Abstract
The process of budding of many enveloped viruses utilizes the cellular ESCRT (endosomal sorting complex required for transport) machinery, that is normally involved in the formation of luminal vesicles of endosomal multivesiculate bodies (MVB). A late step in the MVB pathway involves the recruitment of VPS4, an AAA+ ATPase, to the ESCRT complexes. Our earlier work had shown that the formation of influenza virus-like particles was not inhibited by dominant negative VPS4A. However, it was not known if there was a role of VPS4 and the ESCRT pathway in influenza virus particle budding and this needed to be investigated. It was found that neither siRNA knockdown of VPS4A and VPS4B expression nor the use of cell lines that inducibly express VPS4A or VPS4B, inhibited influenza virus budding. In contrast, and in keeping with more recent data, vesicular stomatitis virus budding was diminished by VPS4 dysfunction.
Keywords: influenza virus budding, influenza virus assembly
Introduction
The final step of enveloped RNA virus replication is assembly and budding, a process that occurs at a host cellular membrane, often the plasma membrane. Viral proteins and the viral genome have to accumulate at the site of assembly on the membrane. The assembly process proceeds with deformation of the membrane, bud formation and pinching off (scission) from the cellular membrane. Originally this process was thought to occur by “push or pull” forces derived from self-assembly of viral proteins (Garoff et al., 1998; Welsch et al., 2007). However, recent studies have shown that many enveloped viruses depend on host cellular machinery to complete their budding more efficiently. The host machinery is the ESCRT (endosomal sorting complex required for transport) pathway, required for the sorting of ubiquitinated proteins to multivesicular bodies (MVB). The ESCRT pathway consists of four ESCRT complexes (ESCRT-0, I, II, and III) and ubiquitinated protein cargo is processed sequentially by each complex. A late step in the MVB pathway involves the recruitment of VPS4, an AAA+ ATPase, to the ESCRT complexes. This leads to budding into the MVB for subsequent degradation within the lysosome (Raiborg and Stenmark, 2009). Topologically, formation of MVPs is equivalent to that of virus budding and the ESCRT pathway has been shown to be utilized by more than 10 families of enveloped RNA viruses (Chen and Lamb, 2008a).
The involvement of the ESCRT pathway in virus budding was firstly discovered for HIV-1. Mutations in the HIV-1 p6 protein, which is part of the GAG polyprotein, abolished virus release from the virus-infected cells (Gottlinger et al., 1991). The critical amino acid sequence in HIV-1 p6 required for virus release was identified as P(T/S)AP and it was termed as a viral “late domain” (L domain) (Huang et al., 1995). Subsequently, three other L domain sequences were identified in various virus families (YXXL (Puffer et al., 1997), PPXY (Xiang et al., 1996), FPIV (Schmitt et al., 2005); where X is any amino acid). L domains usually consist of 4 amino acids and are thought to bind into late domain pockets in various host ESCRT proteins (Lee et al., 2007; Pornillos et al., 2002) such as Tsg101, AIP/Alix, or Nedd4 (Garrus et al., 2001; Harty et al., 2001; Ikeda et al., 2001; Strack et al., 2003). Viruses harboring L domain mutations cannot be recruited into ESCRT complexes and transferred to the assembly site and thus they fail to complete the budding process.
Experiments designed to investigate the budding requirements of the enveloped RNA virus vesicular stomatitis virus (VSV) have yielded mixed results. VSV was one of the first non-retrovirus examples of a virus containing an L domain motif in its matrix (M) protein (Craven et al., 1999; Harty et al., 1999) and it has been shown that PPPY is required for efficient virus replication (Irie et al., 2004a; Jayakar et al., 2000). However, in experiments involving transfection of dominant negative (DN) VPS4A and VSV infection, budding occurred leading to the conclusion that VSV budding was VPS4A independent (Irie et al., 2004b; Chen et al., 2007). This result was contrary to expectations. However, recently when 293 T cells lines were used that stably express VPS4-DN in response to tetracycline induction (Taylor et al., 2007) VSV growth was reduced approximately 18 fold. Furthermore, mutation of the VSV M protein L domain PPPY caused budding to occur in the beads on a string morphology, typical of L domain mutants (Jayakar et al., 2000).
