Abstract
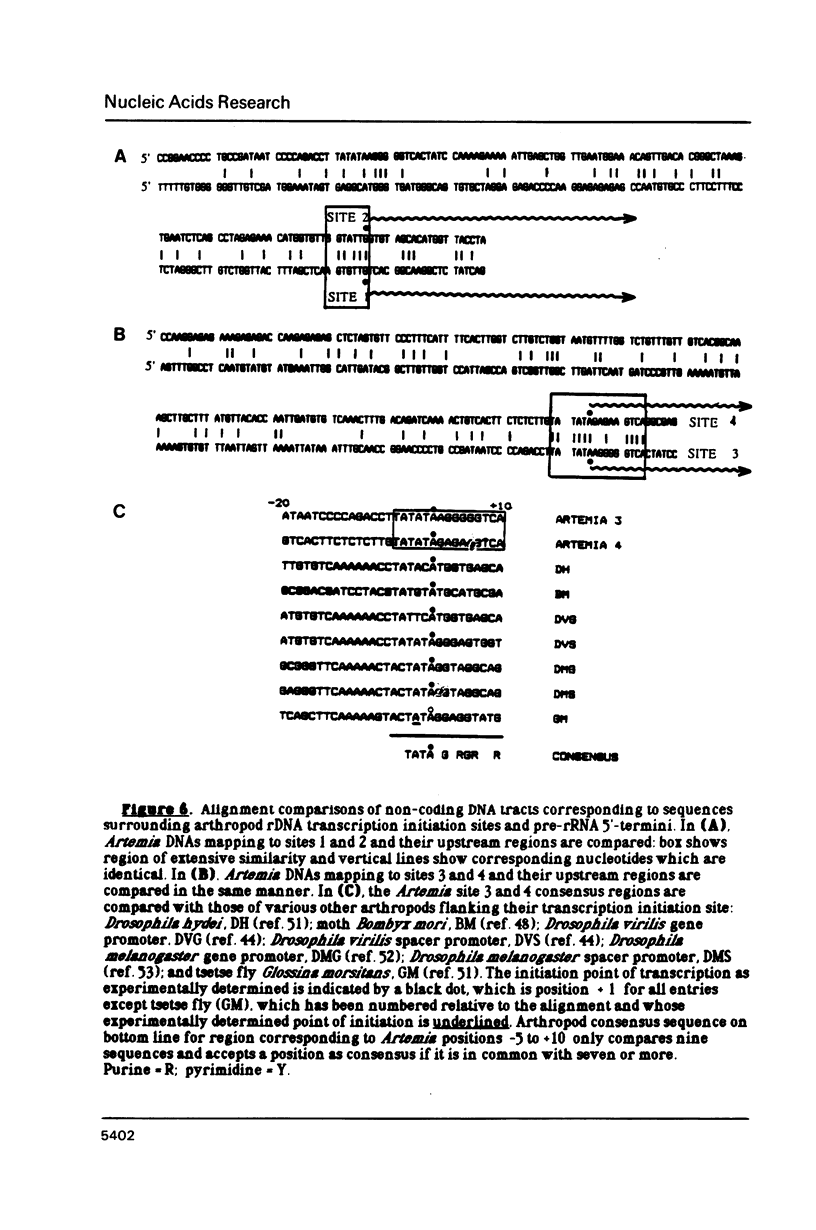
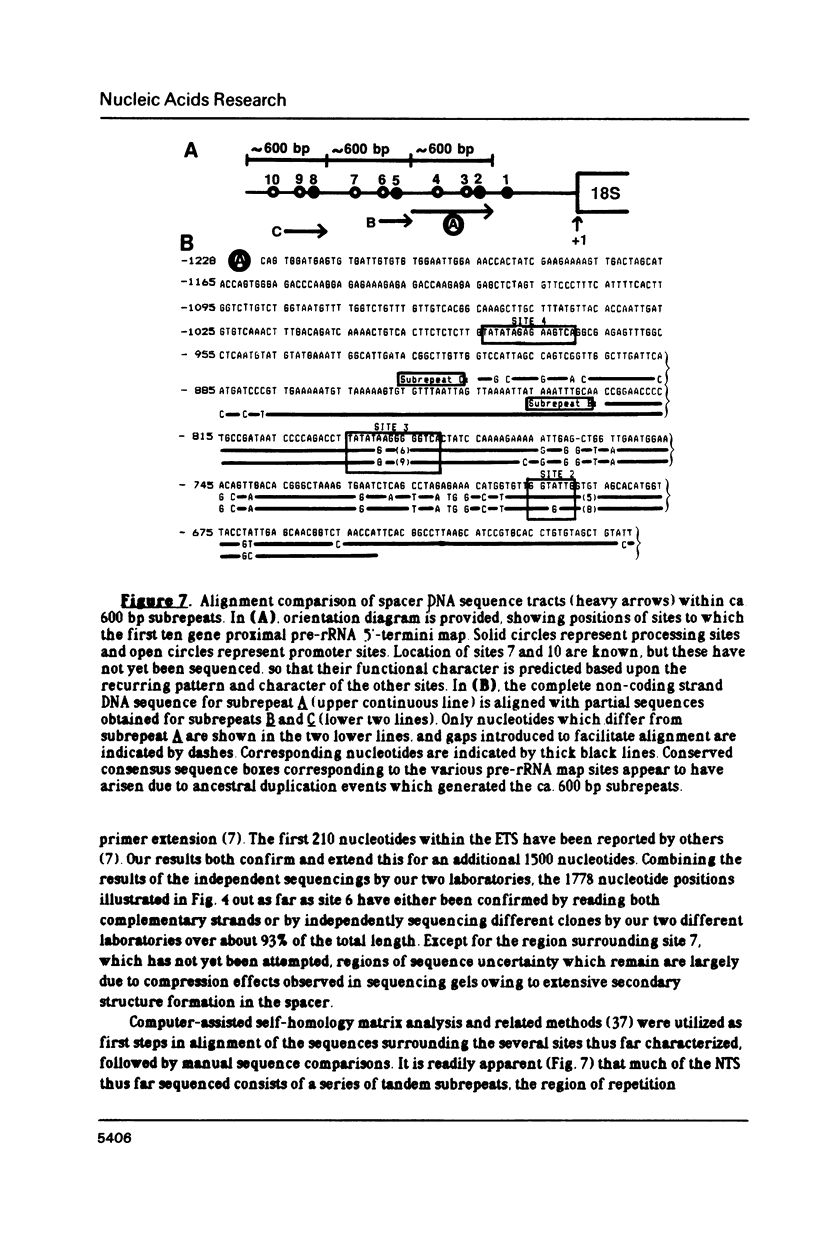
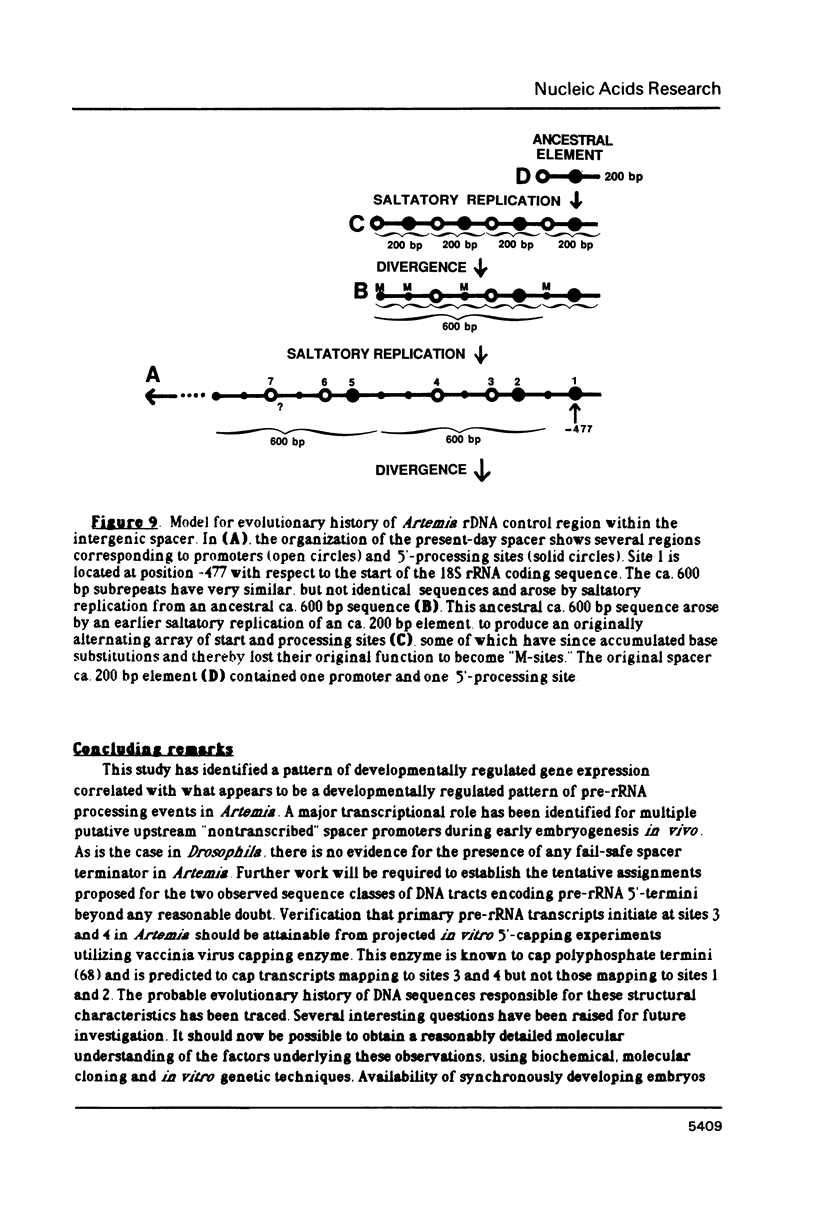
The control of ribosomal RNA (rRNA) gene expression during development can be productively studied by examination of the relationship between promoter structure and function as well as the processing of primary transcripts. Toward this end total cell RNA was extracted from embryos at various stages and probed with cloned rRNA genes using the "dot blot" method. This exercise showed that rRNA gene expression is a stage-specific process and is thus under developmental control. S1 nuclease protection experiments localized fourteen different upstream DNA sites encoding 5'-termini of pre-rRNAs during this synthetic phase of development. There is no indication of any spacer fail-safe terminator function. The S1 approach contributed to the sequencing of several of the sites. Comparative sequence alignments reveal short conserved regions in DNAs corresponding to these sites, which are shown to fall into two structural classes. Sites 3, 4, 6 and 9 are proposed to function in transcription initiation and are found to have the consensus sequence 5'...T-A-T-A-T-Pu-Pu-Pu-G-Pu-Pu-G-T-C-A 3'. Sites 1, 2, 5 and 8 which are proposed to function in 5'-processing have the consensus sequence; 5'...Pu-G-T-Pu-T-T-G 3'. These short sequence conserved regions are hypothesized to serve as recognition signals for proteins within the rDNA transcription initiation complex and for 5'-processing enzymes, respectively. Sequencing of the intergenic spacer region from which a model for spacer evolution is derived shows that tandem ca 600 bp subrepeats explain much of the multiplicity observed within control sites.
