Abstract
The PIV-5 hemagglutinin-neuraminidase (HN) protein is a multifunctional protein with sialic acid binding, neuraminidase and fusion promotion activity. HN is internalized by clathrin-mediated endocytosis and degraded. HN lacks internalization signals in its cytoplasmic tail but a single glutamic acid present at residue 37 at the putative transmembrane/ectodomain boundary is critical. We rescued rPIV-5 with mutations E37D or E37K, which have been shown to impair or abolish HN internalization, respectively. These viruses exhibited growth properties similar to wild-type (wt) virus but are impaired for fitness in tissue culture. Biochemical analysis of HN activities showed differences between HN E37D and HN E37K in fusion promotion and incorporation of HN and F into virions. Furthermore, oligomeric analyses indicate that HN E37 mutants perturb the tetrameric organization of HN, probably by destabilizing the dimer-of-dimers interface.
Introduction
Paramyxoviruses are enveloped, negative-stranded RNA viruses that include many clinically and agriculturally important pathogens such as mumps virus, measles virus, Newcastle disease virus (NDV), Hendra virus, and Nipah virus. Cellular entry by paramyxoviruses is mediated by two glycoproteins present at the surface of the virion. For the paramyxovirus parainfluenza virus 5 (PIV-5), these proteins are the fusion protein (F) and the hemagglutinin-neuraminidase protein (HN). F mediates the fusion of the viral membrane with the cellular plasma membrane at neutral pH. Coexpression of the HN protein enhances this fusion process by lowering the activation energy required for F to mediate fusion (Russell et al., 2001). In addition to its fusion promotion activity, HN also functions in binding the virion to its receptor sialic acid on target cells and possesses receptor-destroying activity (neuraminidase activity) that cleaves sialic acid from the surface of both infected cells and virions. This action is thought to prevent the aggregation of budded virions at the surface of infected cells. The viral matrix (M) protein is a peripheral membrane protein that underlies the lipid bilayer and makes contact with the glycoprotein cytoplasmic tails. The PIV-5 ribonucleoprotein is composed of three proteins: nucleocapsid (NP), phosphoprotein (P), and the large polymerase (L), which together act to transcribe and replicate the genome RNA (Lamb and Parks, 2007). Additionally, PIV-5 contains proteins that assist in evasion of host cell immunity: the small hydrophobic protein (SH), which inhibits tumor necrosis factor alpha signaling and prevents apoptosis in infected cells (He et al., 2001; Lin et al., 2003; Wilson et al., 2006) and the V protein that antagonizes interferon synthesis and signaling (Andrejeva et al., 2004; Didcock et al., 1999).
PIV-5 HN is a type II integral membrane protein that consists of a short N-terminal cytoplasmic tail of 17 residues, a hydrophobic domain of 19 residues that acts as both a signal sequence to target HN to the ER membrane and as a stop-transfer transmembrane (TM) domain, a stalk region of 82 residues, and a large globular head (447 residues) that contains both the receptor binding and destroying activities (Hiebert et al., 1985; Parks and Lamb, 1990). HN exists at the surface of virus-infected cells as a tetramer, consisting of two disulfide-linked dimers that are linked through noncovalent interactions (Ng et al., 1989). The crystal structure of the full-length ectodomain of HN has been solved both in the presence and absence of ligand (Yuan et al., 2005). No electron density was identified for the stalk region; however, biophysical data indicates that the stalk adopts a flexible and rod-like α-helical conformation. Additionally, HN head domain expressed with the stalk forms a tetramer, whereas expression of the HN head domain on its own is monomeric. Thus it is thought that the stalk domain stabilizes the head domain oligomer (Yuan et al., 2008). The enzymatically active head region of HN contains the antigenic sites of the protein and has a typical sialidase/neuraminidase fold, a superbarrel with six antiparallel β strands with a centrally located active site. The crystal structure data shows that the dimer interface within the head region buries an extensive area between the monomers of 1810 Å2. In contrast, the interface between the dimer-of-dimers is much smaller, burying only 657 Å2 and involving ten residues (Yuan et al., 2005). This suggests that the dimer-of-dimers interface may be easier to perturb. The interaction between the dimer-of-dimers interface is not well conserved among paramyxovirus attachment proteins, and its weaker interaction energy could be a feature of the process of F activation.
Despite the fact that PIV-5 HN is a major spike glycoprotein of the budded virion, in virus-infected cells PIV-5 HN is extensively internalized from the cell surface whereas F is not internalized and is stably expressed at the cell surface (Ng et al., 1989). Furthermore, it has been shown that HN is internalized by clathrin-coated pits and enters the endocytic pathway (Leser et al., 1996). When HN was expressed from cDNA using an SV40-recombinant virus it was found that the rate of HN turnover from the cell surface was 6.5–7.0%/min, which is faster than the bulk membrane turnover and comparable with the rate of other endocytosed receptors. Analysis by electron microscopy showed that PIV-5 HN colocalizes with transferrin and gold-conjugated bovine serum albumin, markers for early endosomal/late endosomal compartments and lysosomal compartments, respectively (Leser et al., 1996). Furthermore it was shown by fluorescence microscopy that HN colocalized with lamp-1, indicating that the lysosome is the subcellular compartment where internalized HN becomes localized (Leser et al., 1996).
The observation that PIV-5 HN is internalized from the cell surface was surprising, as the protein lacks a canonical internalization signal in its cytoplasmic tail. The 17 residue cytoplasmic tail of HN does not contain a tyrosine-based internalization sequence, nor does it have other known internalization signals such as a di-leucine, a di-lysine, or a site for monoubiquitination (Bonifacino and Weissman, 1998; Chen et al., 1990; Collawn et al., 1990; Hamer et al., 1997; Itin et al., 1995; Letourneur and Klausner, 1992; Trowbridge et al., 1993). Mutagenesis experiments showed the cytoplasmic tail of HN is dispensable for internalization (Leser et al., 1999). Further, a single glutamic acid (E37), the first residue of the ectodomain at the putative TM/ectodomain boundary, was shown to be critical for HN internalization (Leser et al., 1999). A series of E37 HN mutants were constructed and expressed in cells. One of these mutants, E37D, had a rate of internalization and overall extent of internalization that was significantly less than that of wild-type (wt) HN. Other mutants, such as E37K, were not internalized above background levels. Reversing the first seven amino acids of the ectodomain was also examined, as was substituting leucine for the glutamic acid at position 37 and glutamic acid for the leucine at position 43; all of these combinations diminished HN internalization. Thus it was concluded that the presence of a glutamic acid at position 37 was critical for PIV-5 HN internalization (Leser et al., 1999).
We have examined the role of internalization mutants on the lifecycle of PIV-5 through the rescue of recombinant PIV-5 containing the mutations HN E37D or E37K. These rescued viruses were greatly impaired or defective for HN internalization during cellular infection. We describe the growth characteristics of these viruses together with an analysis of the enzymatic and receptor binding activities of the mutated HN proteins. The ability of these E37 mutants to promote F mediated fusion is also described, and related to findings regarding the oligomeric organization and stabilities of these proteins.
Results
HN internalization in virus-infected cells
Previous work has demonstrated that in PIV-5-infected cells, HN is internalized from the cell surface and degraded by lysosomes (Ng et al., 1989). Analysis of HN expressed from a recombinant SV40 virus indicated that internalization of HN was an intrinsic property of the HN protein, not requiring expression of other PIV-5 proteins (Leser et al., 1996). It was also found that mutation of the charged residue (E37) that defines the ectodomain/TM domain boundary largely abolished internalization (Leser et al., 1996; Leser et al., 1999). To determine if the HN E37 mutants affected internalization in PIV-5-infected cells, HN E37D and HN E37K mutations were recovered in PIV-5 virus using a previously described reverse genetics system (He et al., 1997; Waning et al., 2002). To detect HN degradation, CV-1 cells were infected with wt PIV-5 or with mutant HN E37D or HN E37K viruses. The virus-infected cells were metabolically labeled with [35S]-TranS-label in a pulse chase protocol followed by immunoprecipitation with a mix of monoclonal antibodies and analysis of polypeptides by SDS-PAGE. Two of the antibodies used for immunoprecipitation (HN-1b and HN-4b) have been shown previously to be conformation specific (Ng et al., 1989). Thus, samples taken at 0 time display lower signals than those samples taken after 1 h because the conformational epitopes had not fully matured (Fig. 1).
Figure 1.

Stability of HN E37D and HN E37K in PIV-5-infected cells. CV-1 cells were infected with wt PIV-5 or PIV-5 HN E37D or HN E37K and at 18 h p.i. were labeled with [35S]-TranS-label for 30 min (pulse), the label removed and the cells incubated in DMEM for varying periods ranging from 0–5 h. Cells were lysed in lysis buffer and proteins immunoprecipitated with a mix of HN monoclonal antibodies and polypeptides analyzed by SDS-PAGE as described in Materials and Methods. The time of chase period is indicated above each lane in hours.
