proteins (Rossman et al., 2010). Thus, M2 expressed by transient transfection may mediate in vivo budding from lower-cholesterol regions of the plasma membrane in a manner that is dependent on the amphipathic helix, similar to the in vitro budding of M2 from low-cholesterol GUVs (Figs. 2–4).
Figure 5. M2 Causes Membrane Budding in vivo.
(A) 293T cells were transfected with M2 or M2(AH-Mut) expression plasmids and 24 h post-transfection the supernatant was harvested, concentrated, and M2 content was analyzed by western blot as compared to whole cell lysates. Values indicate average +/− standard deviation of the percentage of M2 found in the supernatant and calculated from a minimum of three repeats. M2-low indicates cells transfected with a 4-fold reduction of DNA to provide comparable M2 expression levels to that of M2(AH-Mut). The vertical line indicates separate blots. (B) Concentrated supernatant from Fig. 5a was analyzed by electron microscopy. Scale bars indicate 100 nm. The inset is an enlargement of a 100nm square region of the panel below.
M2 Localization at the Point of Membrane Scission
For M2 to mediate membrane scission in virus-infected cells, the M2 protein needs to localize specifically to the neck of the budding virion. We have shown previously that M2 is recruited to lipid rafts and to sites of budding during virus infection (Rossman et al., 2010). However, whereas M2 can be generally localized to the base of budding filamentous virions (Fig. 6a, b), the specific localization of M2 cannot be resolved via light microscopy. Thus, we examined the localization of M2 in virus-infected cells by immuno-gold labeling and electron microscopy. We observed that M2 localized to the base of budding filamentous and spherical virions, and a specific localization at the neck of budding viruses could be seen for 59.9% (n=1038) of all immunogold-labeled M2 (Fig. 6c, d), confirming our previous results showing that the majority of M2 in virally-infected cells is found at sites of virus budding (Rossman et al., 2010).
Figure 6. M2 Localizes to the Neck of Budding Virions.
MDCK cells were infected with an MOI of 3 pfu/cell of A/Udorn/72 for 18 h. (A) Virus-infected cells were fixed and processed for immunofluorescent detection of HA (shown in green) and M2 (shown in red). (B) Magnification of images shown in Fig. 6a. Scale bars indicate 10 μm. (C) Virus-infected cells were stained for M2 via immuno-gold labeling and thin sections were analyzed by electron microscopy. (D) Magnification of images shown in Fig. 6c. Scale bars indicate 100 nm. Arrows indicate M2 foci at sites of virus budding. See also Fig. S6.
M2 localization predominantly but not exclusively to the base of virions was also observed on viral filaments that had completed the budding process. Using fluorescent microscopy, it was observed that in a population of filamentous and spherical virions M2 is concentrated at one end of the filamentous particles (Fig. S6a). Quantification showed that 63% of filaments (n=300) had a detectable focus of M2, representing the majority of M2 in the virus. Electron microscopy studies show that one end of filamentous and a focus on spherical virions reacts with antibody to M2 and accounts for 79.8% (n=213) of all immunogold-labeled M2 found on the virion (Fig. S6b). This concentration of M2 is not seen at the free end of budding viral filaments, but instead colocalizes with the site of budding (Fig. 6), suggesting that M2 is incorporated during the completion of budding, at the trailing end of filamentous virions. This is consistent with the role of M2 in mediating membrane scission and virion release.
Recent results have shown that influenza virus budding is dependent on the expression of Rab11 (Bruce et al., 2010). Rab11 is known to function in endosomal budding as well as trafficking of proteins to the apical plasma membrane, thus knockdown of Rab11 could have a direct role in membrane scission or could affect the transport of influenza virus proteins, such as M2. siRNA knock-down of Rab11a/b lead to greater than 85% knock-down in Rab11a protein levels (Fig. S7a) and a statistically-significant 40% reduction in the levels of M2 found on the cell surface, while the levels of HA increased 2 fold (Fig. S7b, c). This difference cannot be attributed to expression differences as the levels of HA and M2 were comparable in control and Rab11 knockdown cells (Fig. S7d). Thus, while a further affect of Rab11 cannot be ruled out, it is likely that Rab11 has an essential role in the transport of M2 to the apical membrane and not a direct role in membrane scission.
