Abstract
Germline transcription precedes class switch recombination. The promoter regions and I exons of these germline transcripts include binding site for activation- and cytokine-induced transcription factors, and the promoter regions/I exons are essential for class switch recombination. Therefore, it is a strong hypothesis that the promoter/I exons regions are responsible for much of cytokine-regulated, gene-specific class switch recombination. We tested this hypothesis by swapping the germline promoter and I exons for the murine γ1 and γ2a H chain genes in a transgene of the entire H chain C region locus. We found that the promoter/I exon for γ1 germline transcripts can direct robust IL-4-induced recombination to the γ2a gene. On the other hand, the promoter/I exon for the γ2a germline transcripts works poorly in the context of the γ1 H chain gene, resulting in expression of γ1 H chains that is less than 1% the wild type level. Nevertheless, the small amount of recombination to the chimeric γ1 gene is induced by IFN-γ. These results suggest that cytokine regulation of CSR, but not the magnitude of CSR, is regulated by the promoter/I exons.
Introduction
Class switch recombination (CSR2) moves a rearranged VDJ exon from physical and functional association with the Cμ coding regions to association with Cγ, Cε, or Cα coding regions. CSR is induced as a consequence of antigen-driven B cell differentiation in vivo, and can be induced in tissue culture by a combination of B cell activators (CD40 ligation, mimicking T cell help, or LPS, via Toll-like receptors) and cytokines (1,2). An activated B cell has the potential to undergo CSR to multiple H chain genes. This decision is important, as different H chain genes encode different effecter functions and thus dictate how different microbes are processed once antibody is bound. In murine B cells, the combination of B cell activators and cytokines determines to which H chain gene CSR will occur. For example, LPS+IL-4 directs CSR to the γ1 gene and LPS+IFN-γ directs CSR to the γ2a gene (3–5).
CSR is preceded by germline transcription of only the H chain gene to which CSR is directed (6,7). Germline transcription is initiated at an I exon upstream of the switch region (the region of DNA in which the CSR deletion begins or ends), and proceeds through the switch region and C region coding exons. The promoter regions of these germline transcripts include the appropriate transcription factor binding sites (1,2). For example, nearly all promoter regions for germline transcripts include LPS- or CD40 ligation-responsive NF-κB sites, the promoter region for γ1 germline transcripts includes Stat6 binding sites, and the promoter region for γ2a germline transcripts includes motifs that resemble interferon-response factor (IRF) binding sites. It is widely hypothesized that the promoter regions for germline transcripts dictate the cytokine-specific induction of both germline transcription and CSR (1,2). Consistent with this idea, deletion of the promoter region and I exon, or the most of the I exon and donor splice site, does eliminate CSR to the corresponding H chain gene (8–10). In addition, substitution of the germline promoter with a constitutive promoter allows cytokine-independent CSR to the corresponding H chain gene (10–12). Deletion and substitution experiments establish that sequences within the promoter regions/I exons are critical for CSR; those sequences could include the transcription start sites, transcription factor binding sites that attract AID, and other potential functions. However, deletion and substitution experiments do not test if these sequences encode the cytokine induction of CSR. Furthermore, investigations of cytokine-regulated, gene specific CSR have focused almost exclusively on the promoter/I exons, ignoring other parts of the heavy chain genes. In fact, a series of experiments testing the role of the switch region, culminating in the replacement of one switch region for another, suggests that some gene-specific CSR may be directed by the switch regions themselves (13). We asked if the promoter regions/I exons, moved into the context of potential regulatory elements in another H chain gene, could direct cytokine-regulated gene-specific CSR. We swapped the promoter regions and I exons for γ1 and γ2a within a transgene for the entire murine H chain C region locus. If the promoter region plus I exon dictates gene-specific recombination, we expected the promoter/Iγ2a region to direct IFN-γ-induced CSR to Cγ1, and the promoter/Iγ1 region to direct IL-4-induced CSR to Cγ2a.
Materials and Methods
Transgenic mice
The starting artificial bacterial chromosome (BAC) was named ARS/Igh81 (Fig. 1, line 1). ARS/Igh81 has two copies of the chicken β–globin insulator and a NotI restriction site inserted 3 kb 5’ of the VDJH2 exon, four bp insertions in Iγ1 and Iγ2a, and a Flag tag inserted three codons 5’ of the carboxy terminus of the secreted version of the γ2a heavy chain. This BAC was targeted three times to (i) delete 2.1 kb of the promoter/Iγ1 region, (ii) substitute 1.8 kb of the promoter/Iγ1 region for the promoter/Iγ2a region, and (iii) substitute 2.2 kb of the promoter Iγ2a region for the promoter/Iγ1 region. In all three targetings, fragments containing the 5’ and 3’ homology regions for each targeting vector were sequenced to verify that no additional point mutations were introduced into the BAC during homologous recombination. When various fragments were brought together, their relative orientation was confirmed by at least two independent restriction digests. Targeting sequences were moved into pSV1.RecA (14) using SalI sites flanking the targeting sequences in an intermediate vector.
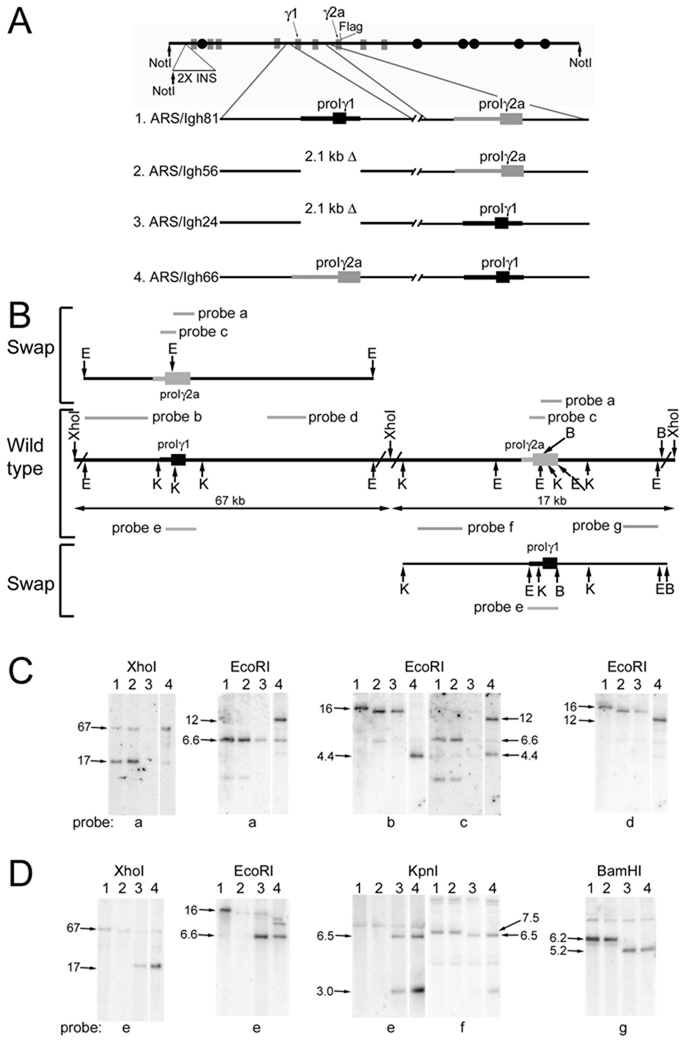
Figure 1.
Verification of the ARS/Igh66 gene structure. A. Construction of heavy chain constant region locus with an exchange of promoter/Iγ1 and promoter/Iγ2a. The structure of the ARS/Igh BAC is shown in the top of the figure. B. In the middle of Part B is shown the DNA around the Iγ1 and Iγ2a exons in the ARS/Igh wild type BAC. Only relevant restriction sites are shown, and some restriction sites are abbreviated: E, EcoRI; K, KpnI; B, BamHI. The locations of various probes, used in Southern hybridization experiments, are shown as grey bars with letter designations. On the upper left of Part B, a schematic depicts the location of the promoter/Iγ2a insertion into the γ1 gene. Since probes “b” and “c” are partly or wholly within the promoter/Iγ2a region, they are shown above the swapped region in their new location. On the lower right of Part B, a schematic depicts the location of the promoter/Iγ1 insertion into the γ2a gene. Since probe “a” is wholly within the promoter/Iγ1 region, it is shown below the swapped region in its new location. C and D. DNA samples (lane numbers are constructs designated in part A) were digested with the restriction enzymes listed above the panels. The XhoI digests were fractionated on a CHEF gel; other digests were fractionated on conventional 0.9% agarose gels. After blotting onto nitrocellulose, the fractionated digests were hybridized to the probes indicated below the panels. Sequences in other γ genes that are related to various probes results in weaker, cross-hybridizing fragments in some lanes. The 1.7 and 1.3 kb KpnI fragments that hybridize to probe “e” in the germline version of the locus have been run off the gel.