Cells infected with wt virus displayed robust fragmentation of HN, with degradation products (40 kD and 34 kD) detectable as early as two h in the chase period (Fig. 1). By five h post-label, approximately 61% of the detectable HN protein was present as degradation products. In contrast, cells infected with either PIV-5 HN E37D or HN E37K viruses did not display high levels of HN degradation. For HN E37D virus-infected cells, degradation products were barely detectable at three h post-label and comprised only 2.7% of detectable HN at five h post-label. Results for HN E37K virus-infected cells were similar with degradation products comprising less than 1% of all detectable HN at the five h time point. The small difference in HN E37D and HN E37K degradation mirrors earlier findings in that the block of HN E37D internalization is not absolutely complete as compared with HN E37K internalization, which is indistinguishable from the bulk membrane turnover (Leser et al., 1999). This analysis shows that mutations at residue E37 in HN block internalization and degradation of the protein in the context of virus-infected cells.
To assess further the stability of HN E37D and HN E37K at the cell surface protein synthesis was inhibited in virus-infected cells at 18 h p.i. by treatment with cycloheximide to prevent transport of newly synthesized HN to the cell surface. At 0 or 4 h after cycloheximide treatment, cells were fixed and labeled using either HN-1b to detect HN or F1a to detect the F protein, and stained using an Alexa Fluor 488-conjugated secondary antibody. At the t=0 time point, HN staining was widespread at the surface of the infected cells and there was no detectable staining above background for the mock infected cells (Fig. 2). After four h of cycloheximide treatment there was less HN present on the surface of the infected cells, though HN protein was detectable (Fig. 2). This is consistent with the immunoprecipitation experiments shown above that indicated that about half of wt HN protein in PIV-5-infected cells was still present in its full-length form at four h post-labeling (Fig. 1). This is also consistent with the data obtained in flow cytometry experiments (Fig. 3). In contrast, cells infected with PIV-5 HN E37D or HN E37K virus did not display a decrease in HN staining (Fig. 2). This indicates HN E37D and HN E37K proteins are stably expressed at the surface of infected cells for at least four h. PIV-5 F protein was stably expressed at the cell surface confirming earlier data (Leser et al., 1996; Ng et al., 1989). This experiment also demonstrates that the stability of HN protein on the cell surface does not affect the stability of F on the cell surface when HN and F are present together in virus-infected cells.
Figure 2.
Cell surface fluorescence of wt HN and E37 mutants in PIV-5-infected cells. PIV-5-infected CV-1 cells at 18 h p.i. (0 h) were incubated with 100 μg/ml cycloheximide to inhibit protein synthesis. At either 0 or 4 h, cells were fixed for indirect fluorescence staining and bound with antibodies specific for either HN (MAb HN-1b) or F (MAb F1a) and then with Alexa Fluor 488-conjugated goat anti-mouse secondary antibody. Fluorescence was detected using a Zeiss Axiovert 200M microscope with Apotome. The top two rows show HN cell surface staining at 0 and 4 h post-treatment, respectively; the bottom two rows show F cell surface staining at 0 and 4 h post-treatment, respectively. Bar, 10 μm.
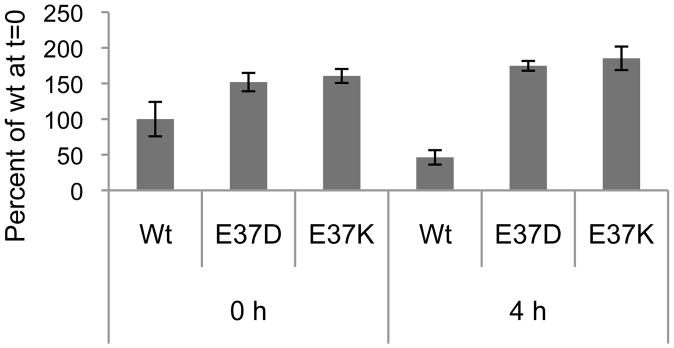
Figure 3.
Detection of wt HN and E37 mutant HNs at the surface of infected cells by flow cytometry. (A) PIV-5-infected CV-1 cells at 18 h p.i. (0 h) were incubated with 100 μg/ml cycloheximide to inhibit protein synthesis. At either 0 or 4 h, cells were fixed for indirect fluorescence staining and bound with antibodies specific for either HN (HN-1b) or F (F1a) and then with a fluoroscein-isothiocyanate-conjugated secondary antibody. The mean fluorescence intensity (MFI) of each sample is expressed as a percentage of the MFI of wt HN at 0 h.
The surface expression level of HN E37D and HN E37K in PIV-5-infected cells was also analyzed by flow cytometry. CV-1 cells were infected for 18 h and treated with cycloheximide as described above. Following cycloheximide treatment for either zero or four h, cells were fixed and stained for either HN or F proteins using either a polyclonal HN antibody or mAb F1a and then analyzed by flow cytometry. The results were expressed as a percentage of the mean fluorescence intensity of the wt HN staining at t=0. Fig. 3 shows that at the zero h time point there is ~50% more HN E37D and E37K present on the surface on virus-infected cells when compared to wt virus-infected cells. At the four h time point, the amount of wt HN staining present on PIV-5-infected cells had decreased ~50%. In comparison, there were similar fluorescence intensities of HN E37D and HN E37K present at both the zero-h and four-h time points (Fig. 3). Flow cytometry also showed that there were similar percentages of virus-infected cells for all viruses used. Parallel experiments were conducted to determine the level of F staining in either wt or E37 mutant virus-infected cells. The data demonstrated that the F protein was stable in virus-infected cells after four h of cycloheximide treatment (data not shown).
Growth properties of viruses containing HN E37D and E37K mutations
To determine the growth rate for rPIV-5 viruses containing mutations at residue E37, MBDK cells were infected at a MOI of 0.01 PFU/cell and cell culture medium was harvested every 24 h for six days. As shown in Fig. 4A, growth of the E37 mutants was indistinguishable from wt PIV-5 with the exception of the 24 h time point, at which the wt virus displayed a titer that was one log higher than either of the two mutant viruses.
Figure 4.
Growth kinetics and plaque formation of PIV-5 HN E37 mutants. (A) Growth curves of PIV-5 E37 mutants. MDBK cells were infected at an MOI of 0.01 pfu/cell and the culture media harvested at various times p.i. Titers at each time point are the average of triplicate experiments. (B) Morphology of plaques formed in BHK-21F PIV-5-infected cells at 4 days p.i.
To analyze further the growth of the E37 PIV-5 mutants, the plaque morphology in BHK-21 cells of each of the mutant viruses was compared to that of wt virus (Fig. 4B). The size and shape of rPIV-5 HN E37D and rPIV-5 HN E37K plaques were similar to those formed in wt PIV-5 virus-infected cells. These data indicate that rPIV-5 HN E37D and rPIV-5 HN E37K have similar growth properties to wt virus.
Fitness assay of viruses containing HN E37D and E37K mutations
To determine whether the decrease or lack of HN internalization in rPIV-5 HN E37D and rPIV-5 HN E37K infected-cells had an effect on the fitness of the virus when compared to wt PIV-5, the mutant E37 viruses were competed against wt PIV-5 in a series of sequence-based viral fitness competition assays. This method is based on one used to analyze mutants of vesicular stomatitis virus (Novella et al., 2004). To study the ratio of mutant virus RNA to wt virus RNA using nucleotide sequencing, a series of standards was developed using plasmid DNAs containing the HN cDNA. Different ratios (weight to weight) of pSV103 plasmid containing the coding region for either the full-length, wt HN or either one of the E37D or E37K mutations (Leser et al., 1999) were mixed together and sequenced using a primer that would cover the nucleotides corresponding to residue 37 in the HN protein. The electropherogram peaks corresponding to each of the mutations and the wt nucleotides (GAG->GAC for E37D and GAG->AAG for E37K) were measured and expressed as a ratio of the height of the mutant peak over the height of the wt peak. This experimentally determined peak ratio was plotted against the actual ratio of mutant to wt plasmid added to the reaction. The resultant graph was used to fit a line, the equation of which could be used to determine the ratio of mutant to wt virus in sequencing reactions of cDNA derived from viral RNA isolated from the cell culture supernatants of virus-infected cells. Figs. 5A and 5B show the sequencing standard curves for HN E37D:wt and HN E37K:wt, respectively. The straight line describes the best fit for the measured data (R2=0.99 for E37D:wt and R2=0.96 for E37K:wt).
Figure 5.
Viral fitness of PIV-5 HN E37 mutants compared to wt PIV-5 in tissue culture. (A) Sequencing standards for HN E37D versus wt HN. Plasmids encoding wt HN and HN E37D were mixed together in various ratios (by plasmid weight) and sequenced. The height of the mutant peak compared to the wt peak on each chromatogram was plotted against the ratio of the plasmid added to the assay and graphed. (B) Sequencing standards for HN E37K versus HN were prepared as described in (A). (C) Sequencing chromatograms of E37 codon for PIV-5 HN E37D and HN E37K versus wt PIV-5 at each infection passage of the passage-based fitness assay. MBDK cells were infected at a mutant to wt virus ratio of 20:1. At 72 h p.i., the media was harvested and used to infect a fresh plate of MBDK for three additional passages. Viral RNA was isolated from each medium collection. The viral RNA was reverse transcribed and sequenced over the E37 position. The relative heights of the chromatogram peaks at each mutation point were used to determine the final ratio of PIV-5 HN E37 mutant virus to wt virus present after each passage, based on the sequencing standards. (D) Quantification of fitness of PIV-5 HN E37D and HN E37K mutants versus wt PIV-5 at each passage number. The ratio of mutant to wt virus after each passage was expressed as a percentage of the original infection ratio and plotted. The slope of the line describes the relative fitness of each PIV-5 HN E37 mutant compared to wt virus. (E) Sequencing chromatograms of E37 codon for PIV-5 HN E37D and HN E37K versus wt PIV-5 at each time point in the time course fitness assay. MDBK cells were infected with PIV-5 at a mutant to wt ratio of 100:1. At the various time p.i. viral RNA was isolated from the medium, reverse transcribed, and sequenced as described in Materials and Methods.