Influenza A virus is an enveloped, negative stranded, segmented RNA virus that buds from the surface of virus-infected cells (reviewed in Barman et al., 2001; Schmitt and Lamb, 2005). The viral matrix protein (M1) underlies the lipid bilayer and makes contact with the cytoplasmic tails of the three integral membrane proteins, hemagglutinin, neuraminidase and the M2 proton selective ion channel. The M1 protein (and all other influenza virus proteins) lacks a recognizable L domain and in an assay for virus-like particles (VLPs) the M1 protein, unlike HIV-1 gag, does not bud on its own. Analysis of VLP particle formation in the presence of VPS4 DN showed that influenza virus VLP formation was not sensitive to VPS4-DN whereas two known control virus VLPs (HIV-1 and parainfluenza virus 5) were sensitive to VPS4-DN expression (Chen et al., 2008b). As influenza virus budding occurs in the presence of the proteosome inhibitor MG-132, indicating ubiquitination of proteins is not required for budding (Hui and Nayak, 2001) and that influenza virus VLP formation occurs in the presence of VPS4-DN, this has lead to the conclusion that influenza virus budding is independent of the ESCRT pathway (Chen and Lamb, 2008a). However, the requirements of the ESCRT pathway for budding in influenza virus-infected cells had not been reported at the time of beginning the experiments described here. Recently, Bruce and coworkers (2009) using some different and also some overlapping approaches concluded that there was no role for the ESCRT pathway in influenza virus budding. Here we confirm a lack of a role of the ESCRT pathway in influenza virus budding.
Materials and Methods
Cells, viruses, and antibodies
293T (human embryonic kidney) and BHK (baby hamster kidney) cells were maintained in Dulbecco's modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS). MDCK cells stably expressing influenza virus M2 protein (M2CK cells) was maintained in DMEM supplemented with 10% FBS, 200 µg/ml Geneticin (G418; InvivoGen, San Diego, CA), and 2 µM amantadine (Sigma-Aldrich [Sigma], St. Louis, MO). Tetracycline-inducible cell lines expressing wild type (wt) or the ATP hydrolysis-deficient VPS4A E228Q mutant that is dominant negative (VPS4A-DN) or VPS4B E225Q mutant that is dominant negative (VPS4B-DN) (Garrus et al., 2001; von Schwedler et al., 2003), both tagged with GFP, were gifts from Dr. Phyllis Hansen (Washington University School of Medicine, St. Louis, MO) via Dr. Margaret Kielian (Albert Einstein College of Medicine, Bronx, NY).
Influenza virus strains A/Udorn/72 and A/WSN/33 were amplified in the allantoic cavity of embryonated chicken eggs and infectious titers were determined by plaque assay using MDCK cells. Recombinant influenza virus A/Udorn/72 possessing an M2 ion channel protein that lacks 27 C-terminal amino acids (Udorn/M2S71) was described previously (Chen et al., 2008b) and the mutant virus was propagated in M2CK cells. Vesicular stomatitis virus (VSV) encoding green fluorescent protein (GFP) instead of the VSV G protein (VSV/ΔG/GFP) was a gift from Dr. J. Rose (Yale University) and the virus was propagated in BHK-21 cells that transiently expressed the VSV-G protein using the expression vector pCAGGS-VSVG.
Goat serum raised to purified influenza A/Udorn/72 (goat anti-Ud) was used to detect HA, NP, and M1 proteins in immunoblotting and immunoprecipitation. Mouse anti-M2 monoclonal antibody 14C2 (Zebedee and Lamb, 1988) was used in immunoblotting to detect M2 protein. Monoclonal antibodies anti-VSVM (23H12: a gift from Douglas S. Lyles) and anti-VSVG (5D4, Sigma) were used for immunoprecipitation. Rabbit anti-VPS4 (Santa Cruz Biotechnology, Santa Cruz, CA) was used in immuno (western) blotting to test the knockdown of endogenous VPS4A and VPS4B.
siRNA and siRNA transfection
Pre-designed Stealth siRNAs against human VPS4A and VPS4B together with control siRNAs were purchased from Invitrogen (Carlsbad, CA). siRNA was transfected into 293T cells twice in consecutive 24 h intervals. Briefly, cells were split into 6-well plates the day before transfection. 4.5 µl/well of 20 µM siRNA stock was mixed with FuGENEHD transfection reagent (Roche Applied Science, Mannheim, Germany) in Opti-MEM (Gibco) and incubated for 15 min at RT. The mixture was added to the culture medium drop-wise to achieve a 30 nM final concentration. A second transfection was done at 24 h post transfection. Cells were harvested at 48 h after the first transfection and knockdown was confirmed by western blotting using anti-VPS4 antisera.