Mutation of the M2 Amphipathic Helix Blocks Membrane Scission
Although the data shown here indicate that M2 localizes to the neck of budding viruses and is capable of mediating budding and membrane scission in vitro as well as in vivo, it does not distinguish between an additive role of the M2 protein in virus budding and a necessary role for the M2 protein in mediating membrane scission. To determine whether the M2 amphipathic helix is required for virus budding and membrane scission, we examined the budding of wt and M2 mutant influenza viruses by thin section electron microscopy. Whereas in wt virus-infected cells the budding of filamentous and spherical virions could be observed at the cell surface (Fig. 7a), cells infected with M2(AH-Mut) virus showed the presence of many virions in the process of budding; however, the scission and release of these virions appeared to be impaired (Fig. 7a, b). These mutant virions on the surface of M2(AH-Mut) virus-infected cells exhibited the classical ‘beads on a string’ morphology found in viruses with late domain mutations that are unable to recruit host ESCRT proteins, and so fail to undergo membrane scission (Fig. 7b) (Yuan et al., 2000). It is important to note that, though the membrane neck connecting each of the attached virions was not always visible, this is most likely due to its presence out of the plane of sectioning. Immuno-gold labeling of M2(AH-Mut) incompletely-budded virions showed that 56.5% (n=929) of all gold particles localized to M2 foci at the base of the budding M2(AH-Mut) virions and also at the constrictions between incompletely budded virions (Fig. 7c). This suggests that M2 is incorporated into the virion as budding nears completion and that mutation of the M2 amphipathic helix leads to incorporation of M2 at the membrane proximal end of each particle; however, M2 is unable to complete membrane scission, resulting in a string of attached particles. The ‘beads on a string’ morphology is also found during infection with a virus that does not produce the M2 protein (ΔM2; Fig. 7a, d), confirming an earlier observation (McCown and Pekosz, 2006). The failed-scission morphology of both M2(AH-Mut) and ΔM2 viruses (Fig. 7b, d), coupled with previous results showing that these viruses have a significant growth defect and are impaired in viral particle release (Cheung et al., 2005; Jackson and Lamb, 2008; Rossman et al., 2010), suggests that M2 plays a necessary role in mediating membrane scission and is required for the efficient release of budding virions.
Figure 7. The M2 Amphipathic Helix is Necessary for Membrane Scission and Virion Release.
MDCK cells were infected with an MOI of 3 pfu/cell of (A) A/Udorn/72, (A-C) A/Udorn/72 M2(AH-Mut) or (D) A/Udorn/72-ΔM2 for 18 h and thin sections were analyzed by electron microscopy. M2 was detected via immunogold labeling in (B). Arrows indicate foci of M2 at points of failed membrane scission. Scale bars indicate 100 nm.
Discussion
Although many steps in the assembly of influenza virus have been proposed, the final pinching off of a budding virion (membrane scission) lacks for a mechanistic explanation. In this study we identify a crucial role for the influenza virus M2 protein in virus budding. We observe that M2 is capable of altering membrane curvature, causing membrane budding and scission in a reduced-cholesterol environment (Fig. 2). During virus assembly and budding, M2 localizes to the neck of budding virions (Fig. 6) where it is required for membrane scission and the release of budding virions (Fig. 7).
Our previous work showed that expression of HA, NA or M2 causes budding in a VLP system, however, expression of both HA and M2 increased the efficiency of VLP release compared to each protein alone (Chen et al., 2007). During virus infection, mutation or deletion of HA eliminates the production of infectious virus, but does not significantly alter the numbers of viral particles that bud from the cell (Chen et al., 2005; Pattnaik et al., 1986). In contrast, mutation or deletion of the M2 protein impairs both infectivity as well as viral particle release, for multiple different influenza virus strains (Chen et al., 2008; Cheung et al., 2005; Iwatsuki-Horimoto et al., 2006; McCown and Pekosz, 2005, 2006; Rossman et al., 2010). In the absence of M2, budding virions still form, however, membrane scission and virion release never occurs (Fig. 7b, d). This suggests that HA may be able to initiate the budding event, but that assembly of other viral proteins may prevent membrane scission until the recruitment of M2.