For construction of ARS/Igh56, with a 2.1 kb deletion of the promoter/Iγ1 region (Fig. 1A, line 2), a targeting vector was produced with a HindIII/BamHI fragment (from the HindIII site about 600 bp 5’ of residue 1 to residue 542 in D78344—the sequence of the BALB/c γ1-γ2b-γ2a region) joined to a BamHI fragment (from 2690 to 3407) in the physiologic orientation. Moving this fragment into ARS/Igh81 deleted 2.1 kb of the promoter/Iγ1 region flanked by BamHI sites. For construction of ARS/Igh24, with a replacement of the promoter/Iγ2a region (grey box and thick line) with the promoter/Iγ1 region (black box and thick line—Fig. 1A, line 3), the 5’ homology region was amplified using GTACGcGgCCgCTCCCCAGTGACCCATG (D78344 residues 41107-41034, with residues mutated, as indicated in lower case, to form a SalI site) and GCTGGGaTcCTCTGCTATACCAGAGGCCTTG (42184-42156, with mutations, as indicated in lower case, to create a BamHI site). The 3’ homology region was amplified using GGATCcTGCACTAGAGATATGGG (44002-44024, with a mutation, as indicated in lower case, to create a BamHI site) and gcggccgcTACCATGGCTCTGTACTACTCACC (residues 42025-42102, with residues added, as indicated in lower case, to create a SalI site). These homology regions were brought together in the physiological orientation with a single, engineered BamHI site in between them. The promoter/Iγ1 region-containing 1.8 BamHI fragment was inserted, in the same gene orientation, at the BamHI site. Moving this targeting vector into ARS/Igh56 replaced the promoter/Iγ2a region with the promoter/Iγ1 region. For construction of ARS/Igh66, with a replacement of promoter/Iγ1 with promoter/Iγ2a (Fig. 1A, line 4), a fragment containing the promoter/Iγ2a region was amplified using CTCCAGGaTCcACTCTACCTACAG (residues 42186-42208, with mutations, as indicated in lower case, to create a BamHI site) and CCTGGaTCCACACCCATATCTCTAG (residues 44459-44436, insertion, as indicated in lower case, to create BamHI site). This fragment was moved following a partial BamHI digest into the BamHI site in the middle of the targeting vector used to construct ARS/Igh56 and delete the promoter/Iγ1 (described above). Moving the resulting targeting vector into ARS/Igh24 replaced the promoter/Iγ1 with promoter/Iγ2a.
The ARS/Igh66 BAC was digested with Not1, and the 230 kb insert fragment (with loss of both the vector and a 6 kb Not1 fragment at the 5’ end of the BAC) was purified and injected into C57BL/6×SJL F2 embryos for the preparation of transgenic mice. Founder mice were back-crossed two or three times to C57BL/6 mice. One line of transgenic mice (line 57) was found to lack the transgenic γ1 gene, and so it was not analyzed further. All work with mice was approved and monitored by the University of Michigan Committee on Use and Care of Animals.
Analysis of CSR
Resting splenic B cells were prepared from spleens of transgenic and C57BL/6 mice using a magnetic bead-based kit (Miltenyi Biotec #130-090-862) that depletes CD43-, CD4-, and Ter119-expressing cells. For RNA analysis, B cells were cultured at 1 million per ml in RPMI supplemented with 10% FBS, penicillin, streptomycin, glutamine, and 20 µM 2ME. The following additions were made in various combinations: 25 µg/ml LPS (Sigma L7261), 100,000 CD40L-expressing Sf21 cells/ml (15), 35 ng/ml recombinant murine IL-4, or 100 units/ml recombinant murine IFN-γ. RNA was prepared after three days of culture. Various transcripts were amplified using primers and detected by incorporation of 32P-dATP during the reaction as described (16). Chimeric IγICγ2a transcripts were amplified using primers GACGGCTGCTTTCACAGCTT (Iγ1) and GCTGGGCCAGGTGCTCGAGGTT (Cγ2a). Chimeric Iγ2aCγ1 transcripts were amplified using GCTGATGTACCTACCTGAGAGAG (Iγ2a) and GCATGATGGGAAGTTCACTGACTG (Cγ1). For antibody secretion, 100,000 B cells were cultured in 1 ml of RPMI, with supplements, cytokines, and B cell activators at the same concentration as above, except that 10,000 CD40L-expressing Sf21 cells/ml were used. Supernatants of these 1 ml cultures were tested for Ig secretion by ELISA after 7 days of culture (16). Supporting data for both RNA and secreted Ig was generated using two or more similar cultures of T-depleted splenocytes, which included activated B cells. These results with T-depleted splenocytes were entirely consistent with the data presented here, except for higher levels of IgG2a and γ2a mRNA from cells cultured with LPS or CD40L alone.
Results
Construction of a H chain locus BAC with a promoter/I exon swap
We utilized a 230 kb transgene that includes a knocked-in VDJ exon (encoding anti-ARS activity, ref. 17), the coding regions for all eight murine H chain C genes, and the entire 3’ enhancer region (Fig. 1A). Both germline transcription and CSR of the transgenic locus are regulated by B cell activators and cytokines like that of the endogenous locus (16). Since the transgene is derived from strain 129 DNA (Igha) and we bred the transgene onto the C57BL/6 background (Ighb), we distinguished the transgene from the endogenous H chain genes by multiple restriction site polymorphisms. We distinguished Ig expression by the transgene from Ig expression of the endogenous genes by allotype specific antibodies (IgG1) or by a Flag epitope we inserted into the carboxy terminus of the secreted form of IgG2a. Using targeted homologous recombination in E. coli (14), we replaced 2.2 kb of the γ2a promoter region and I exon with a 1.8 kb BamHI fragment that includes the γ1 promoter region and I exon. We also replaced 2.1 kb of the promoter/Iγ1 region with the 2.2 kb fragment from the γ2a gene. We included the I exons in this swap, because the only Stat1 binding site (IFN-γ responsive) in the γ2a gene is at the 3’ end of the Iγ2a exon. In addition, the I exon and/or its donor RNA splice site, have been implicated in the regulation of CSR (10,18,19). By a series of Southern blots, we verified the structure of the BAC with the promoter/I exon swap, with no obvious additional rearrangements or deletions (Fig. 1B). For example, probe “a” hybridizes to the 6.6 kb EcoRI fragment that includes the promoter/Iγ2a in ARS/Igh81 and in ARS/Igh56 (Fig. 1C, second set of panels, lanes 1 and 2). Hybridization to the promoter/Iγ2a fragment is lost when this fragment is replaced by promoter Iγ1 in ARS/Igh24 (cross hybridization to the 6.6 kb EcoRI fragment with the promoter/Iγ2b sequences remains, lane 3). Since probe “a” is part of the promoter/Iγ2a fragment, it moves to the γ1 gene in ARS/Igh66. The introduction of an EcoRI sites results in hybridization to a 12 kb fragment in the γ1 gene (lane 4). As a second example, probe “b” (from the γ1 gene) hybridizes to a 16 kb EcoRI wild type fragment (Fig. 1C, middle panel, lane 1). Due to the 2.1 kb promoter/Iγ1 deletion, the hybridizing fragment is 14 kb in ARS/Igh56 and ARS/Igh24 (lanes 2 and 3). The insertion of promoter/Iγ2a brings an EcoRI site into the γ1 gene, and so probe “b” hybridizes to a 4.4 kb fragment in ARS/Igh66 (lane 4). Eight combinations of other digests and probes provided further confirmation of the structure of ARS/Igh66 (Fig 1 C and D).