Trans-complementation assay with replication incompetent viruses
To avoid the virus infection/transfection problem where wt virus replicates in cells that were not transfected with siRNA, we used a strategy in which replication incompetent influenza Udorn/M2S71 and VSV/ΔG/GFP was used in combination with the co-transfection of pCAGGS-M2 and pCAGGS-VSVG, respectively. This approach was established for herpes virus when defective virus replication was limited only in the cells expressing the trans complementing protein of interest by plasmid transfection (Pawliczek and Crump, 2009). In brief, 293T cells were split into gelatin-coated 6-well plates at a density of 0.3×106/well the day before transfection. Cells were transfected two times with siRNA (final concentration, 30 nM) at 24 hours intervals. For the second transfection, expression plasmids for each complementing protein (pCAGGS-M2 or pCAGGS-VSVG) were transfected with the siRNA. Twenty four hours after the second transfection, cells were infected with influenza virus Udorn/M2S71 or VSV/ΔG/GFP at a multiplicity of infection (moi) of 0.33 or 3.3 pfu/cell, respectively. Culture supernatants were harvested at 24 h post-infection (p.i.) (Udorn/M2S71) or 7 h p.i. (VSV/ΔG) and infectious titers were determined as described below.
Virus titration
The infectious titer of wt Udorn and VSV was determined by plaque assay on MDCK and BHK cells, respectively. Titration of influenza Udorn/M2S71 was done by plaque assay using M2CK cells as described previously (Chen et al., 2008b). To determine the infectious unit of VSV/ΔG/GFP, BHK cells were infected with serially diluted virus. Cells were harvested at 24 h p.i. and GFP fluorescence-positive cells were detected by flow cytometry (FACSCalliber, Becton Dickinson, Franklin Lakes, NJ). Infectious units were calculated based on the percentage of GFP positive cells in the population.
Virus budding assay
The budding efficiency of virus particles was determined by a quantitative budding assay as described previously (Chen et al., 2007). A different time schedule was used for VSV and influenza virus due to the different replication rates. In brief, the infected cells were labeled with [35S]-Trans-label at 2 h (VSV) or 6 h (influenza virus) p.i. and then incubated in unlabeled medium (chase) for 4 or 20 h, respectively. The released virions were collected by ultracentrifuge (Beckman, Ti70.1 rotor, 45,000 rpm for 2 h) and lysed in RIPA buffer (1% deoxycholic acid, 1% Triton X-100, 0.1% SDS, 10mM Tris [pH 7.4], and 0.15M NaCl). The cells used in the experiment were also lysed in RIPA buffer. The released virions and cell fractions were subjected to immunoprecipitation. For VSV proteins anti-VSVM (23H8), anti-VSVG (5D4, Sigma) sera were used and immune complexes collected with protein A Sepharose. For influenza virus goat anti-Udorn sera was used and immune complexes collected with protein G Sepharose. Polypeptides were separated by SDS-PAGE on a 15% polyacrylamide gel. Radiolabeled proteins were detected by using Fuji BioImager FLA-5100 and quantified by MultiGauge v3.0 software (Fuji Medical Systems, Stanford, CT)
Results
The establishment of a trans-complementation assay for influenza virus
A possible explanation for the unexpected result that VSV did not utilize the ESCRT pathway, although the M protein contained an L domain, may lie in the method used. The experiment required transfection of VPS4A-DN and VPS4B-DN and 24 h later infection with VSV (Irie et al., 2004b). Whereas all the cells can be infected with VSV it is very much harder to transfect all the cells with DNA. Depending on the transfection efficiency, this experimental system may not be sufficiently sensitive to detect inhibition of the very rapidly growing VSV.