To assess the fitness of the mutants compared to wt virus, two different approaches were used. In the first, either rPIV-5 HN E37D and wt rPIV-5 or rPIV-5 HN E37K and wt rPIV-5 were used to infect MDCK cells at a mutant:wt ratio of 20:1. Additionally, cells were infected individually with wt rPIV-5, rPIV-5 HN E37D, or HN E37K viruses to check that the virus sequence was stable over the time-course of the experiment. Three days p.i., the cell culture supernatant was used to infect a fresh plate of MDBK cells. At the same time, viral RNA was isolated from the supernatant and used to derive cDNA using reverse transcription followed by PCR. The resulting amplicon was subjected to sequencing using the same primer that was used to sequence the plasmids in order to make the standard sequencing curves. This process was repeated for four cycles. Fig. 5C shows examples of electropherograms generated by this process. For each electropherogram, the heights of the mutant and wt peaks were measured. The equation derived from the sequencing standards was then used along with the ratio of the measured peaks to determine the ratio of mutant to wt vRNA present in the cell culture supernatant at each passage. For passage 1 (p1) in the competition of either HN E37D and HN E37K with wt virus, the ratio of mutant to wt remained unchanged at roughly 20:1. However, by p2 the ratio of mutant to wt present in the cell culture supernatants began to decrease. By passage 4, the mutant to wt ratio was nearly 1:1 for HN E37D:wt and had decreased to ~1:3 for HN E37K:wt. The decrease in the mutant to wt virus ratio for each virus is graphed in Fig. 5D. The relative fitness of each mutant virus is expressed as the slope of the line that describes the change in ratio in each passage. The relative fitnesses of the mutant viruses are −0.29 and −0.33 for HN E37D and HN E37K, respectively.
In the second approach to analyzing viral fitness, MBDK cells were infected as above at a 100:1 mutant to wt ratio. Instead of passaging the virus every three days, in this experiment MDBK cells were allowed to remain infected for 8 days. MDBK cells can be infected with PIV-5 with minimal cytopathic effects for long periods of time (Choppin, 1964). At days 5, 6, 7 and 8 days p.i., a small aliquot of cell culture supernatant was removed from the cell culture dish. vRNA was isolated from the supernatant, and cDNA and amplicons for sequencing were prepared as described above. In this experiment, the mutant to wt ratio dropped from 100:1 to 2.1:1 and 4.9:1 for HN E37D and HN E37K, respectively (Fig. 5E). In a similarly designed experiment that lasted for 11 days, the mutant to wt ratio dropped from 100:1 to 2:1 and 4:1 for HN E37D and HN E37K, respectively (data not shown). Taken together, these assays indicate that the HN E37D and HN E37K viruses are less fit as compared to wt virus in tissue culture cells despite having similar growth properties when compared in a multi-step growth curve and by plaque morphology.
As a basis of comparison and as a verification of the technique, a virus that displays a severe growth defect was competed against wt virus in a fitness assay. PIV-5 HN Δ2-13 contains a deletion of twelve amino acids of the HN cytoplasmic tail and has been characterized previously (Schmitt et al., 1999). This virus is replication impaired as judged by its small plaque size, reduced replication rate, and lowered maximum titers when compared with wt PIV-5 virus. Sequencing electropherogram standards were constructed as described above. PIV-5 HN Δ2-13 was co-cultivated at a ratio of 100:1 with wt virus. By day 4, the mutant to wt ratio had dropped to 1:1 and by day 7 the wt had outcompeted the mutant and was present at a 1:5 mutant to wt ratio (data not shown). Thus, the data indicate that the loss of fitness for HN E37D and HN E37K is a subtle loss as compared to the major loss of a greatly impaired mutant virus.
E37 mutations affect the composition of PIV-5 virions
To investigate if the HN mutations affected virion composition, wt PIV-5 virus, HN E37D, and HN E37K viruses were grown in MDBK cells and virions purified. The polypeptide composition of the virions was studied using SDS-PAGE followed by either silver staining (Fig. 6A). The virions contained abundant amounts of HN, NP, F1/F2, P, and M. Viral proteins V, L, and SH were not readily visible by silver staining. The HN mutant viruses had similar compositions to wt virus, with the exception of the amount of F in the PIV-5 HN E37D virus as F1/2 were present in more abundant amounts as compared to wt PIV-5.
Figure 6.

Protein composition of rPIV-5 virions containing HN E37 mutations. (A) Wt PIV-5 and E37 mutant virions were grown in MBDK cells and purified on sucrose density gradients as described in Methods and Materials. Virion polypeptides were analyzed by SDS-PAGE and by silver staining. (B) Polypeptides from [35S]-methionine-labeled virions were analyzed by SDS-PAGE. Radioactivity was quantified on a Fuji FLA-5100 Imager and using MultiGauge v3.0 software.
To investigate further this finding, [35S]-methionine labeled virions were prepared for wt PIV-5, HN E37D and HN E37K viruses (Fig. 6B). The polypeptides of the virions were analyzed by SDS-PAGE and radioactivity quantified using Multigauge software. The abundance of each protein present in the virions was calculated based on 2541 NP proteins per genome RNA (Lamb and Parks, 2007), the known number of methionines in each protein and the quantified radioactivity per protein (Table 1). When compared with wt virus, both PIV-5 HN E37D and HN E37K have similar overall amounts of M protein. However, HN E37D virions have ~68% of the number of HN tetramers as wt virions. The differences for F trimers are even more dramatic; HN E37D virions contain 1.6 times the amount of F as compared to wt virions, confirming the results obtained from silver staining of virus. There were also differences observed for PIV-5 HN E37K virions, though they were not as large. HN E37K virions contained about 89% of the HN tetramers as wt virus, and slightly more F trimers. Overall, these changes affected the ratios of the glycoproteins on the surface of virions. For wt PIV-5 virions, HN and F were present at a 1.6:1 ratio. In contrast, for HN E37D and HN E37K the ratios were 0.7:1 and 1.3:1, respectively. These results suggest that mutations present at residue 37 of HN affect the recruitment of the surface glycoproteins into budding virions. These data also imply that the ratio of HN to F on the surface of virus-infected cells is not maintained in the budded virion (data not shown and Table 1), indicating that the virus preferentially recruits wt HN into the virion.
Table 1.
| HN Wt | HN E37D | HN E37K | |
|---|---|---|---|
| HN Tetramers | 629 | 425 | 559 |
| F Trimers | 389 | 635 | 440 |
| M | 5031 | 4904 | 5005 |
| HN/F Ratio | 1.62 | 0.67 | 1.27 |
To calculate the relative abundance of the F, HN and M proteins in virions [35S]-methionine labeled virions were prepared for wt PIV-5, HN E37D and HN E37K viruses (Fig. 6B). The polypeptides of the virions were analyzed by SDS-PAGE and radioactivity quantified using a Fuji BioImager and Multigauge software. The abundance of each protein present in the virions was calculated based on 2541 NP proteins per genome RNA (Lamb and Parks, 2007), the known number of methionine residues in each protein and the quantified radioactivity per protein. F and HN are calculated as trimers and tetramers to yield the average number of each spike glycoprotein per virion.
Analysis of HN neuraminidase and hemadsorption activities
As neuraminidase activity is required to prevent aggregation of virus on budding at the plasma membrane the neuraminidase activity of purified wt PIV-5 viruses and HN E37D and HN E37K was tested (Fig. 7A). It was found that whereas HN E37D and wt virions have similar neuraminidase activities when compared by overall amounts of virus added to the enzymatic reactions, when the amount of HN tetramers in the preparations was taken into consideration HN E37D has a neuraminidase activity that is over double that of wt HN (p=0.0009). In comparison, PIV-5 HN E37K has a similar activity to wt HN virus (~130% of wt, p=0.09). The kinetics of neuraminidase activity was also tested and the HN mutants were found to have very similar kinetics to that of wt HN (Fig. 7B).
Figure 7.
Receptor destroying and receptor binding activity of HN E37 mutants. (A) Neuraminidase activity of purified PIV-5 virions. Purified virions were mixed with N-acetylneuraminyl lactose at pH 4.5 for 1.5 h at 37 °C. Cleaved substrate was detected using a colorimetric assay as described in Methods and Materials and the absorbance read at 549 nm. Each average is the result of five experiments and is normalized to the amount of HN present in purified virions. (B) Neuraminidase kinetics of purified PIV-5 virions. Purified virions were treated as described in (A) and the amount of cleaved substrate present at various time points was determined by colorimetric assay. (C) Hemadsorption of PIV-5-infected cells. PIV-5-infected CV-1 cells at 18 h p.i. were incubated for 30 min with a 1% suspension of chicken red blood cells (RBCs) in PBS at pH 5.0 or pH 7.0. Bound RBCs were lysed and the absorbance of the lysed RBCs read at 410 nm.