We analyzed four lines of transgenic mice with the promoter/I exon swap, named 46, 55, 78, and 79. By examining the transgene content for twelve DNA segments along the transgene, we verified that all four lines had one or two complete copies of the H chain transgene (Supplemental Fig. 1). We also found that the four lines of transgenic mice produced abundant B cells, and that, like other ARS/Igh transgenes (16), allelic exclusion of the endogenous genes was more than 95% complete (Supplemental Fig. 2). Expression from the transgenic γ2b gene, which is representative of expression from the transgenic γ3 and α genes, for these lines is presented in Supplemental Figure 3.
Expression of chimeric germline transcripts
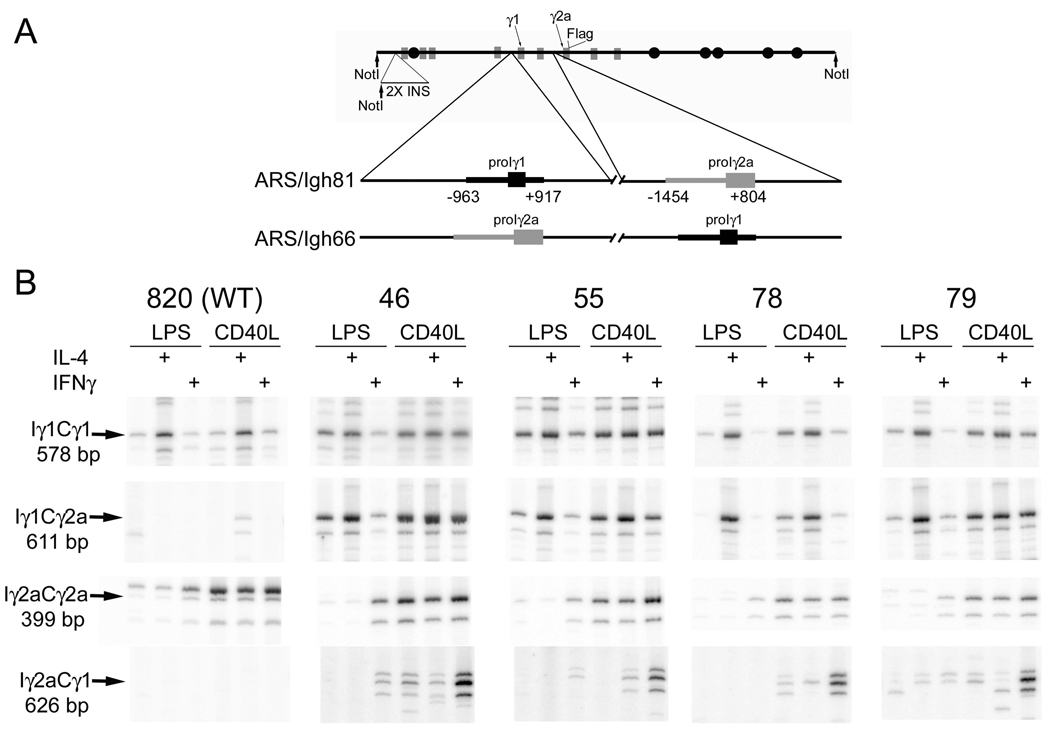
B cells with the ARS/Igh66 transgene (Fig. 2A) would be expected to express chimeric Iγ1Cγ2a and Iγ2aCγ1 germline transcripts. We examined the expression of these chimeric germline transcripts by RT-PCR. We have previously determined that γ1germline transcripts, in B cells with a wild type ARS/Igh transgene, are expressed from both the transgene and germline endogenous γ1 genes (16). In that study, we determined that germline transcripts from the two types of genes are expressed in parallel by digestion of PCR products with a restriction enzyme that allows us to distinguish germline transcripts of the endogenous and transgenes (16). In samples from line 820, with a wild type ARS/Igh transgene (top left panel, Fig. 2B), undigested PCR products of germline transcripts (“Iγ1Cγ1”) from the endogenenous and transgenic γ1 genes migrate together (Fig. 2B, top row of panels). However, since transgenes with the promoter/I exon swap cannot produce transgenic Iγ1Cγ1 transcripts, all of the transcripts for lines 46, 55, 78, and 79 in the top row of panels are derived from the endogenous genes. Endogenous γ1 germline transcripts are induced by LPS+IL-4 and CD40L+IL-4, and induced somewhat by CD40 ligation alone (Fig. 2B, top panels, and ref. 15). Iγ1Cγ2a transcripts from the transgenes with the promoter/I exon swap are expressed in parallel to the endogenous Iγ1Cγ1 germline transcripts (compare the top and second set of panels, Fig. 2B). The chimeric transcripts are not expressed in B cells with a wild type transgene (line 820). The parallel expression of endogenous and line 820 transgenic γ2a germline transcripts is directly demonstrated in left-most panel in the third set (Fig. 2B), as the transgenic product migrates a little slower than the endogenous product, due to a four bp insertion in the transgenic Iγ2a (16). This slower migrating transgenic product is not detected in the four lines with the promoter/I exon swap, as they cannot be expressed; only the product of the endogenous γ2a genes is detected in lines 46, 55, 78, and 79. Both endogenous and line 820 transgenic γ2a transcripts are induced by LPS+IFN-γ (albeit modestly), by CD40L alone, and by CD40L+IFN-γ (third row of panels, Fig. 2B). However, in the context of the transgenic γ1 gene, Iγ2aCγ1 germline transcripts are induced well only by CD40L+IFN-γ (bottom row of panels). As expected, Iγ2aCγ1chimeric transcripts are not detected in RNA from B cells bearing a wild type (no promoter/I exon swap) transgene (line 820). Thus, chimeric germline transcripts are expressed, and their induction is, to a large extent, dictated by the promoter region/I exon. The notable exception is that the promoter/Iγ2a region is a poor inducer of CD40L-induced germline transcripts in the context of the γ1 gene.
Figure 2.
H chain transgene with a swap of the γ1 promoter/I exon and the γ2a promoter/I exon. A. Structure of the ARS/Igh 66 transgene. Coding regions are depicted as grey boxes and enhancer elements as black circles. A 2.4 kb fragment including two copies of the chicken β-globin insulator (“2X INS”), with an engineered NotI restriction site, was inserted 3 kb 5’ of the VDJ exon. See text for further explanation. B. Expression of chimeric germline transcripts from the ARS/Igh66 transgene. cDNA from B cell culture of the indicated transgenic lines, expression of Iγ1Cγ2a, Iγ2aCγ1 transcripts, γ2a germline transcripts, and γ1 germline transcripts. The chimeric germline transcripts were cloned and sequenced, and found to be the predicted products, with splicing from the major splice site of the Iγ1 or Iγ2a exon to the appropriate CH1 acceptor splice site (20,21). Within each transgenic line, the cDNA were first adjusted to be approximately equal for expression of transgenic VDJCμ transcripts (see Fig. 3). The slower migrating, more intense, band in the line 820 Iγ2aCγ2a panel is the transgenic germline transcripts (403 bp) from the wild type transgene. The second slowest band (399 bp) represents germline transcripts of the endogenous γ2a gene (present in all lanes), and the fastest migrating band is an alternative splice product of the γ2a germline transcripts (21).