To attempt to circumvent these potential difficulties, we established a trans complementation system for influenza virus by using a mutant virus that lacked residues in the M2 cytoplasmic tail (Ud/M2S71) that could only replicate efficiently in the presence of wt M2 protein that was provided by transient expression (Chen et al., 2008b). For VSV we used a virus recovered from cloned DNA that had the G glycoprotein gene substituted with a gene expressing GFP (VSV/ΔG/GFP). This virus was trans complemented by transfecting a plasmid expressing G protein. This scheme should overcome the differing efficiencies of infection/transfection. A conceptually related scheme was used for analysis of the requirement of herpes virus for a functional ESCRT-III complex (Pawliczek and Crump, 2009)
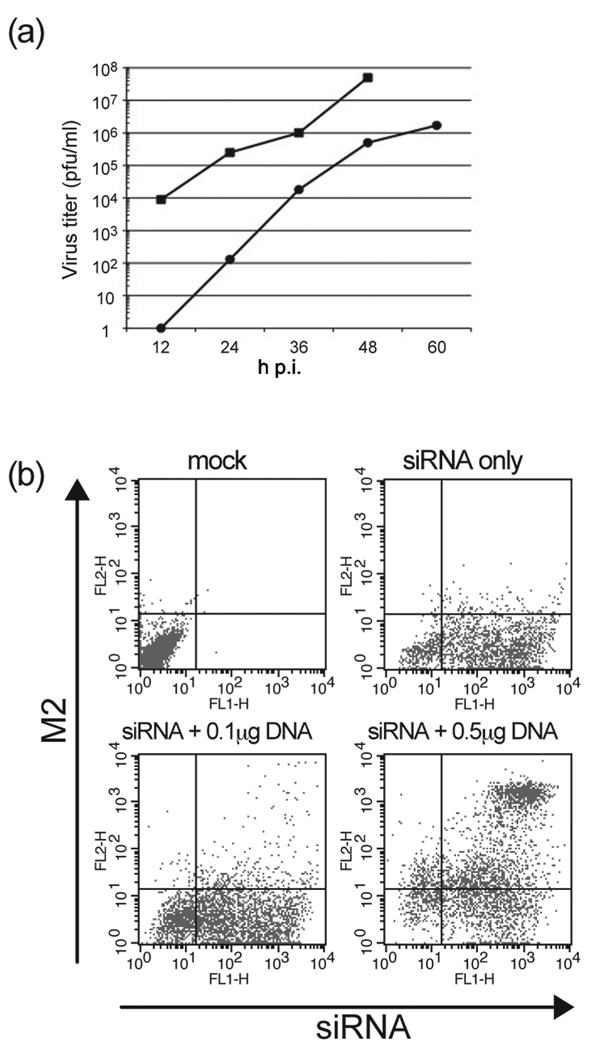
The growth kinetics of Ud/M2S71 was confirmed in the presence of wt M2 protein. M2CK cells were infected with Udorn/M2S71 at a MOI=0.001 pfu/cell and the culture supernatant was harvested at 12 h intervals. As shown in Fig. 1a, Ud/M2S71 showed similar replication kinetics (slope of line) to wt Udorn virus but it grew to a lower titer. As our goal was to observe the effect of knocking down the expression of VPS4 on influenza virus replication, we used HEK 293T cells as these cells can be transfected by both siRNA and an expression plasmid at relatively high efficiency. To determine the efficiency of co-transfection, cells were co-transfected with fluorescent-labeled siRNA and the pCAGGS-M2 expression vector DNA and the cells stained with M2-specific monoclonal antibody 14C2. Fluorescence for siRNA and M2 protein expression were determined by flow cytometry. As shown in Fig 1B, 52.1% of cells in the culture showed positive staining for both siRNA and M2 when we used 0.5 µg plasmid DNA and 30 nM siRNA.
Figure 1. Establishment of an influenza virus trans-complementation assay.
(a) Replication kinetics of influenza A/Udorn/M2S71 mutant virus (●) and wt A/Udorn/72 (■). M2CK cells that constitutively express the M2 protein were infected with Udorn/M2S71 virus or wt Udorn virus at a MOI=0.001 pfu/cell and the supernatant was collected at the indicated time points. Virus titer was determined by plaque assay on M2CK cells. (b) Determination of the efficiency of co-transfection of siRNA and pCAGGS-M2 plasmid. 293T cells were co-transfected with fluorescent-labeled siRNA and pCAGGS-M2. Cells were stained with anti-M2 14C2 and analyzed by flow cytometry to calculate the dual-positive population.
SiRNA against human VPS4A and VPS4B block VSV virus replication but not influenza virus in the trans-complementation assay
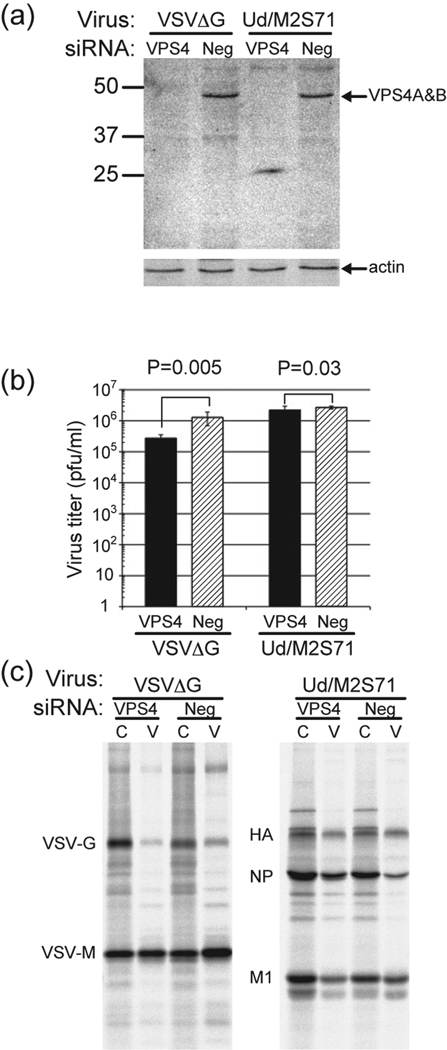
We tested the effect of siRNA to VPS4A and VPS4B on influenza virus and VSV replication using the trans complementation system. The mixture of three different siRNA for each VPS4 isoform could knockdown endogenous VPS4A and VPS4B in 293T cells as examined by western blotting (Fig. 2a). To test the effect of VPS4 knockdown on virus growth plasmid DNA encoding the wt M2 or G protein was co-transfected with the second round of siRNA and 24 h later the cells were infected with VSV/ΔG/GFP or Udorn/M2S71. VPS4 knockdown caused an ~1 log reduction in VSV titer as compared to control siRNA and it was significant (p=0.005), whereas VPS4 knockdown had a small effect on influenza virus replication that was not considered significant (p=0.03) (Fig. 2b). The effect of VPS4 knockdown on virus budding was also assayed by quantifying the amount of virus protein released from cells as compared to the amount of protein remaining in the cells. It was observed for VSV that the release of VSV/ΔG/GFP was a suppressed (Fig. 2c, left panel: 5-fold and 1.5-fold reduction for VSV-G and VSV-M, respectively). On the other hand, there was little effect on Udorn/M2S71 budding (Fig. 2b, right panel): it was noted that the release of HA and M1 was slightly impaired when VPS4 was depleted, whereas NP release was enhanced. A confounding factor in the experiment is the fact that HEK 292 T cells do not support high yields of wt influenza virus budding.