To determine if there is an effect of the E37 mutations on the receptor binding capability of the HN protein, a hemadsorption assay was used. CV-1 cells were infected with wt PIV-5 or the HN E37 mutants and at 18 h p.i. were incubated with chicken erythrocytes for 1 h at 4 °C using two pHs, pH 7.0 and pH 5.0. pH 7.0 is similar to the extracellular environment where receptor binding takes place, and pH 5.0 is close to the pH optimum for the neuraminidase activity of the HN protein (Merz et al., 1981). Comparison of the receptor-binding activities of HN showed little difference between wt HN or either HN E37D or HN E37K at either pH (Fig. 7C).
Analysis of HN fusion promotion activity
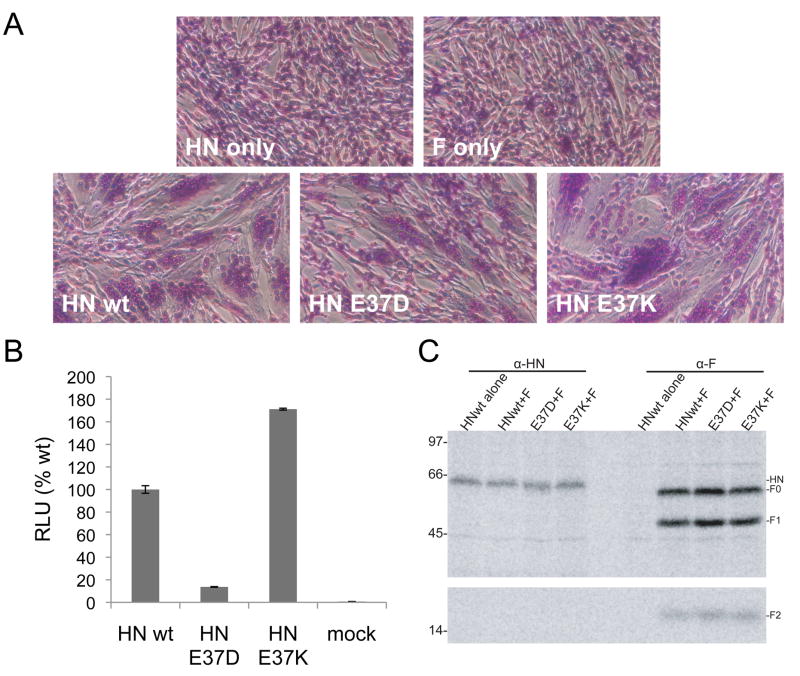
To investigate if E37 mutations impact the ability of HN to promote fusion, BHK-21 cells were transfected with a plasmid expressing PIV-5 F and plasmids expressing wt HN, HN E37D, or HN E37K. Cells co-expressing wt HN and F displayed robust syncytia formation (Fig. 8A). In contrast, cells that co-expressed F and HN E37D displayed syncytia formation, but not as extensively as wt HN and F transfected cells. Cells co-expressing HN E37K and F showed large syncytia formation with extensive cytopathic effects. To quantify these results, the HN E37 mutants were compared to wt HN in a luciferase reporter assay. The results shown in Fig. 8B parallel those obtained in the syncytia assay. Co-expression of F and HN E37D only caused 13.7% percent of wt HN and F fusion. Co-expression of F and HN E37K caused a fusion activity of 171% of wt HN and F fusion. To ensure that F and HN were properly expressed, Vero cells were transfected with PIV-5 F and wt HN and E37 mutants in parallel to the luciferase assay and examined for protein expression by immunoprecipitation. HN E37D and HN E37K were expressed properly in the Vero cells used for luciferase reporter assays (Fig. 8C, left panel). Additionally, wt F was expressed at similar levels when co-transfected with HN constructs in Vero cells (Fig. 8C, right panel). This rules out the notion that the syncytia and luciferase reporter data are a result of differential expression of HN and F in co-transfected cells.
Figure 8.
Fusion promotion activity of HN E37 mutants. (A) Representative micrographs of syncytia formed in BHK-21F cells expressing PIV-5 F and either wt HN or HN E37 mutants at 18 h p.i. (B) Luciferase reporter assay of cell-cell fusion. Vero cells were transfected with an expression plasmid for PIV-5 F and a plasmid expressing luciferase under the control of a T7 promoter together with wt HN or one of the HN E37 mutants. At 18 h p.t., cells were overlaid with the T7 polymerase expressing BSR-T7/5 cells and incubated for an additional 6 h and luciferase activity measured.. The assay was performed in triplicate and the data expressed as a percentage of the fusion promoted in the presence of wt HN. (C) Expression of PIV-5 HN and F in Vero cells. Vero cells were transfected with an expression plasmid for PIV-5 F and a plasmid expressing luciferase under the control of a T7 promoter together with wt HN or one of the HN E37 mutants. At 18 h p.t., cells were radiolabeled for 30 min with [35S]-TranS-label and then incubated for 1.5 h with unlabeled media. Cells were lysed and proteins were immunoprecipitated with either a polyclonal HN antibody or F antibody and separated using SDS-PAGE as described in Materials and Methods.
Mutations at E37 Affect HN Oligomerization
Membrane bound HN and soluble HN consisting of the stalk and head domain is tetrameric. In contrast, the expressed head domain is monomeric (Ng et al., 1989; Russell et al., 1994; Yuan et al., 2005). The HN tetramer consists of a dimer-of-dimers, with each pair of dimers linked covalently by an interchain disulfide bond at residue C111. The X-ray structure of HN shows that the dimer-of-dimers interface is weak (657 Å2), stabilized by non-covalent interactions involving only ten residues. To determine if mutations at E37 perturb the oligomeric organization, metabolically-labeled infected cells were subjected to sucrose gradient density centrifugation in 0.1% Triton X 100. Alternate fractions of the gradient were analyzed by immunoprecipitation and non-reducing SDS-PAGE. HN from wt virus-infected cells sedimented primarily at the dimer-of-dimers position on the gradient (Fig. 9A, left, lanes 17 and 19). A small amount of wt HN was also present in monomer and dimer fractions. In contrast, HN E37D sedimented primarily as a dimer (Fig. 9A, middle, lanes 13 and 15), with a smaller population existing in the dimer-of-dimers fractions (Fig. 9A, middle lanes 17 and 19). HN E37K also sedimented as a dimer on the sucrose gradient (Fig. 9A, right, lanes 13 and 15). Thus, these data suggest HN E37D and HN E37K both exist primarily as dimers. To ascertain if mutations at E37 prevent the HN tetramer from being formed or simply destabilize the dimer-of-dimers interface such that the tetrameric form of the protein falls apart during centrifugation or on exposure to detergent, we investigated the oligomerization of HN in different detergents using blue native PAGE of purified virions followed by western blotting. In the presence of digitonin, wt HN, HN E37D, and HN E37K were all present primarily as tetramers (T) and higher order molecular weight oligomers (Fig 9B, left lanes). However, HN E37D and HN E37K were also present as dimers (D) whereas wt HN was tetrameric. These data suggest that both mutant HN proteins are capable of forming tetramers, but these mutant oligomers fall apart more readily upon exposure to detergent. The effect of other detergents was also investigated. Treatment with Triton X-100 changed the oligomeric state of all three proteins tested, with a majority of the proteins present in either the dimeric or tetrameric form (Fig. 9B, right lanes). However, there were subtle differences exhibited among the proteins. Higher molecular weight oligomers of HN E37D appeared to be resistant to detergent, whereas those present in wt HN and HN E37K were not. Additionally, there appeared to be some other species of HN present for HN E37D and HN E37K, indicating that the mutant monomers may mediate unique protein-protein contacts as compared to wt HN. Thus, mutations present at E37 perturb the structure of HN.
Figure 9.
Oligomerization of PIV-5 HN. (A) Sucrose density ultracentrifugation of PIV-5 HN from infected cells. PIV-5-infected CV-1 cells at 18 h p.i. were labeled for 30 min with [35S]-TranS-label and then incubated for 1.5 h in unlabeled medium. The cells were lysed in Triton X-100 and subjected to ultracentrifugation on a 7.5–22.5% wt/vol sucrose gradient for 19 h at 37,000 rpm. Fractions were collected from the top of the gradient. Alternating fractions were immunoprecipitated for HN using a polyclonal HN antibody and analyzed by SDS-PAGE. Wt HN- left panel, HN E37D- middle panel, HN E37K- right panel. (B) Blue native PAGE (BN-PAGE) of purified PIV-5 virions. Purified PIV-5 virions solubilized in either digitonin or Triton X-100 were fractionated by BN-PAGE on a 4–13% gel using crosslinked glutamate dehydrogenase as a size marker. Separated proteins were transferred to a polyvinylidene difluoride membrane in the presence of SDS and probed with a polyclonal antibody specific to HN followed by the addition of a Cy5-conjugated goat anti-rabbit secondary antibody. Fluorescence was detected on a Fuji FLA-5100 imager. D = dimer; T = tetramer.