Following the HA-mediated initiation of virus budding, M1 may bridge HA and M2, allowing for the recruitment of M2 to the, lipid-raft enriched, sites of virus budding (Chen et al., 2008; Rossman et al., 2010). M2 in this cholesterol-rich environment would be unable to induce membrane scission (Fig. 2) and may induce negative membrane curvature instead (Fig. 2a). This block in scission may stabilize the site of virus budding long enough to allow for recruitment and assembly of the full complement of viral proteins. Virion assembly may eventually deplete the local pool of raft-associated HA, placing M2 at the boundary between the lipid ordered phase of the budding virus and the lipid disordered phase of the bulk plasma membrane (Rossman et al., 2010). Acting at the phase boundary, insertion of the M2 amphipathic helix into the Ld phase may alter the line tension between the lipid phases (Fig. 4). M2-mediated modifications of line tension may provide the driving force for alterations in membrane curvature, as has been previously suggested for other line-tension altering proteins (Allain and Ben Amar, 2006; Liu et al., 2006; Romer et al., 2010). Alternatively, the M2 amphipathic helix may only be able to insert into low-cholesterol membranes due to rigidifying effects of cholesterol. Deep insertion of the amphipathic helix into the membrane may directly induce lipid packing defects, causing alterations in membrane curvature without the need for line tension-derived energy. Alteration of membrane curvature at the neck of the budding virus may be sufficient to cause membrane scission and the release of the budding virus (Figs. 2–3, 6–7). Mutation of the M2 amphipathic helix or alteration of M2 localization though disruption of M1 binding prevents membrane scission and the release of the budding virion, explaining the growth defect observed in both mutant viruses (Chen et al., 2008; McCown and Pekosz, 2006; Rossman et al., 2010).
Our results show a mechanism for influenza virus budding, that of a virus-encoded scission machine. Future investigation will determine if other enveloped viruses whose budding is thought to be ESCRT-independent (reviewed in Chen and Lamb, 2008) also encode their own ESCRT-substitutes, capable of mediating membrane scission and virus release. Our data show that the M2 amphipathic helix is necessary and sufficient to mediate membrane scission, and suggest that M2 may function as a virus-encoded ESCRT substitute, responsible for mediating the budding of influenza virions. Thus, we conclude that M2 is a multifunctional protein that, in addition to its proton-selective ion channel activity, has an essential role in virus budding.
Experimental Procedures
Cell, Viruses and Reagents
Madin-Darby canine kidney (MDCK) cells, M2-MDCK cells, 293T cells, viral stock propagation and viral infections were as previously described (Chen et al., 2008). M2 AH-Mut (previously called M2-Helix) and ΔM2 recombinant viruses have been previously described (Rossman et al., 2010); (Jackson and Lamb, 2008). Antibodies used were: αHA (NR3118, BEI, Manassas, VA) and αM2 (ectodomain mAb 14C2; (Zebedee and Lamb, 1988). Peptides were synthesized by GenScript (Piscataway, NJ). Additional detail is included as supplemental information.
Immunofluorescence Microscopy
MDCK cells grown on glass coverslips were infected as indicated, fixed and stained for HA and M2. Virus supernatant was stained following spotting on a glass coverslip. For Rab11 knock-down, A549 cells were transfected with control siRNA or siRNAs targeting Rab11a and Rab11b, infected as indicated and stained for HA and M2. Fluorescent intensity was quantified for 50–100 cells and averaged. Images were collected on a LSM5 Pascal (Zeiss, Thornwood, NY) confocal microscope.