Figure 2. The effect of siRNA against VPS4A and VPS4B on virus replication in the trans-complementation assay.
(a) Knockdown of endogenous VPS4A and VPS4B proteins in 293T cells after siRNA transfection. 293 T cells were transfected twice at 24 h intervals with siRNA specfic for VPS4A and VPS4B. The cells were then infected with VSV/ΔG virus or influenza Udorn/M2S71 at a MOI of 0.33 for influenza virus and 3.3 for VSV. At 7 h p.i. for VSV and 24 h p.i. for influenza virus cells were lysed and the amount of VPS4A and VPS4B analyzed by western blotting. (b) The effect of VPS4 knockdown on virus titer. Influenza virus was harvested at 24 h p.i. and VSV at 7 h p.i. and their titers determined as described in Materials and Methods. IU/ml = infectious units as VSV titer was determined by GFP expression level. (c) The effect of VPS4 knockdown on virus budding in the trans-complementation assay. Infected cells were labeled with [35S] Trans-label and virus and cells harvested as described in Materials and Methods and polypeptides immunoprecipitated. C = cells and V = virions. Budding efficiency (%) was calculated as amount in virus fraction/(amount in virus fraction + amount in cell lysate). Radioactivity was quantified using a Fuji BioImager.
Inducible cell lines expressing VPS4A and VPS4B DN reduced VSV replication and budding but did not reduce influenza virus replication and budding
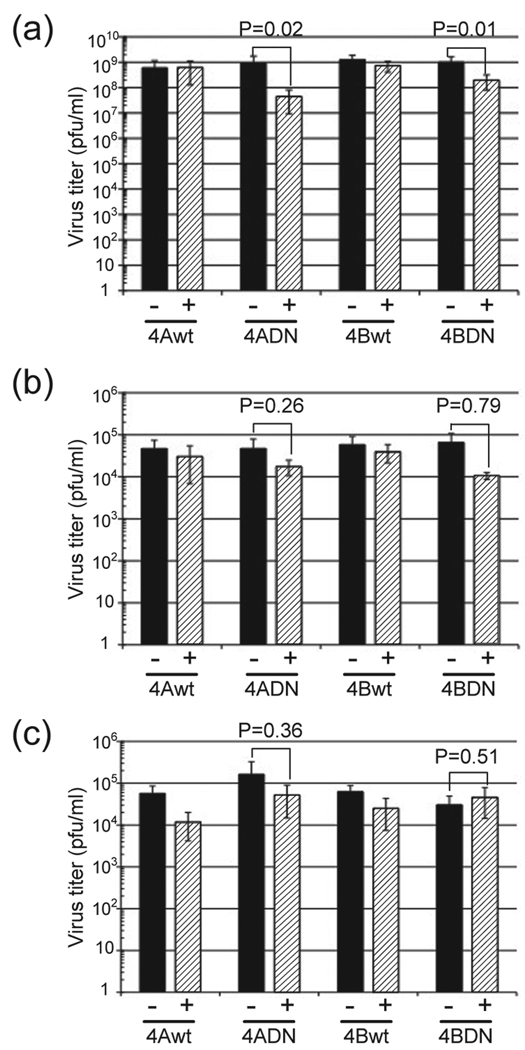
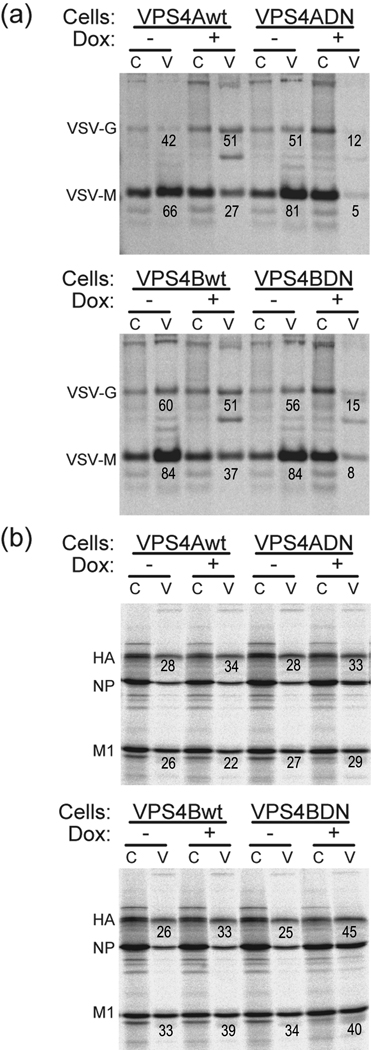
The use of permanent inducible cell lines expressing VPS4A-DN or VPS4B-DN avoids the efficiency of transfection/infection problem. These cell lines had been used successfully to show that VSV did require functional VPS4 activity whereas Semliki Forest virus budding did not require functional VPS4 activity (Taylor et al., 2007). The advantage of the system is that it has been shown that all cells express VPS4A-DN and VPS4B-DN after tetracycline induction (Taylor et al., 2007) and we confirmed this observation. We used the 293T VPS4A-DN and 292T VPS4B-DN inducible cell lines (Taylor et al., 2007) and examined virus replication with or without doxycycline induction. VSV was used as a control virus for the experiment and it was shown that the expression of VPS4A-DN and VPS4B-DN reduced the infectious titer of released virus 22-fold and 5-fold, respectively (Fig. 3a, Table 1), values very similar to those obtained previously (Taylor et al., 