Analysis of the stalk: the HN tetramerization domain
It has been reported previously that the HN stalk (residues 56-115) is a tetramer in solution (Yuan et al., 2008). Based on the results of the oligomerization of the full-length wt HN protein and the E37 mutants, we were interested to determine if the E37 mutants impacted the oligomerization and secondary structure of the HN stalk. We expressed HN stalk residues 34 through 115 containing an N-terminal 6X His-tag in bacteria and purified the stalk as described in Materials and Methods. The E37K stalk construct was very poorly expressed and is not considered further. Initially, the N-terminal His-tag was removed from each of the stalk constructs using enterokinase. However, this made the stalk peptides more susceptible to proteolytic degradation even after gel filtration. It was found that the protein yield and stability was increased greatly by omitting the enterokinase cleavage step. Fig. 10A shows purified wt and E37D stalk constructs. Circular dichroism spectroscopy was used to investigate the secondary structure of the HN stalk peptides. The spectra were analyzed using the computer program CDSSTR and it was determined that the resulting structural data were of good quality (NRMSD<0.1). Wt stalk and E37D stalk displayed similar CD spectra that were typical of α-helical profiles (Fig. 10B). Analysis using the CDSSTR program revealed that both wt stalk and HN E37D stalk had similar secondary structure contents (α-helical contents of 0.83 and 0.84, respectively).
Figure 10.
Expression and secondary structure of HN stalk constructs. (A) SDS-PAGE analysis of HN stalk constructs (residues 34-115) for HN wt and HN E37D. Samples were either not reduced (left lanes) or reduced (right lanes). (B) Representative CD spectra of HN wt and HN E37D stalk. (C) Differential scanning fluorimetry of HN wt and E37D stalk. HN stalk was mixed with SYPRO Orange and subjected to a temperature gradient of 25–90 °C. Fluorescence was measure using a Bio-Rad iCycler iQ Real-Time PCR Detection System. Shown is the negative first derivative of the Tm curve.
To determine the stability of each of the HN stalk constructs, differential scanning fluorimetry was used to monitor the thermal unfolding of each of the polypeptides. The increase in fluorescence of SYPRO Orange, a dye with affinity for hydrophobic parts of a protein, was obtained as the temperature of the stalk peptides was raised from 25 to 95 °C. The melting point was obtained by determining the minimum of the negative first derivative of the raw fluorescence data from the melting curve. The melting points of wt and E37D stalk were found to be 62.5 °C and 61.7 °C, respectively (Fig. 10C). Thus, both wt and E37D stalk constructs have similar thermal stabilities.
Discussion
It has been demonstrated previously that the HN protein of PIV-5 is rapidly internalized from the surface of cells through clathrin-mediated uptake and enters the endocytic pathway, where it is rapidly degraded (Leser et al., 1996; Ng et al., 1989). The cytoplasmic tail of PIV-5 HN lacks a known canonical signal for internalization and deletion of the cytoplasmic tail does not affect endocytosis. Studies have shown that the signal for HN internalization resides in the first seven amino acids of the presumptive ectodomain of the protein; a glutamic acid (E37) located at the transmembrane domain/ectodomain boundary was shown to be critical for internalization (Leser et al., 1999). Mutation of E37 to aspartic acid (E37D) led to an internalization phenotype that was intermediate when compared with wt HN, both in the rate and extent of uptake from the cell surface, whereas mutation of E37 to other residues (e.g. E37K) resulted in a loss of the internalization phenotype.
Our previous studies have used HN expression from viral vectors (SV40) or plasmid cDNA. To elucidate the role of E37 of PIV-5 HN in the context of a native viral life cycle, recombinant PIV-5 viruses that contained either the mutation HN E37D or HN E37K were rescued using a reverse genetics system (He et al., 1997; Waning et al., 2002). A detailed study of properties of these viruses was performed.
In comparison to wt HN that is internalized and degraded, HN expressed by the recombinant viruses HN E37D and HN E37K were stable in pulse-chase experiments and greater amounts of HN E37D and HN E37K were expressed at the plasma membrane than wt HN. However, the single-step growth curves of the HN E37D and HN E37K viruses were very similar to that of wt PIV-5 with the exception of the 24 h time point when PIV-5 showed about 1 log greater infectivity. Unlike a PIV-5 mutant that is greatly debilitated in growth, such as HN Δ2-13 that contains a deletion of the HN cytoplasmic tail (Schmitt et al., 1999) and forms small fuzzy plaques, the PIV-5 HN E37D and HN E37K viruses yielded wt plaque morphology and size. Fitness assays showed that PIV-5 HN E37D and HN E37K were biologically less fit in tissue culture than wt virus, although the loss of fitness of these viruses was only subtle in comparison to a greatly debilitated virus, HN Δ2-13. There is not a useful animal model to study the pathogenicity of PIV-5 and thus it is unknown whether PIV-5 HN E37D and HN E37K would exhibit a much greater loss of fitness in an animal model.
Investigation of biochemical properties of PIV-5 HN E37D and HN E37K showed different phenotypes of the two HN mutants. Analysis of the purified virion polypeptide composition showed that PIV-5 HN E37D incorporated fewer HN tetramers into virions than wt virions (Table 1) but concomitantly incorporated an increased amount of F trimers. PIV-5 HN E37K had slightly less HN tetramers in virions than wt virus and had slightly more F trimers. Thus, for HN E37D despite high levels of HN expression at the cell surface, less HN becomes assembled into budded virions. Neuraminidase activity, based on the number of HN tetramers per virion showed that PIV-5 E37D exhibited a 2-fold higher level of activity than wt virus whereas PIV-5 E37K had a similar level of neuraminidase activity to wt virus. Receptor binding (hemadsorption) activity for the HN E37D and HN E37K viruses was equivalent but given the difference in HN tetramers on the surface of the cell, the assay may be affected by steric hindrance of the bulky red blood cells used to measure hemadsorption. Fusion activity of PIV-5 HN E37D was impaired in a reporter-based fusion assay, whereas that of HN E37 K had increased over wt HN. However, there is more HN E37K expressed at steady-state on the surface of cells than wt HN due to its lack of internalization. It has recently been demonstrated that increased amounts of HN protein present in concert with the F protein increases the amount of cell to cell fusion observed, thus it is likely that the increase in fusion observed with HN E37K is due to this fact (Connolly et al., 2009).
The effect of the mutations on the biochemical properties of HN E37D and HN E37K do not lead to a simple explanation. However, it seems possible that the mutations which are located at the presumed junction of the stalk and the transmembrane (TM) domain affect the stability of the HN tetramer. Consistent with this is the observation that HN E37D and HN E37K migrate as tetramers and some dimers on blue native gel electrophoresis using digitonin as detergent, but when Triton X-100 was used as detergent HN E37D and HN E37K showed more dimer formation. Similarly in an analysis of the sedimentation pattern of wt HN, HN E37D and HN E37K on sucrose gradients containing 0.1% Triton X-100 as detergent, whereas wt HN sedimented as a tetramer (dimer-of-dimers), the two mutant HN molecules sedimented predominantly as a dimer. Taken together these data suggest that the HN E37D and HN E37K mutations have an effect on the stability of the tetramer, which is observed on detergent solubilization of HN. The effect on stability of the HN tetramer may not be identical for HN E37D and HN E37K as it was observed that there are differences in the amounts of dimer and tetramer between mutants (Fig. 9A & B). These subtle differences could lead to the differences in the biochemical properties of the mutants.
It has been found previously that mutations in the HN TM domain affect the stability of the HN tetramer. Substitution of serine at residue 31 with residues having bulky side chains led to HN sedimenting as a dimer (Parks and Pohlmann, 1995). Further mutation of HN tyrosine residue 36 also resulted in an HN protein that sedimented as a mixture of dimers and tetramers (Parks and Pohlmann, 1995). Thus, mutations in the HN TM domain and the TM domain – ectodomain boundary affect the stability of the tetramer.
It has been found that the HN stalk domain (residues 56-115) forms a tetramer and is predominantly α-helical (Yuan et al., 2008). Analysis of an HN stalk containing the E37D mutation did not show a difference in α-helicity or thermal stability. However, if the stalk is a dimer of dimer of helices that do not form a four-helix bundle, then a decrease in stability of the oligomer may not be detected in the absence of the ectodomain and TM domain.
The mechanism by which the HN is internalized via clathrin-coated pits is not known. We speculate that the HN stalk is recognized by a cellular protein that contains an internalization signal and acts as an adaptor for internalization. In this scenario we propose that the HN E37 mutations affect the recognition of the adaptor protein hence preventing internalization.
Materials and Methods
Cells, virus, and plasmids
CV-1, MDBK, and Vero cells were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS). BHK-21 cells were maintained in DMEM supplemented with 10% FBS and 10% tryptose phosphate buffer. HeLa-CD4-LTR-βgal cells were grown in DMEM containing 10% FBS, 200 μg/ml G418, 100 μg/ml hygromycin B, and 20 mM HEPES (pH 7.4). BSR T7/5 cells were maintained in DMEM containing 10% FBS, with the addition of 500 μg/ml G418 every third passage. PIV-5 (isolate W3A) was grown in MDBK cells. Recombinant PIV-5 viruses were obtained using a reverse genetics system described previously (He et al., 1997; Waning et al., 2002). Viral titers were determined by plaque assay on BHK-21 cells. Expression plasmids pCAGGS-HN and pCAGGS-F have been described previously (Paterson et al., 2000). pCAGGS-HN mutants were constructed by four-primer PCR followed by cloning into pCAGGS. The nucleotide sequence of HN in all plasmids and recovered viruses was verified using an Applied Biosystems 3100-Avant automated DNA sequencer (Life Technologies Corp, Carlsbad, CA). pT7-luciferase was obtained from Promega Corp (Madison, WI).