Electron Microscopy
LUVs were treated as indicated and stained with phosphotungstic acid before imaging. For cryo-electron microscopy, LUVs were imbedded in vitreous ice using an FEI Vitrobot (Hillsboro, Or) and imaged using a Gatan (Pleasanton, CA) cryo-holder. For analysis of virus-infected cells, MDCK cells were treated as indicated and subjected to pre-embedding αM2 immunogold labeling followed by thin section examination as previously described (Leser et al., 1996). Samples were imaged on a JEOL 1230 (Tokyo, Japan) electron microscope.
Protein Purification
M2 and M2(AH-Mut) were expressed using the Bac-N-Blue baculovirus expression system (Invitrogen) and purified as previously described (Tosteson et al., 1994).
Large Unilamellar Vesicles
LUVs were prepared by extrusion from a 4:1:2 molar ratio of POPC:POPG:Cholesterol (Avanti Polar Lipids, Alabaster, AL) or a 4:1 molar ratio of POPC:POPG using purified wt M2 or M2 mutant protein where indicated.
Giant Unilamellar Vesicles
GUVs were electroformed using LUV lipid ratios with 0.5 mol % of TopFluor-Cholesterol (Avanti) added for visualization. Phase separated GUVs were electroformed at 60 °C using a 2:2:1 or 10:10:1 molar ratio of DOPC:SM:Cholesterol, incorporating 0.5 mol % of TopFluor-Cholesterol and Rho-PE (Avanti). GUVs containing M2 protein were prepared through dehydration of M2-containing LUVs as previously described (Girard et al., 2004; Streicher et al., 2009).
Plasma Membrane Spheres
Plasma membrane spheres were prepared as previously described (Keller et al., 2009; Lingwood et al., 2008), transfected with M2 or M2(AH-Mut)-pCAGGS and incubated in PMS buffer for 12 h. For visualization PMSs were incubated with labeled CTxB, stained for M2 and imaged at 37 °C.
Budding Assay
293T cells were transfected with M2 or M2(AH-Mut)-pCAGGS. 24 h post-transfection supernatant was collected and concentrated. Supernatant and cell lysates were analyzed by SDS-PAGE and immunoblotting as previously described (Chen et al., 2008).
Highlights.
The influenza virus M2 protein contains a highly-conserved amphipathic helix.
The M2 amphipathic helix is sufficient for ESCRT-independent budding in vitro.
M2 localizes to the neck of budding virions.
Mutation of the M2 amphipathic helix inhibits membrane scission and virion release.
Supplementary Material
Acknowledgments
We thank members of the Lamb laboratory for helpful discussions and critical reading of the manuscript. The electron microscopy was performed in the Northwestern University Biological Imaging Facility. This research was supported in part by a grant R01 AI-20201 from the National Institute of Allergy and Infectious Diseases. J.S.R. is an Associate and R.A.L. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allain JM, Ben Amar M. Budding and fission of a multiphase vesicle. Eur Phys J E. 2006;20:409–420. doi: 10.1140/epje/i2006-10030-4. [DOI] [PubMed] [Google Scholar]
- Antonny B. Membrane deformation by protein coats. Curr Opin Cell Biol. 2006;18:386–394. doi: 10.1016/j.ceb.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Barman S, Adhikary L, Chakrabarti AK, Bernas C, Kawaoka Y, Nayak DP. Role of transmembrane domain and cytoplasmic tail amino acid sequences of influenza a virus neuraminidase in raft association and virus budding. J Virol. 2004;78:5258–5269. doi: 10.1128/JVI.78.10.5258-5269.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DA, Rose JK. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 1992;68:533–544. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- Bruce EA, Digard P, Stuart AD. The rab11 pathway is required for influenza a virus budding and filament formation. J Virol. 2010;84:5848–5859. doi: 10.1128/JVI.00307-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce EA, Medcalf L, Crump CM, Noton SL, Stuart AD, Wise HM, Elton D, Bowers K, Digard P. Budding of filamentous and non-filamentous influenza A virus occurs via a VPS4 and VPS28-independent pathway. Virology. 2009;390:268–278. doi: 10.1016/j.virol.2009.05.016. [DOI] [PubMed] [Google Scholar]