The promoter/Iγ1 dictates robust IL-4-induction of Cγ2a post-switch transcripts
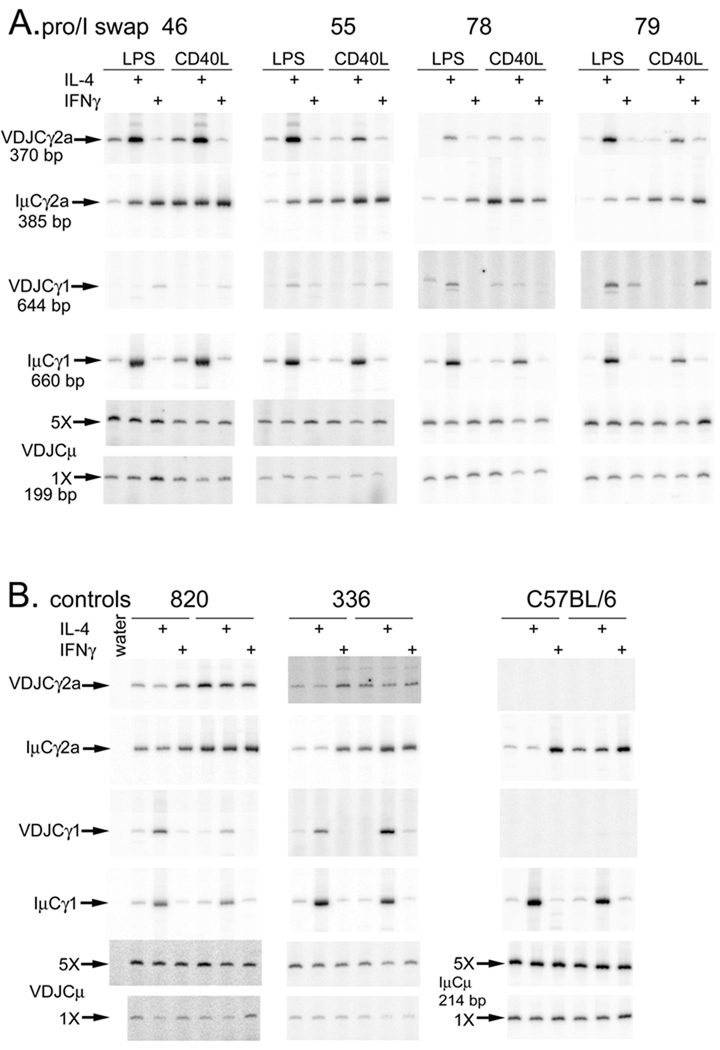
We tested CSR of the ARS/Igh66 transgene in tissue culture in two ways. First, we tested expression of transgene specific VDJCγ1 or VDJCγ2a transcripts by RT-PCR. To equalize cDNA samples from various induction regimens for total transgene expression, we adjusted the amount of cDNA so that transgenic VDJCμ expression was approximately equal (Fig. 3AB, bottom panels). In Supplemental Fig. 4A, we present a comparison of VDJCμ RNA expression to RNA expression of a housekeeping gene, HPRT. Like the chimeric Iγ1Cγ2a germline transcripts, transgenic VDJCγ2a transcripts are induced by IL-4 (Fig. 3A, top row of panels). For comparison, we also tested the expression of IμCγ1 transcripts. Even though the excluded endogenous genes may have no in-frame VDJ exon, they will switch their H chain genes, and express IμCγ transcripts (22). Apparently, the majority of the IμCγ1 transcripts derive from the endogenous genes, as they follow the well-documented IL-4 induction (Fig. 3A, fourth panel from the top), as do post-switch VDJCγ1 transcripts from the wild type H chain transgenes in lines 820 and 336 (Fig. 3B, third panel from the top).
Figure 3.
Analysis of cytokine-induced post-switch RNA expression. cDNA from B cell cultures of the indicated transgenic lines, cultured with the indicated combinations of activators and cytokines, was tested by RT-PCR for various post-switch transcripts. Part B is arranged like Part A--within each transgenic line the left three lanes are from cells cultured with LPS and the right three lanes are from cells cultured with CD40L. Within each transgenic line, cDNAs were first adjusted to be approximately equal for expression of transgenic VDJCμ transcripts (bottom two panels for promoter/I exon swap mice—Part A, or wild type and nontransgenic mice —Part B). VDJ and Iμ transcripts were tested using the same relative amounts of cDNA. In Part B, since C57BL/6 B cells do not express transgenic transcripts, we used IμCμ transcripts to demonstrate that these samples included intact RNA.
The promoter/Iγ2a is a poor inducer of Cγ1 post-switch transcripts
Post-switch IμCγ2a transcripts from endogenous genes (Fig. 3A, second set of panels from the top), or VDJCγ2a transcripts from wild type transgenes (Fig. 3B, top set of panels), are induced modestly by LPS+IFN-γ relative to LPS alone or LPS+IL-4. As we reported, the CD40L expressed by insect cells is a potent inducer of CSR to γ2a (much stronger than anti-CD40 antibodies—ref. 23), and so we observe as much or more IμCγ2a transcripts in CD40L alone or in CD40L+IFN-γ compared to LPS+IFN-γ (Fig. 3A, second set of panels). This expression pattern is not transferred by insertion of the γ2a promoter/I exon into the γ1 gene; expression of the chimeric γ1 is poor, regardless of induction regimen (Fig. 3A, third row of panels from the top). Occasionally we observed good induction of post-switch VDJCγ1 transcripts in B cells treated with CD40L+IFN-γ (for example, line 79 in Fig. 3A), but this was the exception.
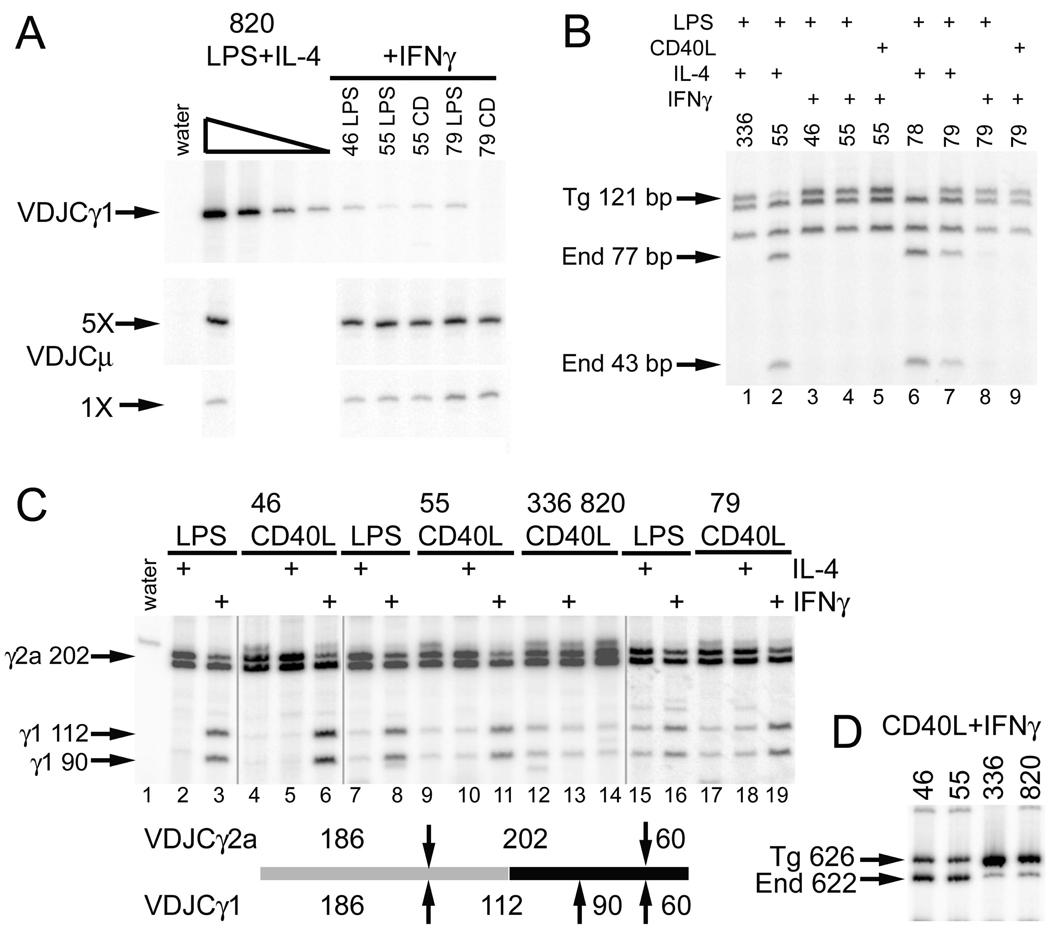
To characterize better the induction of the VDJCγ1 transcripts, under control of the promoter Iγ2a region, we first normalized several samples for approximately equal expression of VDJCμ (Fig. 4A, second and third row of panels). We compared the RT-PCR products from transgenes with the promoter swap to those from a cDNA dilution series VDJCγ1 cDNA from a wild type transgene (line 820). The quantity of VDJCγ1 PCR products from transgenes with the promoter swap was equal to or less than a 125-fold dilution (0.8%) of the cDNA from the wild type transgene. Hence, comparing samples with equal transgenic VDJCμ expression, γ1 expression from transgenes with the promoter swap was less than 1% of γ1 expression from wild type transgenes (Fig. 4A). We detected some of the best VDJCγ1 expression in transgenic cells cultured in LPS+IL-4 (Fig. 3A, third row of panels, lines 55, 78, and 79), the condition in which the endogenous γ1 gene would be accessible to the activation-induced cytidine deaminase. We had observed trans-recombination of the transgenic VDJ with endogenous Cγ genes in other studies (16). We determined if the LPS+IL-4-induced VDJCγ1 products indeed used an endogenous Cγ1 gene by testing for a polymorphic MboI site in the CH2 region (Fig. 4B). Virtually all LPS+IL-4-induced VDJCγ1 transcripts use the transgenic Cγ1 in wild type (line 336, lane 1) transgenes, as do all of the IFN-γ-induced VDJCγ1 transcripts in lines 46, 55, and 79 (lanes, 3, 4, 5, 8, and 9, Fig. 4B). On the other hand, the vast majority of LPS+IL-4-induced VDJCγ1 transcripts from lines 55 and 78 with the promoter swap use the endogenous Cγ1 (lanes 2 and 6). About one-half of the LPS+IL-4-induced VDJCγ1 transcripts in line 79 use the endogenous Cγ1 (lane 7). In general, IL-4-induced CSR, as estimated by the expression of VDJCγ1 transcripts, is not directed to the transgenic Cγ1 gene driven by the γ2a promoter; a more complex recombination event between the transgenic and endogenous locus is preferred. Alternatively, it is a formal possibility that these molecules derive from a trans-splicing event between the transgenic VDJCμ transcript and the endogenous germline transcripts (24,25). However, since these two types of transcripts are also found in B cells with the wild type transgene, one might expect to find the same trans-spliced transcripts in wild type cells. This latter prediction is not confirmed (Fig. 4D, lane 1)
Figure 4.