Expression of HN and F proteins
CV-1 cells were infected with PIV-5 in DMEM containing 1% bovine serum albumin (BSA) at a multiplicity of infection (MOI) of 3 plaque forming units (PFU) for one h in 3.5 cm or 6 cm dishes at 37 °C. Following incubation, the infection medium was replaced with DMEM containing 2% FBS and returned to 37 °C for an additional 18 h. For transfections, pCAGGS-HN, pCAGGS-F, and pT7-luciferase were expressed in BHK-21F or Vero cells seeded in 3.5 cm dishes using the Lipofectamine PLUS system (Invitrogen, Carlsbad, CA) in OptiMEM. Vero cells were incubated for four h, after which the media was replaced with DMEM containing 2% FBS and incubated for an additional 18 h.
Immunoprecipitation and SDS-PAGE
Infected or transfected cells were starved in cysteine and methionine deficient DMEM for one h, followed by labeling with 50 μCi [35S]-TranS-label in the same medium for 30 min. Cells were then incubated (chased) for the indicated amounts of time in complete DMEM supplemented with 2 mM methionine and cysteine. Cells were lysed in cold radioimmunoprecipitation assay (RIPA) buffer (Paterson and Lamb, 1993) containing protease inhibitors, 50 mM iodoacetamide, and 2 mM PMSF and clarified in a Beckman TLA100 at 55,000 rpm for 10 min at 4 °C. Clarified lysates were incubated with the appropriate antibody overnight at 4 °C, followed by the addition of protein A sepharose beads for 30 min at 4 °C. Antigen-antibody complexes were washed three times with RIPA containing 0.3 M NaCl, two times with RIPA containing 0.15 M NaCl, and once with 50 mM Tris-HCl pH 7.4, 0.25 mM EDTA, and 0.15 M NaCl. Proteins were boiled in protein lysis buffer containing 2.5% dithiothreitol and separated on 15% acrylamide gels. Detection and quantification of radioactivity was performed using a Fuji FLA-5100 with Multi Gauge v3.0 software (Fuji Medical Systems, Stamford, CT).
Fluorescence Microscopy
CV-1 cells seeded on coverslips in 3.5 cm dishes were infected with PIV-5 at an MOI of 3 PFU/cell. At 18 h p.i., coverslips were fixed in PBS containing 1% formaldehyde for 10 min at room temperature. Fixed coverslips were then moved to 4 °C and incubated with HN-1b antibody (1:100 dilution) (Ng et al., 1989; Randall et al., 1987) or mAb F1a (1:100 dilution) in PBS containing 1% BSA. Cells were washed extensively with PBS to remove unbound antibody and incubated with Alexa Fluor 488-conjugated goat anti-mouse antibody (Invitrogen, Carlsbad, CA). To detect the internalization of PIV-5 HN, some coverslips were treated with 100 μg/ml cycloheximide for 4 h at 37 °C in DMEM containing 2% FBS. Following cycloheximide treatment, cells were fixed and treated as described above. Coverslips were mounted on glass slides using Prolong Gold (Invitrogen) and viewed using an Axiovert 200M microscope with Apotome (Zeiss, Thornwood, NY). Images were collected using an AxioCam digital camera and Zeiss Axiovision software.
Flow Cytometry
Monolayers of CV-1 cells were infected with PIV-5 at a MOI of 3 pfu/cell in triplicate in 6 well plates and 18 h p.i. were fixed in PBS containing 1% formaldehyde. Plates were then moved to 4 °C and incubated with either a polyclonal HN antibody, mAb HN-1b, or mAb F1a, as indicated, in PBS containing 1% BSA for one h. Monolayers were washed extensively with PBS to remove unbound antibody and then incubated with a fluoroscein-isothiocyanate-conjugated secondary antibody. Cells were washed again with PBS and resuspended in PBS containing 0.5% formaldehyde. The fluorescence intensity of 10,000 cells was determined using a FACSCalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ) Cells that required cycloheximide treatment were prepared as described above, followed by fixation.
Growth Curves and Plaque Assays
MBDK cells were infected with PIV-5 as described above at an MOI of 0.01 pfu/cell in 6 cm dishes. Medium was removed and assayed for viral titer at varying time points. For plaque assays BHK-21F cells were infected with dilutions of PIV-5 in DMEM containing 1% BSA for 1 h at 37 °C. Following infection, monolayers were overlaid with medium containing DMEM, 2% FBS, and 1% low-melting temperature agarose. Upon solidification, dishes were returned to 37 °C and incubated for 96 h, at which time the agarose was removed and the cells were fixed. Plaques were visualized following a 10 min Giemsa stain treatment.
Viral Fitness Competition Assay
The relative fitness of mutant viruses compared to wt PIV-5 was determined using a virus competition assay. MDBK cells were infected at a mutant to wt ratio of 20:1 (MOI of 0.02 to 0.001 PFU/cell, respectively) as described above. At 72 h p.i., the cell medium was collected, clarified by low speed centrifugation to remove cell debris, and used to infect a fresh population of MDBK cells at an MOI of 0.01 PFU/cell. This was repeated for a total of four cycles. To determine the abundance of mutant virus compared to wt, viral RNA was isolated from the cell supernatant using the QIAamp Viral RNA Mini Kit (Qiagen, Valencia, CA). Viral RNA was reverse transcribed and amplified using SuperScript III One–Step PCR System (Invitrogen), with the primer 1 (5′-CCAATTGTAGAGGTAGAGCGAC-3′) and primer 2 (5′-GAAAGCTACAAGGAGAGCAGGAAAC-3′). Resulting amplicons were sequenced using primer 1 using an Applied Biosystems 3100-Avant automated DNA sequencer.
To determine the ratio of mutant to wt RNA in a sample by comparing chromatogram peak heights, a sequencing standard curve was developed. For each mutant, this was accomplished by mixing mutant and wt plasmids together in the following ratios: 20:1, 10:1, 5:1, 2:1, 1:1, 1:2, 1:5, and 1:10. These plasmid mixtures were sequenced using primer 1 and the heights of the chromatogram peaks at the mutation site were measured. The ratio of the measured peak heights for the sequenced mutant versus wt plasmids was plotted against the original ratio of the plasmid mixture. A best-fit line was obtained to describe the resulting curve. The equation of this line was used to determine the final mutant to wt ratio in the viral competition assay.
The alternative fitness assay was approached using a variation of the standardization method described above. MDBK cells were infected as described with a mutant to wt ratio of 100:1. At 5, 6, 7, and 8 days p.i., medium (140 μl) was removed from the infected dishes and subjected to vRNA purification, RT-PCR, and sequencing as described above. To ensure that there were no sequence reversions back to wt, MDBK cells were also infected with wt PIV-5, PIV-5 HN E37D, or PIV-5 HN E37K alone and monitored as described above.
PIV-5 Virion Purification and Labeling
MDBK cells in 10 cm dishes were infected at an MOI of 0.01 PFU/cell with PIV-5. 6 days p.i., the cell supernatant was cleared of cellular debris by low speed centrifugation. The clarified supernatant was centrifuged in a Ti45 rotor at 38,500 rpm for 45 min at 4 °C in an Optima L-80 XP Ultracentrifuge (Beckman Coulter, Brea, CA). The pelleted virus was resuspended in 1 ml NTE buffer (10 mM Tris-HCl, pH 8.0, 0.5 M NaCl, 1 mM EDTA) and overlaid on a 15–60% sucrose gradient made with NTE. The final volume of the gradient was 36 ml. The gradients were centrifuged in a SW32 rotor at 24,000 rpm for 1 h at 4 °C. The bands that contained purified virus were collected with a syringe, diluted in NTE buffer, and pelleted in a Ti70 rotor at 40,000 rpm for 45 min at 4 °C. The pelleted, purified virus was resuspended in 500 μl of NTE, flash frozen, and stored at −80 °C. The concentration of the purified virus was determined by BCA assay (Pierce, Rockford, IL) using BSA as a standard. The protein composition of the purified virus was verified by SDS-PAGE followed by Coomassie blue R250 staining and silver staining.
For [35S]-labeled purified virus, MDBK cells were infected as above. 48 h p.i., the medium was removed and replaced with a medium that contained 20% of the normal amount of methionine found in DMEM together with [35S]-methionine (50 μCi/ml). Following 2 days of incubation in the [35S]-labeled medium, the plates were supplemented with methionine in order to bring the final concentration of methionine in the medium to the level in complete DMEM. After an additional 48 h of incubation at 37 °C, the supernatant was harvested and virus was purified as described above.