2007). We tested influenza virus replication under these conditions, and it was found that the infectious titer of influenza virus was not affected using either strain A/Udorn/72 (H3N2) or strain A/WSN/33 (H1N1) (Fig. 3b, 3c, Table 1). The effect of VPS4A-DN and VPS4B-DN on virus budding was also examined and it was shown that VPS4A-DN and VPS4B-DN decreased VSV-M release 20-fold and 10-fold respectively (Fig. 4a). In contrast, the expression of VPS4A-DN and VPS4B-DN did not affect influenza virus budding (Fig. 4b). These results suggest that functional ESCRT machinery is important for efficient VSV budding but that influenza virus budding occurs by a mechanism that is independent of the ESCRT pathway.
Figure 3. The effect of VPS4 DN expression on virus replication.
293T/VPS4 inducible cells with (+) or without (−) doxycycline induction were infected with (a) Wt VSV; (b) influenza virus A/Udorn/72, or (c) A/WSN/33. Culture supernatant was harvested at 7 hp.i. (VSV) or 24 h p.i. (influenza virus) and virus infectious titer was determined by plaque assay.
Table 1.
The effect of VPS DN expression on virus replication.
| Dox | VPS4Awt | VPS4ADN | VPS4Bwt | VPS4BDN | |
|---|---|---|---|---|---|
| VSV | − | 6.0×108 (5.5) |
9.2×108 (8.0) |
1.2×108 (6.1) |
1.0×109 (8.0) |
| + | 6.2×108 (4.9) |
4.5×107 (3.5) |
7.3×108 (3.3) |
1.9×108 (1.1) |
|
| UDORN | − | 4.7×104 (2.6) |
4.7×104 (3.1) |
5.8×104 (3.2) |
6.6×104 (4.0) |
| + | 3.0×104 (2.3) |
1.7×104 (0.7) |
3.2×104 (1.8) |
4.0×104 (0.1) |
|
| WSN | − | 5.7×104 (2.8) |
1.6×104 (1.6) |
6.3×104 (2.4) |
3.0×104 (1.8) |
| + | 1.2×104 (0.7) |
5.2×104 (3.7) |
2.5×104 (1.8) |
4.6×104 (3.2) |
Virus titer was shown as pfu/ml.
Standard deviation is indicated in parenthesis.
The results are the average value of three independent experiments.
Figure 4. The effect of VPS4 DN expression on virus release.
293T/VPS4 inducible cells with (+) or without (−) doxycycline induction were infected with (a) Wt VSV or (b) influenza virus A/Udorn/72. The infected cells were labeled with [35S] Trans-label and chased in cold medium as described in Materials and Methods. Virus proteins were immunoprecipitated and the release efficiency of virions was calculated from the intensity of the polypeptide bands and is indicated beneath each polypeptide species. Budding efficiency was determined as described in the legend to Fig. 2. Two separate experiments are shown in panels (a) and (b).