The promoter/Iγ2a induces small amounts of IFN-γ-induced CSR to the γ1 gene. A. Quantitative comparison of VDJCγ1 expression. Samples were first balanced for VDJCμ expression (lower panels), and then tested for VDJCγ1 expression. cDNA from wild type line 820 was tested in four five-fold dilutions. B. Expression of transgenic VDJ with endogenous Cγ1. VDJCγ1 RT-PCR products were digested with Mbo1. The endogenous Cγ1 gene has an extra MboI site in CH2. C. Induction of CSR to γ1 by IFN-γ. Post-switch VDJCγ transcripts were amplified using a primer that hybridizes to both Cγ1 and Cγ2a. VDJCγ1 and VDJCγ2a were then distinguished by digestion with MboI (up or down arrows) as illustrated below the lanes. The vertical grey lines note that these data were derived from three independent RT-PCR experiments: one using the cDNA from line 46 cells activated with CD40L, with or without cytokines, one using cDNA from line 79 cells, and one using cDNA from the other samples. D. Reduced amount of Iγ2aCγ2a transcripts in B cells with the promoter/I exon swap. Germline transcripts were amplified using an Iγ2a primer and a primer that hybridizes to both Cγ1 and Cγ2a. Transgenic Iγ2aCγ1 products migrate slower than the endogenous Iγ2aCγ2a products due to a four bp insertion in the transgenic Iγ2a exon.
These results suggest that the induction of γ1 post-switch transcripts, if directed by the promoter/Iγ2a region, is regulated by IFN-γ. To perform an independent test of this idea, we amplified VDJCγ post-switch transcripts from various cDNA samples with a primer that hybridized to both Cγ1 and Cγ2a. We distinguished VDJCγ1 from VDJCγ2a products by digestion with MboI (Fig. 4C). This approach does not distinguish transgenic and endogenous VDJCγ1 since the PCR ends in CH1, 5’ of the MboI polymorphism in CH2. Whereas the approach in Fig. 3 and Fig. 4A tests the absolute amount of VDJCγ1 transcripts, this experiment tests the amount of combined transgenic and endogenous VDJCγ1 relative to the amount of VDJCγ2a in the same sample. Since the RT-PCR can go to saturation, this approach yields a sensitive test of the cytokine regulation of γ1 versus γ2a. As expected, with induction of B cells with wild type transgenes by CD40L or CD40L+IFN-γ, there are more VDJCγ2a products than VDJCγ1 products (lanes 12–14, Fig. 4C). In cDNA from B cells with the transgenic promoter/I exon swap (46, 55, and 79), more VDJCγ1 products appear after induction with IFN-γ than after induction with IL-4 (Fig. 4C, compare lanes 3, 6, 8, 11, 16, and 19 to 2, 5, 7, 10, 15, and 18). From the results in Fig. 4B, we know that the vast majority of these IFN-γ-induced VDJCγ1 products are derived from the transgene. Consistent with the results in Fig. 3, in B cells with the promoter/I exon swap, most of the products are VDJCγ2a after induction with IL-4 (lanes 2, 5, 7, 10, 15, and 18). Even though some of these samples include VDJCγ1 products derived from the endogenous C genes (Fig. 4B), the amount of these VDJCγ1 products is very small compared to the VDJCγ2a products induced by IL-4 from the chimeric γ2a gene (for example, Fig. 4C, lane 7). Therefore, even though the levels of γ1 post-switch transcripts in transgenes with the promoter/I exon swap are very small, their expression is IFN-γ dependent.
We considered the possibility that CSR to the chimeric γ1 gene is inefficient due to poor germline transcription. We devised an RT-PCR that would measure the quantity of chimeric Iγ2aCγ1 transcripts by a direct comparison to the endogenous Iγ2aCγ2a germline transcripts, which should be at the same levels in all transgenic cells. As we have reported (23), transgenic, wild type Iγ2aCγ2a (Igha allele) germline transcripts are more abundant (8.6-fold for line 820 and 15-fold for line 336) than endogenous (Ighb allele) germline transcripts (Fig. 4D). On the other hand, the chimeric Iγ2aCγ1 germline transcripts are slightly reduced in quantity (0.8-fold) compared to endogenous transcripts in the same cells (lines 46 and 55, Fig. 4D). Therefore, germline transcripts from the promoter/Iγ2a, in the context of the γ1 gene, are present at only 5–10% of the level of wild type germline transcripts, which may explain, in part, why CSR to the chimeric gene is inefficient.