Neuraminidase Activity
Determination of neuraminidase activity of purified virions was performed essentially as described by Warren (Warren, 1959). 20 μg of purified virus was incubated in glass tubes in buffer containing 80 mM sodium acetate, pH 4.5 and 80 μg of N-acetylneuraminyl lactose (Sigma, St. Louis, MO) in a final volume of 500 μl for 1.5 h at 37 °C. Periodate was then added to a final concentration of 4 mM and incubated for 30 min at 37 °C. Following incubation, 80 μl of a 2% (w/v) sodium arsenite solution was added and the tubes rocked gently until the yellow color disappeared. 800 μl of 0.1 M thiobarbituric acid was added to the sample and boiled for 10 min in a water bath. The reactions were transferred to ice immediately, and 2 ml of acidified butanol was added to the sample. Tubes were centrifuged at 1,500 rpm for 10 min and the butanol phase was extracted and absorbance read at 549 nm.
Hemadsorption
Monolayers of CV-1 cells in 24 well plates were infected in sextuplicate with PIV-5 as described above at an MOI of 10 PFU/cell. At 18 h p.i., cells were washed gently three times with ice cold PBS containing calcium and magnesium (PBS+). Infected cells were then incubated at 4 °C for 30 min with a solution of 1% chicken red blood cells (RBCs) suspended in PBS+. Following receptor binding, cells were washed gently five times with ice cold PBS+ to remove unbound RBCs. Bound RBCs were lysed in 200 μl of lysis buffer (17 mM Tris-HCl, 0.145 M NH4Cl). The absorbance of the lysed RBCs was read on a Spectramax M5 (Molecular Devices, Sunnyvale, CA) at an absorbance of 410 nm. The pH of the PBS+ was adjusted to either pH 5.0 or pH 7.0 depending on the experiment.
Syncytia Formation
BHK-21F cells were transfected as described above using 1 μg each of pCAGGS-HN and pCAGGS-F plasmid. At 18 h post-transfection (p.t.), cells were fixed and syncytia visualized using a Hema3 stain (Fisher Scientific, Pittsburgh, PA) according to the manufacturer’s instructions. Micrographs were taken using an inverted phase-contrast microscope (Diaphot, Nikon, Melville, NY) connected to a digital camera (DCS 760, Kodak, Rochester, NY)
Luciferase Reporter Assay
Vero cell monolayers in 6 well plates were transfected in triplicate with 1 μg each of pCAGGS-HN, pCAGGS-F, and pT7-luciferase, a plasmid that expresses firefly luciferase under the control of the T7 promoter. At 18 h p.t., BSR-T7/5 cells that express T7 polymerase were overlaid onto the Vero cells and incubated for 6 h at 37 °C. Cells were then lysed in 2X Reporter Lysis Buffer (Promega) and clarified by centrifugation. The luciferase activity of each sample was determined by adding 150 μl of lysate to 150 μl of luciferase assay substrate in an Lmax luminescence microplate reader (Molecular Devices, Sunnyvale, CA).
Sucrose Density Gradient Centrifugation
PIV-5-infected CV-1 cells in 6 cm dishes were labeled as described above and then incubated for 1.5 h. Cells were lysed in 750 μl of MNT buffer (20 mM morpholino-ethanesulfonic acid, 30 mM Tris, 100 mM NaCl; pH 5.0) containing 1% Triton X-100 and 1% aprotinin. Lysates were clarified as described above and the supernatant layered over a continuous 7.5–22.5% wt/vol sucrose gradient made in MNT buffer containing 0.1% Triton X-100 that overlaid a 0.75 ml 60% sucrose cushion. The final volume of the gradient was 12 ml. The gradients were centrifuged at 37,000 rpm for 19 h at 20 °C. Fractions (0.5 ml) were collected using an Auto Densi-Flow fraction collector (LabConco, Kansas City, MO). Alternate fractions, beginning with the top fraction, were prepared for immunoprecipitation by the addition of 2X RIPA buffer containing protease inhibitors, iodoacetamide, and PMSF. Proteins were immunoprecipitated as described above using a polyclonal HN antibody, followed by protein A sepharose treatment and RIPA washes. Proteins were separated using an 8% acrylamide gel.
Blue Native PAGE
Blue native PAGE was performed essentially as described by Wittig et al. (Wittig et al., 2006). Briefly, 20 μg of purified virions were resuspended in a final volume of 33 μl of solubilization buffer A. Detergents were added in final concentrations of 1.2% for digitonin or 0.6% for Triton X-100. Virions were solubilized for 20 min at 37 °C after which glycerol was added at a final concentration of 15% and Coomassie blue G-250 was added at a final concentration of 0.05 %. Solubilized viral proteins were loaded on a 4–13% native acrylamide gel. Separated proteins were transferred to a polyvinylidene fluoride membrane in the presence of 0.01% SDS. Proteins were detected by the addition of a polyclonal HN antibody and a Cy5-conjugated goat anti-rabbit antibody followed by visualization using a Fuji FLA-5100.
Purification of HN Stalk
Plasmids expressing residues 34-115 of the PIV-5 HN protein fused to an N-terminal His-Tag were constructed using the pET30 Ek/LIC Vector Kit (EMD Chemicals, Gibbstown, NJ) according to the manufacturer’s instructions. pCAGGS-HN and its corresponding mutant plasmids were used to generate the insert into the pET30 plasmid. The nucleotide sequence of all plasmids insert segments were verified by DNA sequencing.
Origami 2 cells (EMD Chemicals, Gibbstown, NJ) carrying pET30-HNstalk from an overnight culture were diluted 1:20 and grown in M9 minimal medium supplemented with kanamycin (30 μg/ml) and tetracycline (12.5 μg/ml) until the OD600 was between 0.5–0.6. At that point, IPTG was added at a final concentration of 1 mM and the cells were incubated at 37 °C for an additional 4 h. Cells were pelleted using centrifugation and frozen at −20 °C. Pellets were thawed and resuspended in buffer (10 mM Tris-HCl pH 7.4, 2.5 mM MgCl2, 0.5 mM CaCl2) containing 1% Triton X-100 and protease inhibitors and incubated at 25 °C for 30 min. DNase was added and the lysate was incubated an additional 30 min at 25 °C, followed by sonication on ice for 2.5 min using a probe sonicator. Lysates were clarified using a JA-14 rotor for 30 min at 10,000 rpm at 4 °C. The supernatant was filtered using a 0.45 μm filter and brought to a final concentration of 50 mM Tris-HCl pH 8.0 and 250 mM NaCl. HN stalk was purified from the supernatant by passage over a pre-equilibrated Ni-NTA (Qiagen, Valencia, CA) column using the manufacturer’s instructions. The purified protein was concentrated by centrifugal ultrafiltration (Millipore, Billerica, MA) and further purified by size-exclusion chromatography on a Pharmacia Superdex-200 column (10×300 mm) pre-equilibrated with a buffer of 50 mM Tris-HCl pH 7.4 and 250 mM NaCl. The column was calibrated with a gel filtration standard (Bio-Rad, Hercules, CA). Protein concentration was determined by measuring absorbance at 280 nm using an extinction coefficient of 8480 M−1 cm−1 based on predictions obtained from the ExPASy server (Swiss Institute of Bioinformatics, Lausanne, Switzerland).
Circular Dichroism
Purified protein was diluted in buffer (10 mM sodium phosphate, pH 7.4). The CD spectra was obtained using a Jasco model 715 spectropolarimeter (Easton, MD) using a quartz cuvette with a path length of 2.0 mm. Data were collected every 0.2 nm with a bandwidth of 2.0 nm and a response time of 2 s. The sensitivity was set at 100 mdeg. Baseline scans of buffer were subtracted from the sample spectra. Each spectrum represents the average of five reads. α-helical content was determined using the CDSSTR program (Johnson, 1999; Sreerama and Woody, 2000).
Differential Scanning Fluorimetry
Differential scanning fluorimetry (DSF) was carried out essentially as described by Niesen et al (Niesen et al., 2007). Purified HN stalk at a final concentration of 60 μM was diluted in a buffer containing 50 mM Tris-HCl pH 7.4 and 250 mM NaCl together with SYPRO Orange (Invitrogen, Carlsbad, CA) at a final concentration of 50X. The final volume of the reaction was 50 μl. SYPRO Orange was supplied as a 5,000X solution in 100% (v/v) dimethyl sulfoxide (DMSO) and was diluted 1:100 in buffer before addition to the reaction in order to prevent high concentrations of DMSO from contacting and damaging the protein. The reactions were prepared in triplicate in iQ 96-well PCR plates (Bio-Rad, Hercules, CA). The melting point of the protein was determined using a Bio-Rad iCycler iQ Real-Time PCR Detection System (Bio-Rad, Hercules, CA) using a program of 3 min at 25 °C, followed by a temperature scan from 25 to 95 °C at 1 °C/min. The minimum of the negative first derivative of the fluorescence curve was used to determine the melting point of each protein.