Discussion
Previously in an investigation of the requirements for influenza virus VLP formation we showed that VLP production was not inhibited by expression of dominant negative VPS4A or VPS4B. VLP formation of HIV-1 as a control virus was inhibited by >95% by expression of DN VPS4A or VPS4B (Chen et al., 2007). However, it was important to confirm these observations in influenza virus-infected cells. Simple siRNA knockdown of VPS4A and VPS4B was not feasible due to the efficiency of transfection/infection problem and the rapid (6 h) single-step growth curve of influenza virus. Thus, we resorted to the siRNA co-transfection/trans complementation method. VSV was used as a control virus for the experiment and it was shown that the expression of VPS4A-DN and VPS4B-DN reduced the infectious titer of released virus from 9.2×108 pfu/ml to 4.5×107 pfu/ml and 1.0×109 pfu/ml to 1.9×108 pfu/ml, respectively. These reduction values (22-fold and 5-fold reduction) were very similar to those obtained previously (Taylor et al., 2007). Although the siRNA trans-complementation data indicated a reduction in VSV titer that was statistically significant, there was no effect on influenza virus that was statistically significant. Examination of VSV particle formation was also slightly decreased by the siRNA trans complementation method whereas influenza particle formation was largely unaffected. Use of the inducible cell lines expressing VPS4A-DN and VPS4B-DN showed inhibition of VSV titer and of virus particles released from cells. In contrast, the cell lines expressing VPS4A-DN and VPS4B-DN did not affect influenza virus titer or particle release.
After our work was initiated Digard and colleagues (Bruce et al., 2009) using some different and also some overlapping approaches published a study on the requirements of influenza virus budding on VPS4. They found that the influenza virus M1 protein did not colocalize with VPS4 at the sub-cellular level and that trafficking of HA and M1 appeared normal when endosomal sorting was impaired by expression of inactive VPS4. Furthermore, they also used inducible cell lines expressing dominant negative VPS4A and they found the titer of influenza virus released was unaffected by expression of the VPS4A-DN protein. Bruce and coworkers (2009) concluded that they could see no role for the ESCRT pathway in influenza virus budding.
Taking all the data together VSV budding depends on a functional VPS4 protein whereas influenza virus and influenza virus VLP formation occurs independent of a functional VPS4 protein. Thus, the major question to be addressed is whether there exists another alternative cellular budding pathway that could be hijacked by influenza virus or whether virus proteins can perform budding functions. In this regard, it is noted that when HA is expressed, provided there is a source of exogenous neuraminidase activity, HA buds and is released in vesicles. NA and M2 also bud from cells when expressed alone and M1 budding requires expression of HA and NA and is augmented by expression of M2. Expression of the cytoplasmic tails of HA and NA are required for efficient budding of M1 (Chen et al., 2007). Thus, it seems possible that influenza virus has evolved its own mechanism for budding virus.
Acknowledgements
We thank Dr. John K. Rose for VSVΔG virus stock and Dr. Margaret Kielian for the VPS4A-DN and VPS4B-DN cell lines. This research was supported by National Institutes of Health Research Grant AI-20201. R.W. was an Associate and R.A.L. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barman S, Ali A, Hui EK, Adhikary L, Nayak DP. Transport of viral proteins to the apical membranes and interaction of matrix protein with glycoproteins in the assembly of influenza viruses. Virus Res. 2001;77:61–69. doi: 10.1016/s0168-1702(01)00266-0. [DOI] [PubMed] [Google Scholar]
- Bruce EA, Medcalf L, Crump CM, Noton SL, Stuart AD, Wise HM, Elton D, Bowers K, Digard P. Budding of filamentous and non-filamentous influenza A virus occurs via a VPS4 and VPS28-independent pathway. Virology. 2009;390:268–278. doi: 10.1016/j.virol.2009.05.016. [DOI] [PubMed] [Google Scholar]