CSR as measured by secreted protein
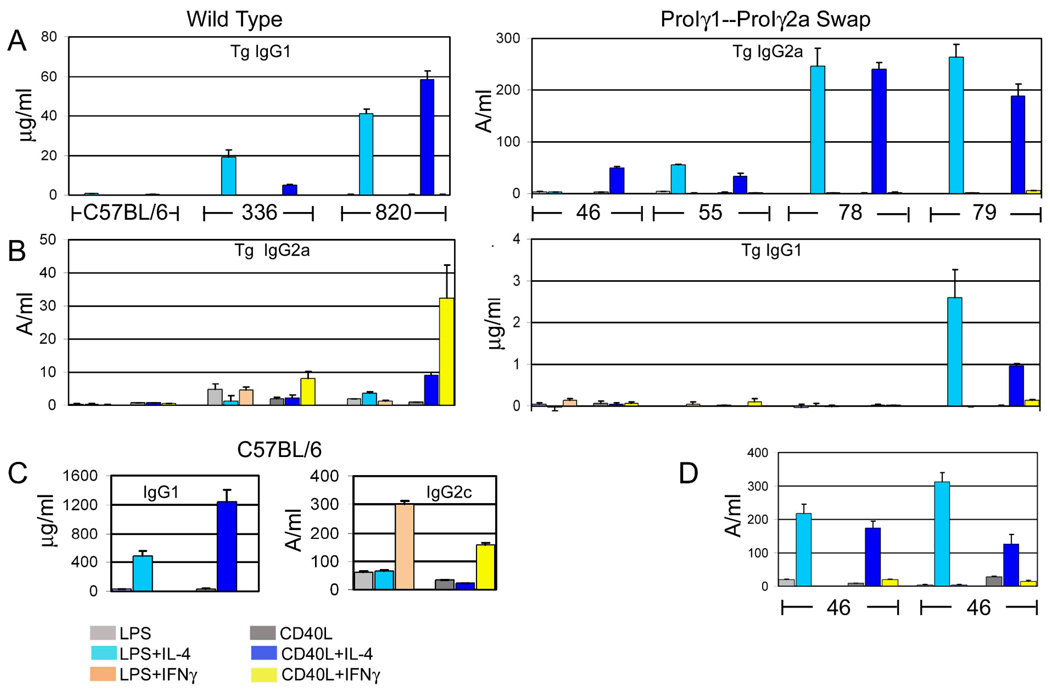
We also tested CSR in tissue culture by expression of transgene-specific IgG1a or Flag-tagged IgG2a. The results reproduced the expression pattern of post-switch transgenic VDJCγ transcripts described above. Transgenic IgG1a (in wild type lines 820 and 336) or total IgG1 (in C57BL/6 nontransgenic B cells) was induced by LPS + IL-4 or CD40L + IL-4, but not by B cell activators alone or with IFN-γ. Transgenic Flag-tagged IgG2a (wild type ARS/Igh transgenes) or IgG2c (C57BL/6) was induced by IFN-γ (Fig. 5). In the mice with the swap of promoter/Iγ1 and promoter/Iγ2a, transgenic IgG2a secretion was now induced by IL-4, in apparently greater quantities than from wild type transgenes3. B cells from wild type lines 336 and 820, cultured with LPS or CD40L and IFN-γ secreted 1.3 to 32.5 units of transgenic IgG2a. B cells from transgenic mice with the promoter/I exon swap, cultured with activators and IL-4, secreted from 33.4 to 264 units of transgenic IgG2a. Line 46 cells cultured with LPS + IL-4 secreted only 2.8 units of transgenic IgG2a in the experiment in Fig. 5A, but in two experiments with T-depleted splenocytes secreted 218 and 311 units of transgenic IgG2a after culture with LPS+IL-4 (Fig. 5D). In mice where the γ2a promoter/I exon drives expression of the Cγ1 gene, transgenic IgG1a expression was very low (near the negative control levels), and barely induced by CD40L+IFN-γ. B cells from line 79, cultured with activators and IL-4, reproducibly secreted transgenic IgG1a, but these amounts were less than 15% that of wild type transgenic B cells.
Figure 5.
Analysis of secreted Ig expression. Resting splenic B cells from the indicated transgenic mice were cultured with various combinations of LPS, CD40L-expressing insect cells, IL-4, and IFN-γ. Supernatant fluids from these cultures were tested by ELISAs specific for transgenic IgG1 (A and B), transgenic IgG2a (A and B), total IgG1 (C), or total IgG2c (C). Data are presented as the means of three or four replicates from one set of cultures with SD error bars. Lines 336, 46, and 55 were tested in the same experiment; C57BL/6 and lines 820, 78, and 79 were tested in different experiments, perhaps accounting for the overall lower Ig expression in lines 336, 46, and 55. D. Transgenic Flag+ IgG2a produced by T-depleted splenocytes from line 46. The left six bars and the right six bars are from two independent experiments.
Discussion
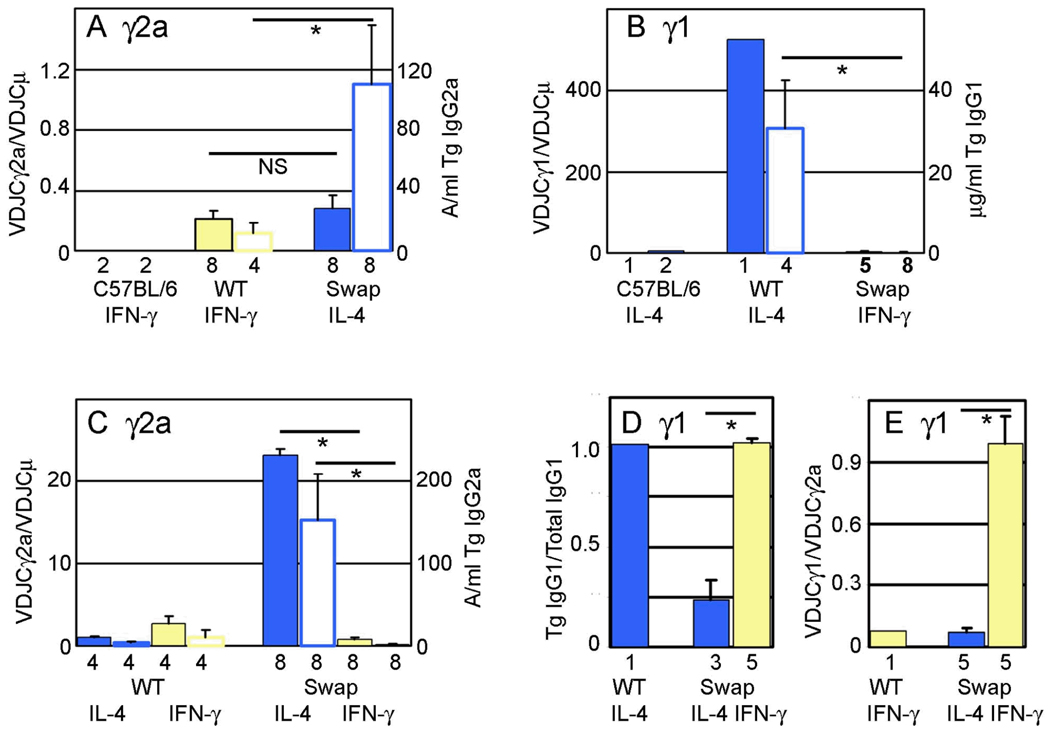
The murine γ1 gene, induced by IL-4, is the most robustly activated γ H chain gene. With optimal induction, more B cells switch to γ1 surface expression than to γ2a expression (26,27). In general, more IgG1 is secreted than IgG2a after B cell induction in vitro (5). Although it is difficult to compare Northern blots with different probes, or different RT-PCR reactions, it is apparent that γ1 germline transcripts are more abundant than γ2a germline transcripts (28). In Fig. 6, we summarize the data presented in Figs. 3–5, by pooling data for mice with the same transgenic construct and treated with the same cytokine. (The individual data points leading to Fig. 6 can be found in Supplemental Table 1.) This summary indicates that the 1.8 kb promoter/Iγ1 fragment we transferred to the transgenic γ2a gene carried with it the expression characteristics associated with the endogenous γ1 gene. The transgenic γ2a expression, in transgenes where it is controlled by the γ1 promoter/I exon, is equal to (as estimated by RT-PCR of mRNA) or greater than (as estimated by secreted IgG2a) than expression of the wild type γ2a gene (Fig. 6A). The discrepancy in the expression level of γ2a gene expression may lie in differences between the two assays. Compared to ELISA, the semi-quantitative RT-PCR is less responsive to changes in the range of two- to four-fold, since the signal is proportional to the log of the mRNA concentration. Second, the intrinsic inaccuracy of the ratio of two band densities of RT-PCR products is more than the inaccuracy of the amount of IgG2a secreted in a culture. At a minimum, the chimeric γ2a gene is expressed at levels similar to the wild type γ2a gene, and is now induced by IL-4, not IFN-γ (Fig. 6C). These results suggest that any regulatory elements in the large (8 kb) γ1 switch region (13,29) play a relatively minor role in IL-4-induced CSR to γ1 relative to elements
Figure 6.