Full text
PDF














Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews M. T., Vaughn J. C., Perry B. A., Bagshaw J. C. Interspersion of histone and 5S RNA genes in Artemia. Gene. 1987;51(1):61–67. doi: 10.1016/0378-1119(87)90474-4. [DOI] [PubMed] [Google Scholar]
- Bach R., Allet B., Crippa M. Sequence organization of the spacer in the ribosomal genes of Xenopus clivii and Xenopus borealis. Nucleic Acids Res. 1981 Oct 24;9(20):5311–5330. doi: 10.1093/nar/9.20.5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagshaw J. C., Bernstein R. S., Bond B. H. DNA-dependent RNA polymerases from Artemia salina: decreasing polymerase activities and number of polymerase II molecules in developing larvae. Differentiation. 1978 Jan 13;10(1):13–21. doi: 10.1111/j.1432-0436.1978.tb00940.x. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Boseley P., Moss T., Mächler M., Portmann R., Birnstiel M. Sequence organization of the spacer DNA in a ribosomal gene unit of Xenopus laevis. Cell. 1979 May;17(1):19–31. doi: 10.1016/0092-8674(79)90291-5. [DOI] [PubMed] [Google Scholar]
- Cizewski V., Sollner-Webb B. A stable transcription complex directs mouse ribosomal RNA synthesis by RNA polymerase I. Nucleic Acids Res. 1983 Oct 25;11(20):7043–7056. doi: 10.1093/nar/11.20.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark L. S., Bowen S. T. The genetics of Artemia salina. VII. Reproductive isolation. J Hered. 1976 Nov-Dec;67(6):385–388. doi: 10.1093/oxfordjournals.jhered.a108758. [DOI] [PubMed] [Google Scholar]
- Clos J., Normann A., Ohrlein A., Grummt I. The core promoter of mouse rDNA consists of two functionally distinct domains. Nucleic Acids Res. 1986 Oct 10;14(19):7581–7595. doi: 10.1093/nar/14.19.7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross N. C., Dover G. A. Tsetse fly rDNA: an analysis of structure and sequence. Nucleic Acids Res. 1987 Jan 12;15(1):15–30. doi: 10.1093/nar/15.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diels L., De Baere R., Vandenberghe A., De Wachter R. The sequence of 5S ribosomal RNA of the crustacean Artemia salina. Nucleic Acids Res. 1981 Oct 10;9(19):5141–5144. doi: 10.1093/nar/9.19.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donelson J. E., Wu R. Nucleotide sequence analysis of deoxyribonucleic acid. VI. Determination of 3'-terminal dnucleotide sequences of several species of duplex deoxyribonucleic acid using Escherichia coli deoxyribonucleic acid polymerase I. J Biol Chem. 1972 Jul 25;247(14):4654–4660. [PubMed] [Google Scholar]
- Financsek I., Mizumoto K., Mishima Y., Muramatsu M. Human ribosomal RNA gene: nucleotide sequence of the transcription initiation region and comparison of three mammalian genes. Proc Natl Acad Sci U S A. 1982 May;79(10):3092–3096. doi: 10.1073/pnas.79.10.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Financsek I., Mizumoto K., Muramatsu M. Nucleotide sequence of the transcription initiation region of a rat ribosomal RNA gene. Gene. 1982 May;18(2):115–122. doi: 10.1016/0378-1119(82)90109-3. [DOI] [PubMed] [Google Scholar]
- Gallego M. E., Díaz-Guerra M., Cruces J., Sebastián J., Renart J. Characterization of two types of rRNA gene repeat units from the crustacean Artemia. Gene. 1986;48(1):175–182. doi: 10.1016/0378-1119(86)90363-x. [DOI] [PubMed] [Google Scholar]
- Gross R. H. A DNA sequence analysis program for the Apple Macintosh. Nucleic Acids Res. 1986 Jan 10;14(1):591–596. doi: 10.1093/nar/14.1.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I., Roth E., Paule M. R. Ribosomal RNA transcription in vitro is species specific. Nature. 1982 Mar 11;296(5853):173–174. doi: 10.1038/296173a0. [DOI] [PubMed] [Google Scholar]
- Haynes J. R., Rosteck P., Jr, Schon E. A., Gallagher P. M., Burks D. J., Smith K., Lingrel J. B. The isolation of the beta A-, beta C-, and gamma-globin genes and a presumptive embryonic globin gene from a goat DNA recombinant library. J Biol Chem. 1980 Jul 10;255(13):6355–6367. [PubMed] [Google Scholar]