- Chen BJ, Lamb RA. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology. 2008a;372:221–232. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BJ, Leser GP, Jackson D, Lamb RA. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J. Virol. 2008b;82:10059–10070. doi: 10.1128/JVI.01184-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BJ, Leser GP, Morita E, Lamb RA. Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid-derived virus-like particles. J. Virol. 2007;81:7111–7123. doi: 10.1128/JVI.00361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven RC, Harty RN, Paragas J, Palese P, Wills JW. Late domain function identified in the vesicular stomatitis virus M protein by use of rhabdovirus-retrovirus chimeras. J. Virol. 1999;73:3359–3365. doi: 10.1128/jvi.73.4.3359-3365.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garoff H, Hewson R, Opstelten DJ. Virus maturation by budding. Microbiol. Mol. Biol. Rev. 1998;62:1171–1190. doi: 10.1128/mmbr.62.4.1171-1190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Cote M, Rich RL, Myszka DG, Sundquist WI. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- Gottlinger HG, Dorfman T, Sodroski JG, Haseltine WA. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. U. S. A. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harty RN, Brown ME, McGettigan JP, Wang G, Jayakar HR, Huibregtse JM, Whitt MA, Schnell MJ. Rhabdoviruses and the cellular ubiquitin-proteasome system: a budding interaction. J. Virol. 2001;75:10623–10629. doi: 10.1128/JVI.75.22.10623-10629.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harty RN, Paragas J, Sudol M, Palese P. A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J. Virol. 1999;73:2921–2929. doi: 10.1128/jvi.73.4.2921-2929.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Orenstein JM, Martin MA, Freed EO. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui EK, Nayak DP. Role of ATP in influenza virus budding. Virology. 2001;290:329–341. doi: 10.1006/viro.2001.1181. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Ikeda A, Longnecker R. PY motifs of Epstein-Barr virus LMP2A regulate protein stability and phosphorylation of LMP2A-associated proteins. J. Virol. 2001;75:5711–5718. doi: 10.1128/JVI.75.12.5711-5718.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie T, Licata JM, Jayakar HR, Whitt MA, Bell P, Harty RN. Functional analysis of late-budding domain activity associated with the PSAP motif within the vesicular stomatitis virus M protein. J. Virol. 2004a;78:7823–7827. doi: 10.1128/JVI.78.14.7823-7827.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie T, Licata JM, McGettigan JP, Schnell MJ, Harty RN. Budding of PPxY-containing rhabdoviruses is not dependent on host proteins TGS101 and VPS4A. J. Virol. 2004b;78:2657–2665. doi: 10.1128/JVI.78.6.2657-2665.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayakar HR, Murti KG, Whitt MA. Mutations in the PPPY motif of vesicular stomatitis virus matrix protein reduce virus budding by inhibiting a late step in virion release. J. Virol. 2000;74:9818–9827. doi: 10.1128/jvi.74.21.9818-9827.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Joshi A, Nagashima K, Freed EO, Hurley JH. Structural basis for viral late-domain binding to Alix. Nat. Struct. Mol. Biol. 2007;14:194–199. doi: 10.1038/nsmb1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawliczek T, Crump CM. Herpes simplex virus type 1 production requires a functional ESCRT-III complex but is independent of TSG101 and ALIX expression. J. Virol. 2009;83:11254–11264. doi: 10.1128/JVI.00574-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pornillos O, Alam SL, Davis DR, Sundquist WI. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat. Struct. Biol. 2002;9:812–817. doi: 10.1038/nsb856. [DOI] [PubMed] [Google Scholar]
- Puffer BA, Parent LJ, Wills JW, Montelaro RC. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 1997;71:6541–6546. doi: 10.1128/jvi.71.9.6541-6546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- Schmitt AP, Lamb RA. Influenza virus assembly and budding at the viral budozone. Adv. Virus Res. 2005;64:383–416. doi: 10.1016/S0065-3527(05)64012-2. [DOI] [PubMed] [Google Scholar]
- Schmitt AP, Leser GP, Morita E, Sundquist WI, Lamb RA. Evidence for a new viral late-domain core sequence, FPIV, necessary for budding of a paramyxovirus. J. Virol. 2005;79:2988–2997. doi: 10.1128/JVI.79.5.2988-2997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- Taylor GM, Hanson PI, Kielian M. Ubiquitin depletion and dominant-negative VPS4 inhibit rhabdovirus budding without affecting alphavirus budding. J. Virol. 2007;81:13631–13639. doi: 10.1128/JVI.01688-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schwedler UK, Stuchell M, Muller B, Ward DM, Chung HY, Morita E, Wang HE, Davis T, He GP, Cimbora DM, Scott A, Krausslich HG, Kaplan J, Morham SG, Sundquist WI. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- Welsch S, Muller B, Krausslich HG. More than one door - Budding of enveloped viruses through cellular membranes. FEBS Lett. 2007;581:2089–2097. doi: 10.1016/j.febslet.2007.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Cameron CE, Wills JW, Leis J. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J. Virol. 1996;70:5695–5700. doi: 10.1128/jvi.70.8.5695-5700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebedee SL, Lamb RA. Influenza A virus M2 protein: monoclonal antibody restriction of virus growth and detection of M2 in virions. J. Virol. 1988;62:2762–2772. doi: 10.1128/jvi.62.8.2762-2772.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]