Summary of heavy chain gene expression and regulation. In Parts A, B, and C, two bars are shown for each transgenic construct for a given cytokine treatment. The filled bar of each pair presents cDNA expression data (scale on the left y axis), and the open bar of each pair presents secreted Ig data (scale on the right y axis). The mean (with SEM bars if three or more samples were included) was determined by pooling data for all lines with the same transgenic construct from both LPS and CD40L cultures. The number of data points used is shown below each bar. Statistical significance is shown by a line above two bars and an asterisk (p<0.02, Mann-Whitney two-tailed test). A. Level of expression of the γ2a gene. Normalized γ2a expression was calculated as the density (from ImageQuant analysis) of the VDJCγ2a PCR fragment divided by the density of the VDJCμ fragment for individual cDNA samples (from Supplemental Fig. 4). Data was pooled from only those cultures with the appropriate cytokine added for maximal expression, as indicated below each pair of bars. The primary data for IgG2a secretion is found in Fig. 5. B. Level of expression of the γ1 gene, calculated as in Part A. The primary data is found in Figs. 4A and 5. C. Cytokine regulation of γ2a gene expression. IL-4 induction ratios were calculated as the VDJCγ2a band density/VDJCμ band density from cultures with activator+IL-4 divided by the VDJCγ2a band density/VDJCμ band density from cultures with LPS or CD40L only (primary data in Fig. 3). IFN-γ induction ratios were calculated similarly. IL-4 and IFN-γ induction ratios for secreted IgG2a were calculated by dividing the expression level in activator + cytokine by the expression level in activator only (primary data in Fig. 5). D. Cytokine regulation of γ1 gene expression. For various mice with the same transgenic construct the ratio of transgenic VDJCγ1 to total VDJCγ1 expression was calculated from fragment densities in Fig. 4B. Means were determined for a wild type transgene (with IL-4) and for transgenes with the promoter/I exon swap (both IL-4 and IFN-γ). E. IFN-γ induction of γ1 gene expression. For various mice with the same transgenic construct the ratio of transgenic VDJCγ1 to VDJCγ2a expression was calculated from fragment densities in Fig. 4C. Means were determined for a wild type transgene (with IFN-γ) and for transgenes with the promoter/I exon swap (both IL-4 and IFN-γ).
In stark contrast, when transferred to the γ1 gene, the γ2a promoter/I exon directs expression of the γ1 gene that is less than 1% of a wild type transgenic γ1 gene (Fig. 6B). In B cells with the promoter swap, even though the absolute level of induction the γ1 heavy chain gene by IFN-γ is small, the specificity of the induction as compared to that by IL-4 (Fig. 6D) or compared to the induction of the γ2a gene by IFN-γ (Fig. 6E) is substantial. Transgenic line 79 was exceptional in that B cells expressed some transgenic IgG1a after activation with both IL-4 and IFN-γ (Figs. 3 and 4). This may be due to an unusual transgene to transgene joint. Analysis of the transgene structure in line 79 revealed one truncated copy of the transgene that joined sequences near the γ2b hinge exon to the 3’ end of another transgene copy, in a tail to tail configuration (Supplemental Fig. 5). This junction would bring the 3’ enhancers close to transgenic γ1 gene, without any intervening H chain genes to compete with the 3’ enhancers, resulting in significant and atypical expression.
There are at least three potential reasons why the promoter/Iγ2a fails to activate CSR to the γ1 gene, while the promoter/γ1 transfers robust CSR to the γ2a gene. First, the γ1 promoter may be intrinsically stronger. The γ1 promoter/I exon may carry its own relatively strong promoter/enhancer elements (30). Consistent with the intrinsic strength of the γ1 promoter, the γ1 gene is affected the least of any heavy chain gene by deletions of various 3’ enhancers (31–33).
Second, while many of the regulatory elements for the γ1 gene may be concentrated in its promoter/I exon, normal expression of the γ2a gene may be the result of a collaboration of many elements, some of which lie outside the 2.2 kb promoter/Iγ2a fragment we transferred to the γ1 gene. Whatever regulation is encoded by the 2.2 kb promoter/Iγ2a fragment cannot interact with putative regulatory elements in the γ1 gene; the chimeric γ1 gene is essentially not expressed. In regards to these two potential factors, strength of the γ1 promoter/I exon and concentration of regulatory elements in the γ1 promoter/I exon, we would speculate that γ1 is the exceptional gene and that other heavy chain genes are more like γ2a. This would predict that cytokine-induced, gene-specific CSR to γ3, γ2b, ε, and α would be regulated by a combination of disperse, and individually less potent, elements.
Third, a change in linear distance, or gene order, relative to the 3’ regulatory region may affect the use of the two promoter/I exons. The promoter/I exon swap moves the γ1 promoter/I exon closer to the 3’ enhancer region, while it moves the γ2a promoter/I exon further away from the 3’ enhancers. On the one hand, chromosome looping within the heavy chain locus must, to some extent, override any effect of linear distance (34). On the other hand, mutations in H chain genes can alter the expression of upstream genes; the 3’ enhancers have some preference for the most proximal, strong promoter (12,33). The rearrangement of the strong γ1 promoter to the more 3’ enhancer-proximal γ2a gene may (like insertions of other strong promoters—ref. 33) inhibit CSR to more upstream genes. Since the γ1 promoter is active after CD40 ligation (15), it may “absorb” all of the 3’ enhancer activity, preventing induction of the γ2a promoter by CD40L alone. When B cells are treated with CD40L+IFN-γ, the γ1 promoter is relatively less active, which may allow some interaction of the 3’ enhancers with the γ2a promoter in the context of the Cγ1 gene (Figs. 2B and 4BC). It is noteworthy that the activity of the chimeric promoter/Iγ2a-Cγ1 gene is similar to that of the wild type promoter/Iγ2a-Cγ2a gene in a transgene with a 3’ enhancer deletion; germline transcription is reduced to 5–10% of wild type, and CSR is reduced to about 1% of wild type (16,35).
Supplementary Material
Acknowledgements
We thank Dr. Cheong-Hee Chang for her insightful comments on the manuscript. We acknowledge Wanda Filipiak, Galina Gavrilina, and Maggie Van Keuren for preparation of transgenic mice and the Transgenic Animal Model Core of the University of Michigan’s Biomedical Research Core Facilities.
Footnotes
This work was supported by AI076057. Core support was provided by The University of Michigan Cancer Center, NIH grant number CA46592.
Abbreviations: CSR, class switch recombination; ARS, arsonate; End, endogenous; I, intervening (germline transcript exon); Tg, transgenic; WT, wild type.
Note that we have presented the transgenic IgG2a expression data for the ARS/Igh66 mice next the transgenic IgG1a expression data for the wild type mice, and vice-versa. The scales are also very different for the wild type and promoter-I exon swap mice.