- Hori H., Osawa S. Evolutionary change in 5S RNA secondary structure and a phylogenic tree of 54 5S RNA species. Proc Natl Acad Sci U S A. 1979 Jan;76(1):381–385. doi: 10.1073/pnas.76.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohorn B. D., Rae P. M. Nontranscribed spacer sequences promote in vitro transcription of Drosophila ribosomal DNA. Nucleic Acids Res. 1982 Nov 11;10(21):6879–6886. doi: 10.1093/nar/10.21.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long E. O., Rebbert M. L., Dawid I. B. Nucleotide sequence of the initiation site for ribosomal RNA transcription in Drosophila melanogaster: comparison of genes with and without insertions. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1513–1517. doi: 10.1073/pnas.78.3.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden B. E., Moss M., Salim M. Nucleotide sequence of an external transcribed spacer in Xenopus laevis rDNA: sequences flanking the 5' and 3' ends of 18S rRNA are non-complementary. Nucleic Acids Res. 1982 Apr 10;10(7):2387–2398. doi: 10.1093/nar/10.7.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Hardison R. C., Lacy E., Lauer J., O'Connell C., Quon D., Sim G. K., Efstratiadis A. The isolation of structural genes from libraries of eucaryotic DNA. Cell. 1978 Oct;15(2):687–701. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McClean D. K., Warner A. H. Aspects of nucleic acid metabolism during development of the brine shrimp Artemia salina. Dev Biol. 1971 Jan;24(1):88–105. doi: 10.1016/0012-1606(71)90048-0. [DOI] [PubMed] [Google Scholar]
- McDonell M. W., Simon M. N., Studier F. W. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J Mol Biol. 1977 Feb 15;110(1):119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- Miller K. G., Sollner-Webb B. Transcription of mouse rRNA genes by RNA polymerase I: in vitro and in vivo initiation and processing sites. Cell. 1981 Nov;27(1 Pt 2):165–174. doi: 10.1016/0092-8674(81)90370-6. [DOI] [PubMed] [Google Scholar]
- Monroy G., Spencer E., Hurwitz J. Characteristics of reactions catalyzed by purified guanylyltransferase from vaccinia virus. J Biol Chem. 1978 Jun 25;253(12):4490–4498. [PubMed] [Google Scholar]
- Morgan G. T., Roan J. G., Bakken A. H., Reeder R. H. Variations in transcriptional activity of rDNA spacer promoters. Nucleic Acids Res. 1984 Aug 10;12(15):6043–6052. doi: 10.1093/nar/12.15.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T. A transcriptional function for the repetitive ribosomal spacer in Xenopus laevis. Nature. 1983 Mar 17;302(5905):223–228. doi: 10.1038/302223a0. [DOI] [PubMed] [Google Scholar]
- Murtif V. L., Rae P. M. In vivo transcription of rDNA spacers in Drosophila. Nucleic Acids Res. 1985 May 10;13(9):3221–3239. doi: 10.1093/nar/13.9.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa H., Cohen P. P. Studies on ribonucleic acid synthesis in tadpole liver during metamorphosis induced by thyroxine. II. Analysis of ribonucleic acid fractions. J Biol Chem. 1967 Feb 25;242(4):642–649. [PubMed] [Google Scholar]
- Nelles L., Fang B. L., Volckaert G., Vandenberghe A., De Wachter R. Nucleotide sequence of a crustacean 18S ribosomal RNA gene and secondary structure of eukaryotic small subunit ribosomal RNAs. Nucleic Acids Res. 1984 Dec 11;12(23):8749–8768. doi: 10.1093/nar/12.23.8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribnow D. Nucleotide sequence of an RNA polymerase binding site at an early T7 promoter. Proc Natl Acad Sci U S A. 1975 Mar;72(3):784–788. doi: 10.1073/pnas.72.3.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt S. C., Reeder R. H. Effect of intercalating agents on RNA polymerase I promoter selection in Xenopus laevis. Mol Cell Biol. 1984 Dec;4(12):2851–2857. doi: 10.1128/mcb.4.12.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder R. H., Roan J. G., Dunaway M. Spacer regulation of Xenopus ribosomal gene transcription: competition in oocytes. Cell. 1983 Dec;35(2 Pt 1):449–456. doi: 10.1016/0092-8674(83)90178-2. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Roberts M. P., Vaughn J. C. Ribosomal RNA sequence conservation and gene number in the larval brine shrimp. Biochim Biophys Acta. 1982 May 31;697(2):148–155. doi: 10.1016/0167-4781(82)90070-7. [DOI] [PubMed] [Google Scholar]