Acknowledgments
We are grateful to David Waning and Anthony Schmitt who originally made the HN E37D and HN E37K constructs, respectively. We are grateful to George Leser for many discussions about HN internalization experiments. The Northwestern University Keck Biophysics Facility was used for the circular dichroism measurements and the facility is supported by a Cancer Center Support Grant (NCI CA060553). This research was supported in part by a Public Health Services grant AI-23173 from the National Institute of Allergy and Infectious Diseases. J.G.R. was supported by predoctoral training grant T32 AI060523 from the National Institute of Allergy and Infectious Diseases. R.A.L. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andrejeva J, Childs KS, Young DF, Carlos TS, Stock N, Goodbourn S, Randall RE. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc Natl Acad Sci USA. 2004;101:17264–17269. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Weissman AM. Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu Rev Cell Dev Biol. 1998;14:19–57. doi: 10.1146/annurev.cellbio.14.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WJ, Goldstein JL, Brown MS. NPXY, a sequence often found in cytoplasmic tails, is required for coated pit-mediated internalization of the low density lipoprotein receptor. J Biol Chem. 1990;265:3116–3123. [PubMed] [Google Scholar]
- Choppin PW. Multiplication of a myxovirus (SV5) with minimal cytopathic effects and without interference. Virology. 1964;23:224–233. doi: 10.1016/0042-6822(64)90286-7. [DOI] [PubMed] [Google Scholar]
- Collawn JF, Stangel M, Kuhn LA, Esekogwu V, Jing SQ, Trowbridge IS, Tainer JA. Transferrin receptor internalization sequence YXRF implicates a tight turn as the structural recognition motif for endocytosis. Cell. 1990;63(5):1061–1072. doi: 10.1016/0092-8674(90)90509-d. [DOI] [PubMed] [Google Scholar]
- Connolly SA, Leser GP, Jardetzky TS, Lamb RA. Bimolecular complementation of paramyxovirus fusion and hemagglutinin-neuraminidase proteins enhances fusion: implications for the mechanism of fusion triggering. J Virol. 2009;83:10857–10868. doi: 10.1128/JVI.01191-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didcock L, Young DF, Goodbourn S, Randall RE. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J Virol. 1999;73:9928–9933. doi: 10.1128/jvi.73.12.9928-9933.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer I, Haft CR, Paccaud JP, Maeder C, Taylor S, Carpentier JL. Dual role of a dileucine motif in insulin receptor endocytosis. J Biol Chem. 1997;272:21685–21691. doi: 10.1074/jbc.272.35.21685. [DOI] [PubMed] [Google Scholar]
- He B, Lin GY, Durbin JE, Durbin RK, Lamb RA. The SH integral membrane protein of the paramyxovirus simian virus 5 is required to block apoptosis in MDBK cells. J Virol. 2001;75:4068–4079. doi: 10.1128/JVI.75.9.4068-4079.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Paterson RG, Ward CD, Lamb RA. Recovery of infectious SV5 from cloned DNA and expression of a foreign gene. Virology. 1997;237:249–260. doi: 10.1006/viro.1997.8801. [DOI] [PubMed] [Google Scholar]
- Hiebert SW, Paterson RG, Lamb RA. Hemagglutinin-neuraminidase protein of the paramyxovirus simian virus 5: Nucleotide sequence of the mRNA predicts an N-terminal membrane anchor. J Virol. 1985;54:1–6. doi: 10.1128/jvi.54.1.1-6.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itin C, Kappeler F, Linstedt AD, Hauri HP. A novel endocytosis signal related to the KKXX ER-retrieval signal. EMBO J. 1995;14:2250–2256. doi: 10.1002/j.1460-2075.1995.tb07219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WC. Analyzing protein circular dichroism spectra for accurate secondary structures. Proteins. 1999;35(3):307–12. [PubMed] [Google Scholar]
- Lamb RA, Parks GD. Paramyxoviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5. Wolters Kluwer/Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 1449–1496. [Google Scholar]
- Leser GP, Ector KJ, Lamb RA. The paramyxovirus simian virus 5 hemagglutinin-neuraminidase glycoprotein, but not the fusion glycoprotein, is internalized via coated pits and enters the endocytic pathway. Mol Biol Cell. 1996;7:155–172. doi: 10.1091/mbc.7.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leser GP, Ector KJ, Ng DT, Shaughnessy MA, Lamb RA. The signal for clathrin-mediated endocytosis of the paramyxovirus SV5 HN protein resides at the transmembrane domain-ectodomain boundary region. Virology. 1999;262:79–92. doi: 10.1006/viro.1999.9890. [DOI] [PubMed] [Google Scholar]
- Letourneur F, Klausner RD. A novel di-leucine motif and a tyrosine-based motif independently mediate lysosomal targeting and endocytosis of CD3 chains. Cell. 1992;69:1143–1157. doi: 10.1016/0092-8674(92)90636-q. [DOI] [PubMed] [Google Scholar]
- Lin Y, Bright AC, Rothermel TA, He B. Induction of apoptosis by paramyxovirus simian virus 5 lacking a small hydrophobic gene. J Virol. 2003;77:3371–3383. doi: 10.1128/JVI.77.6.3371-3383.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz DC, Prehm P, Scheid A, Choppin PW. Inhibition of the neuraminidase of paramyxoviruses by halide ions: a possible means of modulating the two activities of the HN protein. Virology. 1981;112:296–305. doi: 10.1016/0042-6822(81)90635-8. [DOI] [PubMed] [Google Scholar]
- Ng DT, Randall RE, Lamb RA. Intracellular maturation and transport of the SV5 type II glycoprotein hemagglutinin-neuraminidase: Specific and transient association with GRP78-BiP in the endoplasmic reticulum and extensive internalization from the cell surface. J Cell Biol. 1989;109:3273–3289. doi: 10.1083/jcb.109.6.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;2(9):2212–21. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- Novella IS, Ball LA, Wertz GW. Fitness analyses of vesicular stomatitis strains with rearranged genomes reveal replicative disadvantages. J Virol. 2004;78:9837–9841. doi: 10.1128/JVI.78.18.9837-9841.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks GD, Lamb RA. Folding and oligomerization properties of a soluble and secreted form of the paramyxovirus hemagglutinin-neuraminidase glycoprotein. Virology. 1990;178:498–508. doi: 10.1016/0042-6822(90)90347-t. [DOI] [PubMed] [Google Scholar]
- Parks GD, Pohlmann S. Structural requirements in the membrane-spanning domain of the paramyxovirus HN protein for the formation of a stable tetramer. Virology. 1995;213:263–270. doi: 10.1006/viro.1995.1569. [DOI] [PubMed] [Google Scholar]
- Paterson RG, Lamb RA. The molecular biology of influenza viruses and paramyxoviruses. In: Davidson A, Elliott RM, editors. Molecular Virology: A Practical Approach. IRL Oxford University Press; Oxford: 1993. pp. 35–73. [Google Scholar]
- Paterson RG, Russell CJ, Lamb RA. Fusion protein of the paramyxovirus SV5: destabilizing and stabilizing mutants of fusion activation. Virology. 2000;270:17–30. doi: 10.1006/viro.2000.0267. [DOI] [PubMed] [Google Scholar]
- Randall RE, Young DF, Goswami KKA, Russell WC. Isolation and characterization of monoclonal antibodies to simian virus 5 and their use in revealing antigenic differences between human, canine and simian isolates. J Gen Virol. 1987;68:2769–2780. doi: 10.1099/0022-1317-68-11-2769. [DOI] [PubMed] [Google Scholar]
- Russell CJ, Jardetzky TS, Lamb RA. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 2001;20:4024–4034. doi: 10.1093/emboj/20.15.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AP, He B, Lamb RA. Involvement of the cytoplasmic domain of the hemagglutinin-neuraminidase protein in assembly of the paramyxovirus simian virus 5. J Virol. 1999;73:8703–8712. doi: 10.1128/jvi.73.10.8703-8712.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreerama N, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal Biochem. 2000;287(2):252–60. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- Trowbridge IS, Collawn JF, Hopkins CR. Signal-dependent membrane protein trafficking in the endocytic pathway. Annu Rev Cell Biol. 1993;9:129–161. doi: 10.1146/annurev.cb.09.110193.001021. [DOI] [PubMed] [Google Scholar]
- Waning DL, Schmitt AP, Leser GP, Lamb RA. Roles for the cytoplasmic tails of the fusion and hemagglutinin-neuraminidase proteins in budding of the paramyxovirus simian virus 5. J Virol. 2002;76:9284–9297. doi: 10.1128/JVI.76.18.9284-9297.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L. The thiobarbituric acid assay of sialic acids. J Biol Chem. 1959;234(8):1971–5. [PubMed] [Google Scholar]
- Wilson RL, Fuentes SM, Wang P, Taddeo EC, Klatt A, Henderson AJ, He B. Function of small hydrophobic proteins of paramyxovirus. J Virol. 2006;80(4):1700–1709. doi: 10.1128/JVI.80.4.1700-1709.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittig I, Braun HP, Schagger H. Blue native PAGE. Nat Protoc. 2006;1(1):418–28. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- Yuan P, Leser GP, Demeler B, Lamb RA, Jardetzky TS. Domain architecture and oligomerization properties of the paramyxovirus PIV 5 hemagglutinin-neuraminidase (HN) protein. Virology. 2008;378:282–291. doi: 10.1016/j.virol.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P, Thompson T, Wurzburg BA, Paterson RG, Lamb RA, Jardetzky TS. Structural studies of the parainfluenza virius 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure. 2005;13:1–13. doi: 10.1016/j.str.2005.02.019. [DOI] [PubMed] [Google Scholar]