- Bui M, Whittaker G, Helenius A. Effect of M1 protein and low pH on nuclear transport of influenza virus ribonucleoproteins. J Virol. 1996;70:8391–8401. doi: 10.1128/jvi.70.12.8391-8401.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JG, Martin-Serrano J. The ESCRT machinery: new functions in viral and cellular biology. Biochem Soc Trans. 2009;37:195–199. doi: 10.1042/BST0370195. [DOI] [PubMed] [Google Scholar]
- Chazal N, Gerlier D. Virus entry, assembly, budding, and membrane rafts. Microbiol Mol Biol Rev. 2003;67:226–237. doi: 10.1128/MMBR.67.2.226-237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BJ, Lamb RA. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology. 2008;372:221–232. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BJ, Leser GP, Jackson D, Lamb RA. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J Virol. 2008;82:10059–10070. doi: 10.1128/JVI.01184-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BJ, Leser GP, Morita E, Lamb RA. Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid-derived virus-like particles. J Virol. 2007;81:7111–7123. doi: 10.1128/JVI.00361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BJ, Takeda M, Lamb RA. Influenza virus hemagglutinin (H3 subtype) requires palmitoylation of its cytoplasmic tail for assembly: M1 proteins of two subtypes differ in their ability to support assembly. J Virol. 2005;79:13673–13684. doi: 10.1128/JVI.79.21.13673-13684.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik L, Kozlov MM, Zimmerberg J. Lipids in biological membrane fusion. J Membr Biol. 1995;146:1–14. doi: 10.1007/BF00232676. [DOI] [PubMed] [Google Scholar]
- Cheung TK, Guan Y, Ng SS, Chen H, Wong CH, Peiris JS, Poon LL. Generation of recombinant influenza A virus without M2 ion-channel protein by introduction of a point mutation at the 5′ end of the viral intron. J Gen Virol. 2005;86:1447–1454. doi: 10.1099/vir.0.80727-0. [DOI] [PubMed] [Google Scholar]
- de Meyer F, Smit B. Effect of cholesterol on the structure of a phospholipid bilayer. Proc Natl Acad Sci USA. 2009;106:3654–3658. doi: 10.1073/pnas.0809959106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey CE. The actions of melittin on membranes. Biochim Biophys Acta. 1990;1031:143–161. doi: 10.1016/0304-4157(90)90006-x. [DOI] [PubMed] [Google Scholar]
- Drin G, Antonny B. Amphipathic helices and membrane curvature. FEBS Lett. 2010;584:1840–1847. doi: 10.1016/j.febslet.2009.10.022. [DOI] [PubMed] [Google Scholar]
- Egashira M, Gorbenko G, Tanaka M, Saito H, Molotkovsky J, Nakano M, Handa T. Cholesterol modulates interaction between an amphipathic class A peptide, Ac-18A-NH2, and phosphatidylcholine bilayers. Biochemistry. 2002;41:4165–4172. doi: 10.1021/bi011885+. [DOI] [PubMed] [Google Scholar]
- Girard P, Pecreaux J, Lenoir G, Falson P, Rigaud JL, Bassereau P. A new method for the reconstitution of membrane proteins into giant unilamellar vesicles. Biophys J. 2004;87:419–429. doi: 10.1529/biophysj.104.040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatsuki-Horimoto K, Horimoto T, Noda T, Kiso M, Maeda J, Watanabe S, Muramoto Y, Fujii K, Kawaoka Y. The cytoplasmic tail of the influenza A virus M2 protein plays a role in viral assembly. J Virol. 2006;80:5233–5240. doi: 10.1128/JVI.00049-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D, Lamb RA. The influenza A virus spliced messenger RNA M mRNA3 is not required for viral replication in tissue culture. J Gen Virol. 2008;89:3097–3101. doi: 10.1099/vir.0.2008/004739-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Leser GP, Zhang J, Lamb RA. Influenza virus hemagglutinin and neuraminidase cytoplasmic tails control particle shape. EMBO J. 1997;16:1236–1247. doi: 10.1093/emboj/16.6.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser HJ, Lingwood D, Levental I, Sampaio JL, Kalvodova L, Rajendran L, Simons K. Order of lipid phases in model and plasma membranes. Proc Natl Acad Sci USA. 2009;106:16645–16650. doi: 10.1073/pnas.0908987106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller H, Lorizate M, Schwille P. PI(4,5)P2 degradation promotes the formation of cytoskeleton-free model membrane systems. Chemphyschem. 2009;10:2805–2812. doi: 10.1002/cphc.200900598. [DOI] [PubMed] [Google Scholar]