- Rungger D., Achermann H., Crippa M. Transcription of spacer sequences in genes coding for ribosomal RNA in Xenopus cells. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3957–3961. doi: 10.1073/pnas.76.8.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U., Trendelenburg M. F., Krohne G., Franke W. W. Lengths and patterns of transcriptional units in the amplified nucleoli of oocytes of Xenopus laevis. Chromosoma. 1977 Mar 16;60(2):147–167. doi: 10.1007/BF00288462. [DOI] [PubMed] [Google Scholar]
- Simeone A., Boncinelli E. 5'-Cleavage site of D. melanogaster 18 S rRNA. FEBS Lett. 1984 Feb 27;167(2):249–253. doi: 10.1016/0014-5793(84)80136-2. [DOI] [PubMed] [Google Scholar]
- Skinner J. A., Ohrlein A., Grummt I. In vitro mutagenesis and transcriptional analysis of a mouse ribosomal promoter element. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2137–2141. doi: 10.1073/pnas.81.7.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Tower J. Transcription of cloned eukaryotic ribosomal RNA genes. Annu Rev Biochem. 1986;55:801–830. doi: 10.1146/annurev.bi.55.070186.004101. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Wilkinson J. A., Roan J., Reeder R. H. Nested control regions promote Xenopus ribosomal RNA synthesis by RNA polymerase I. Cell. 1983 Nov;35(1):199–206. doi: 10.1016/0092-8674(83)90222-2. [DOI] [PubMed] [Google Scholar]
- Sommerville J. RNA polymerase I promoters and transcription factors. Nature. 1984 Jul 19;310(5974):189–190. doi: 10.1038/310189a0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Southern E. Gel electrophoresis of restriction fragments. Methods Enzymol. 1979;68:152–176. doi: 10.1016/0076-6879(79)68011-4. [DOI] [PubMed] [Google Scholar]
- Tautz D., Dover G. A. Transcription of the tandem array of ribosomal DNA in Drosophila melanogaster does not terminate at any fixed point. EMBO J. 1986 Jun;5(6):1267–1273. doi: 10.1002/j.1460-2075.1986.tb04356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursi D., Vandenberghe A., De Wachter R. The sequence of the 5.8 S ribosomal RNA of the crustacean Artemia salina. With a proposal for a general secondary structure model for 5.8 S ribosomal RNA. Nucleic Acids Res. 1982 Jun 11;10(11):3517–3530. doi: 10.1093/nar/10.11.3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn J. C., Whitman D. J., Bagshaw J. C., Helder J. C. Molecular cloning and characterization of ribosomal RNA genes from the brine shrimp. Biochim Biophys Acta. 1982 May 31;697(2):156–161. doi: 10.1016/0167-4781(82)90071-9. [DOI] [PubMed] [Google Scholar]
- Veldman G. M., Brand R. C., Klootwijk J., Planta R. Some characteristics of processing sites in ribosomal precursor RNA of yeast. Nucleic Acids Res. 1980 Jul 11;8(13):2907–2920. doi: 10.1093/nar/8.13.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman G. M., Klootwijk J., van Heerikhuizen H., Planta R. J. The nucleotide sequence of the intergenic region between the 5.8S and 26S rRNA genes of the yeast ribosomal RNA operon. Possible implications for the interaction between 5.8S and 26S rRNA and the processing of the primary transcript. Nucleic Acids Res. 1981 Oct 10;9(19):4847–4862. doi: 10.1093/nar/9.19.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbeet M. P., Klootwijk J., van Heerikhuizen H., Fontijn R., Vreugdenhil E., Planta R. J. Molecular cloning of the rDNA of Saccharomyces rosei and comparison of its transcription initiation region with that of Saccharomyces carlsbergensis. Gene. 1983 Jul;23(1):53–63. doi: 10.1016/0378-1119(83)90216-0. [DOI] [PubMed] [Google Scholar]
- Wandelt C., Grummt I. Formation of stable preinitiation complexes is a prerequisite for ribosomal DNA transcription in vitro. Nucleic Acids Res. 1983 Jun 11;11(11):3795–3809. doi: 10.1093/nar/11.11.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner A. H., MacRae T. H., Wahba A. J. The use of Artemia salina for developmental studies: preparation of embryos, tRNA, ribosomes and initiation factor 2. Methods Enzymol. 1979;60:298–311. doi: 10.1016/s0076-6879(79)60028-9. [DOI] [PubMed] [Google Scholar]