References
- 1.Stavnezer J. Molecular processes that regulate class-switching. Curr.Topics Micro.Immunol. 2000;245:127–168. doi: 10.1007/978-3-642-59641-4_6. [DOI] [PubMed] [Google Scholar]
- 2.Cogne M, Birshtein BK. Regulation of class switch recombination. In: Honjo T, Alt FW, Neuberger MS, editors. Molecular Biology of B Cells. London: Academic Press; 2004. pp. 289–305. [Google Scholar]
- 3.Bergstedt-Lindqvist S, Sideras P, MacDonald HR, Severinson E. Regulation of Ig class secretion by soluble products of certain T-cell lines. Immun.Rev. 1989;78:25–50. doi: 10.1111/j.1600-065x.1984.tb00475.x. [DOI] [PubMed] [Google Scholar]
- 4.Layton JE, Vitetta ES, Uhr JW, Krammer PH. Clonal analysis of B cells induced to secrete IgG by T-cell derived lymphokines. J.Expt.Med. 1984;160:1850–1863. doi: 10.1084/jem.160.6.1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Snapper CM, Paul WE. Interferon-γ and B cell stimulatory factor-1 reciprocally regulate Ig isotype production. Science. 1987;236:944–946. doi: 10.1126/science.3107127. [DOI] [PubMed] [Google Scholar]
- 6.Stavnezer-Nordgren J, Sirlin S. Specificity of immunoglobulin heavy chain switch correlates with activity of germline heavy chain genes prior to switching. EMBO J. 1986;5:95–102. doi: 10.1002/j.1460-2075.1986.tb04182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yancoupoulos GD, DePinho RA, Zimmerman KA, Lutzker SG, Rosenberg N, Alt FW. Secondary rearrangement events in pre-B cells: VHDJH replacement by a LINE-1 sequence and directed class switching. EMBO J. 1986;5:3259–3266. doi: 10.1002/j.1460-2075.1986.tb04637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jung S, Rajewsky K, Radbruch A. Shutdown of class switch recombination by deletion of a switch region control element. Science. 1993;259:984–987. doi: 10.1126/science.8438159. [DOI] [PubMed] [Google Scholar]
- 9.Zhang J, Bottaro A, Li S, Stewart V, Alt FW. A selective defect in IgG2b switching as a result of targeted mutation of the Iγ2b promoter and exon. EMBO J. 1993;12:3529–3537. doi: 10.1002/j.1460-2075.1993.tb06027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seidl KJ, Bottaro A, Vo A, Zhang J, Davidson L, Alt FW. An expressed neor cassette provides required functions of the Iγ2b exon from class switching. Int.Immunol. 1998;10:1683–1692. doi: 10.1093/intimm/10.11.1683. [DOI] [PubMed] [Google Scholar]
- 11.Bottaro A, Lansford R, Xu L, Zhang J, Rothman P, Alt FW. S region transcription per se promotes basal IgE class switch recombination but additional factors regulate the efficiency of the process. EMBO J. 1994;13:665–674. doi: 10.1002/j.1460-2075.1994.tb06305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qui G, Harriman GR, Stavnezer J. Iα exon-replacement mice synthesize a spliced HPRT-Cα transcript which may explain their ability to switch to IgA. Inhibition of switching to IgG in these mice. Int.Immunol. 1999;11:37–46. doi: 10.1093/intimm/11.1.37. [DOI] [PubMed] [Google Scholar]
- 13.Bhattacharya P, Wuerffel R, Kenter AL. Switch region identity plays an important role in Ig class switch recombination. J.Immunol. 2010;184:6242–6248. doi: 10.4049/jimmunol.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang XW, Model P, Heintz N. Homologous recombination based modification in Esherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome. Nature Biotech. 1997;15:859–865. doi: 10.1038/nbt0997-859. [DOI] [PubMed] [Google Scholar]
- 15.Warren W, Berton MT. Induction of germ-line γ1 and ε Ig gene expression in murine B cells: interleukin 4 and the CD40 ligand-CD40 interaction provide distinct but synergistic signals. J.Immunol. 1995;155:5637–5646. [PubMed] [Google Scholar]
- 16.Dunnick WA, Collins JT, Shi J, Westfield G, Fontaine C, Hakimpour P, Papavasilliou FN. Switch recombination and somatic hypermutation are controlled by the heavy chain 3' enhancer region. J.Expt.Med. 2009;206:2613–2623. doi: 10.1084/jem.20091280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Durdik J, Gerstein RM, Rath S, Robbins PF, Nisonoff A, Selsing E. Isotype switching by a microinjected mu immunoglobulin heavy chain gene in transgenic mice. Proc.Natl.Acad.Sci.USA. 1989;86:2346–2350. doi: 10.1073/pnas.86.7.2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lorenz M, Jung S, Radbruch A. Switch transcripts in immunoglobulin class switching. Science. 1995;267:1825–1828. doi: 10.1126/science.7892607. [DOI] [PubMed] [Google Scholar]
- 19.Hein K, Lorenz MGO, Siebenkotten G, Petry K, Christine R, Radbruch A. Processing of switch transcripts is required for targeting of antibody class switch recombination. J.Expt.Med. 1998;188:2369–2374. doi: 10.1084/jem.188.12.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu M, Stavnezer J. Structure of germline immunoglobulin heavy-chain γ1 transcripts in interleukin 4 treated mouse spleen cells. Dev.Immunol. 1990;1:11–17. doi: 10.1155/1990/47659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collins JT, Dunnick W. Germline transcripts of the murine immunoglobulin γ2a gene: structure and induction by IFN-γ. Int.Immunol. 1993;5:885–891. doi: 10.1093/intimm/5.8.885. [DOI] [PubMed] [Google Scholar]
- 22.Li SC, Rothman PB, Zhang J, Chan C, Hirsh D, Alt FW. Expression of Iμ-Cγ hybrid germline transcripts subsequent to immunoglobulin heavy chain class switching. Int.Immunol. 1994;6:491–497. doi: 10.1093/intimm/6.4.491. [DOI] [PubMed] [Google Scholar]
- 23.Collins JT, Shi J, Burrell BE, Bishop DK, Dunnick WA. Induced expression of murine γ2a by CD40 ligation independently of IFN-γ. J.Immunol. 2006;177:5414–5419. doi: 10.4049/jimmunol.177.8.5414. [DOI] [PubMed] [Google Scholar]
- 24.Chen Y-W, Word CJ, Dev V, Uhr JW, Vitetta ES, Tucker PW. Double isotype production by a neoplastic B cell line. II. Allelically excluded production of µ and γ1 chains without CH gene rearrangement. J.Expt.Med. 1986;164:562–579. doi: 10.1084/jem.164.2.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimizu A, Nussenzweig MC, Han H, Sanchez M, Honjo T. Trans-splicing as a possible molecular mechanism for multiple isotype expression of the immunoglobulin gene. J.Expt.Med. 1991;173:1385–1393. doi: 10.1084/jem.173.6.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hasbold J, Hong JSY, Kehry MR, Hodgkin PD. Integrating signals from IFN-γ and IL-4 by B cells: Positive and negative effects on CD40 ligand-induced proliferation, survival, and division-linked isotype switching to IgG1, IgE, and IgG2a. J.Immunol. 1999;163:4175–4181. [PubMed] [Google Scholar]
- 27.Eccleston J, Schrader CE, Yuan K, Stavnezer J, Selsing E. Class switch recombination efficiency and junction microhomology patterns in Msh2-, Mlh1-, and Exo1-deficient mice depend on the presence of mu switch region tandem repeats. J.Immunol. 2009;183:1222–1228. doi: 10.4049/jimmunol.0900135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Severinson E, Fernandez C, Stavnezer J. Induction of germ-line immunoglobulin heavy chain transcripts by mitogens and interleukins prior to switch recombination. Eur.J.Immunol. 1990;20:1079–1084. doi: 10.1002/eji.1830200520. [DOI] [PubMed] [Google Scholar]
- 29.Misaghi S, Garris CS, Sun Y, Nguyen A, Zhang J, Sebrell A, Senger K, Yan D, Lorenzo MN, Heldens S, Lee WP, Xu M, Wu J, DeForge L, Sai T, Dixit VM, Zarrin AA. Increased targeting of donor switch region and IgE in Sγ1-deficient B cells. J.Immunol. 2010;185:166–173. doi: 10.4049/jimmunol.1000515. [DOI] [PubMed] [Google Scholar]
- 30.Xu M, Stavnezer J. Regulation of transcription of immunoglobulin germ-line γ1 RNA: analysis of the promoter/enhancer. EMBO J. 1992;11:145–155. doi: 10.1002/j.1460-2075.1992.tb05037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cogne M, Lansford R, Bottaro A, Zhang J, Gorman J, Young F, Cheng H-L, Alt FW. A class switch control region at the 3' end of the immunoglobulin heavy chain locus. Cell. 1994;77:737–747. doi: 10.1016/0092-8674(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 32.Manis JP, van der Stoep N, Tian M, Ferrini R, Davidson L, Bottaro A, Alt FW. Class switching in B cells lacking 3' immunoglobulin heavy chain enhancers. J.Expt.Med. 1998;188:1421–1431. doi: 10.1084/jem.188.8.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seidl KJ, Manis JP, Bottaro A, Zhang J, Davidson L, Kisselgof A, Oettgen H, Alt FW. Position-dependent inhibition of class-switch recombination by PGK-neor cassettes inserted into the immunoglobulin heavy chain constant region locus. Proc.Natl.Acad.Sci.USA. 1999;96:3000–3005. doi: 10.1073/pnas.96.6.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wuerffel R, Wang L, Grigera F, Manis J, Selsing E, Perlot T, Alt FW, Cogne M, Pinaud E, Kenter AL. S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity. 2008;27:711–722. doi: 10.1016/j.immuni.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vincent-Fabert C, Fiancette R, Pinaud E, Truffinet V, Cogne N, Cogne M, Denizot Y. Genomic deletion of the whole IgH 3' regulatory region (hs3a, hs1,2, hs3b, and hs4) dramatically affects class switch recombination and Ig secretion to all isotypes. Blood. 2010;116:1895–1898. doi: 10.1182/blood-2010-01-264689. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.