- Lamaziere A, Burlina F, Wolf C, Chassaing G, Trugnan G, Ayala-Sanmartin J. Non-metabolic membrane tubulation and permeability induced by bioactive peptides. PLoS One. 2007;2:e201. doi: 10.1371/journal.pone.0000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb RA, Pinto LH. The proton selective ion channels of influenza A and B viruses. In: Kawaoka Y, editor. Contemporary Topics in Influenza Virology. Wymondham, Norfolk, UK: Horizon Scientific Press; 2005. pp. 65–93. [Google Scholar]
- Lee MT, Hung WC, Chen FY, Huang HW. Mechanism and kinetics of pore formation in membranes by water-soluble amphipathic peptides. Proc Natl Acad Sci USA. 2008;105:5087–5092. doi: 10.1073/pnas.0710625105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leser GP, Ector KJ, Lamb RA. The paramyxovirus simian virus 5 hemagglutinin-neuraminidase glycoprotein, but not the fusion glycoprotein, is internalized via coated pits and enters the endocytic pathway. Mol Biol Cell. 1996;7:155–172. doi: 10.1091/mbc.7.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leser GP, Lamb RA. Influenza virus assembly and budding in raft-derived microdomains: a quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology. 2005;342:215–227. doi: 10.1016/j.virol.2005.09.049. [DOI] [PubMed] [Google Scholar]
- Lingwood D, Ries J, Schwille P, Simons K. Plasma membranes are poised for activation of raft phase coalescence at physiological temperature. Proc Natl Acad Sci USA. 2008;105:10005–10010. doi: 10.1073/pnas.0804374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Kaksonen M, Drubin DG, Oster G. Endocytic vesicle scission by lipid phase boundary forces. Proc Natl Acad Sci USA. 2006;103:10277–10282. doi: 10.1073/pnas.0601045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Polishchuk AL, Ohigashi Y, Stouffer AL, Schon A, Magavern E, Jing X, Lear JD, Freire E, Lamb RA, et al. Identification of the functional core of the influenza A virus A/M2 proton-selective ion channel. Proc Natl Acad Sci USA. 2009;106:12283–12288. doi: 10.1073/pnas.0905726106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCown MF, Pekosz A. The influenza A virus M2 cytoplasmic tail is required for infectious virus production and efficient genome packaging. J Virol. 2005;79:3595–3605. doi: 10.1128/JVI.79.6.3595-3605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCown MF, Pekosz A. Distinct domains of the influenza a virus M2 protein cytoplasmic tail mediate binding to the M1 protein and facilitate infectious virus production. J Virol. 2006;80:8178–8189. doi: 10.1128/JVI.00627-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen PA, Soto CS, Polishchuk A, Caputo GA, Tatko CD, Ma C, Ohigashi Y, Pinto LH, DeGrado WF, Howard KP. pH-induced conformational change of the influenza M2 protein C-terminal domain. Biochemistry. 2008;47:9934–9936. doi: 10.1021/bi801315m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noton SL, Medcalf E, Fisher D, Mullin AE, Elton D, Digard P. Identification of the domains of the influenza A virus M1 matrix protein required for NP binding, oligomerization and incorporation into virions. J Gen Virol. 2007;88:2280–2290. doi: 10.1099/vir.0.82809-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattnaik AK, Brown DJ, Nayak DP. Formation of influenza virus particles lacking hemagglutinin on the viral envelope. J Virol. 1986;60:994–1001. doi: 10.1128/jvi.60.3.994-1001.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto LH, Dieckmann GR, Gandhi CS, Papworth CG, Braman J, Shaughnessy MA, Lear JD, Lamb RA, DeGrado WF. A functionally defined model for the M2 proton channel of influenza A virus suggests a mechanism for its ion selectivity. Proc Natl Acad Sci USA. 1997;94:11301–11306. doi: 10.1073/pnas.94.21.11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto LH, Lamb RA. The M2 proton channels of influenza A and B viruses. J Biol Chem. 2006;281:8997–9000. doi: 10.1074/jbc.R500020200. [DOI] [PubMed] [Google Scholar]
- Pinto LH, Lamb RA. Controlling influenza virus replication by inhibiting its proton channel. Mol Biosyst. 2007;3:18–23. doi: 10.1039/b611613m. [DOI] [PubMed] [Google Scholar]
- Romer W, Pontani LL, Sorre B, Rentero C, Berland L, Chambon V, Lamaze C, Bassereau P, Sykes C, Gaus K, et al. Actin dynamics drive membrane reorganization and scission in clathrin-independent endocytosis. Cell. 2010;140:540–553. doi: 10.1016/j.cell.2010.01.010. [DOI] [PubMed] [Google Scholar]
- Rossman JS, Jing X, Leser GP, Balannik V, Pinto LH, Lamb RA. Influenza virus M2 ion channel protein is necessary for filamentous virion formation. J Virol. 2010;84:5078–5088. doi: 10.1128/JVI.00119-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell CJ, Jardetzky TS, Lamb RA. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 2001;20:4024–4034. doi: 10.1093/emboj/20.15.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell JR, Chou JJ. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451:591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder C, Heider H, Moncke-Buchner E, Lin TI. The influenza virus ion channel and maturation cofactor M2 is a cholesterol-binding protein. Eur Biophys J. 2005;34:52–66. doi: 10.1007/s00249-004-0424-1. [DOI] [PubMed] [Google Scholar]
- Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- Streicher P, Nassoy P, Barmann M, Dif A, Marchi-Artzner V, Brochard-Wyart F, Spatz J, Bassereau P. Integrin reconstituted in GUVs: a biomimetic system to study initial steps of cell spreading. Biochim Biophys Acta. 2009;1788:2291–2300. doi: 10.1016/j.bbamem.2009.07.025. [DOI] [PubMed] [Google Scholar]
- Takeda M, Leser GP, Russell CJ, Lamb RA. Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion. Proc Natl Acad Sci USA. 2003;100:14610–14617. doi: 10.1073/pnas.2235620100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Gao PF, Pinto LH, Lamb RA, Cross TA. Initial structural and dynamic characterization of the M2 protein transmembrane and amphipathic helices in lipid bilayers. Protein Sci. 2003;12:2597–2605. doi: 10.1110/ps.03168503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosteson MT, Pinto LH, Holsinger LJ, Lamb RA. Reconstitution of the influenza virus M2 ion channel in lipid bilayers. J Membr Biol. 1994;142:117–126. doi: 10.1007/BF00233389. [DOI] [PubMed] [Google Scholar]
- Venkatachalapathi YV, Phillips MC, Epand RM, Epand RF, Tytler EM, Segrest JP, Anantharamaiah GM. Effect of end group blockage on the properties of a class A amphipathic helical peptide. Proteins. 1993;15:349–359. doi: 10.1002/prot.340150403. [DOI] [PubMed] [Google Scholar]
- Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan B, Campbell S, Bacharach E, Rein A, Goff SP. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J Virol. 2000;74:7250–7260. doi: 10.1128/jvi.74.16.7250-7260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebedee SL, Lamb RA. Influenza A virus M2 protein: monoclonal antibody restriction of virus growth and detection of M2 in virions. J Virol. 1988;62:2762–2772. doi: 10.1128/jvi.62.8.2762-2772.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Lamb RA. Characterization of the membrane association of the influenza virus matrix protein in living cells. Virology. 1996;225:255–266. doi: 10.1006/viro.1996.0599. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.