Abstract
Craniofacial anomalies (CFA) are the most frequent human congenital disease and a major cause of infant mortality and childhood morbidity. Although CFA appear to arise from a combination of genetic factors and environmental influences, the underlying gene defects and pathomechanisms for the majority of CFA are currently unknown. Here we reveal an unknown role for the E3 ubiquitin ligase Wwp2 in regulating craniofacial patterning. Mice deficient for Wwp2 develop malformations of the craniofacial region. Wwp2 is present in cartilage where its expression is controlled by Sox9. Our studies demonstrate that Wwp2 influences craniofacial patterning through its interactions with Goosecoid (Gsc), a paired-like homeobox transcription factor that plays an important role in craniofacial development. We show that Wwp2 associated Gsc is a transcriptional activator of the key cartilage regulatory protein Sox6. Wwp2 interacts with Gsc to facilitate its mono-ubiquitination, a post-translational modification required for optimal transcriptional activation of Gsc. Our results identify the first physiological pathway regulated by Wwp2 in vivo as well as identify a unique non-proteolytic mechanism through which the Wwp2 controls craniofacial development.
Ubiquitin is an important post-translational modifier with profound effects on various aspects of protein biology through altering turnover rate, subcellular localization and/or the activity of target proteins1, 2. Ubiquitin is conjugated to target proteins through an enzymatic process mediated by E3 ubiquitin ligases, like Wwp2. As a member of the Nedd4-family of E3 ligases, Wwp2 contains a N-terminal C2 domain, a WW domain, and a C-terminal catalytic HECT domain3. The HECT domain of Wwp2 contains an active cysteine residue that mediates ubiquitin transfer from E2 enzymes to itself and then to target proteins. Wwp2 can facilitate the conjugation of ubiquitin to a number of proteins in vitro4–6. However, the in vivo function of Wwp2 is currently unknown. This is generally true for most members of mammalian Nedd4 family of E3 ligases since several studies have demonstrated key roles for these proteins in regulating specific signaling pathways in vitro yet there is a dearth of in vivo data to indicate that these ligases regulate specific physiological processes. Therefore, the ability to demonstrate a role for Wwp2 in vivo would aid in emphasizing the biological relevance of the Nedd4 family of E3 ubiquitin ligases.
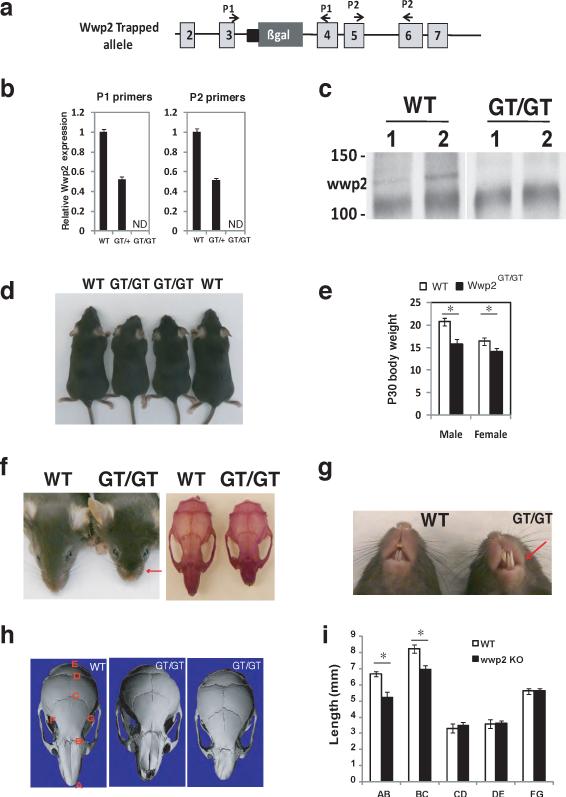
To address what physiological processes Wwp2 regulates, we generated mice deficient in Wwp2. We utilized gene-trap technology to generate Wwp2-null mice using embryonic stem cells that contained the bacterial β-galactosidase (LacZ) gene inserted in intron 3–4 of the Wwp2 locus (Fig. 1a). The insertion of LacZ at this location results in complete ablation of Wwp2 transcript and protein levels in mice homozygous for the gene-trapped allele (Wwp2Gt/Gt) (Fig. 1b–c). Analysis of litters born to male and female mice heterozygous for the Wwp2 gene trap allele (Wwp2Gt/+) revealed that mature Wwp2Gt/Gt mice are runted when compared to age and sex-matched wild-type (WT) controls (Fig. 1d–e). Wwp2Gt/Gt mice display abnormal cranial facial development that is characterized by the presence of a domed skull and a shortened snout (Fig. 1f). While we observed 100% penetrance in the development of craniofacial abnormalities in Wwp2Gt/Gt mice, the severity of the phenotype was variable. A subset of Wwp2Gt/Gt mice developed a more profound craniofacial malformation characterized by a misaligned jaw and a twisted snout (Fig. 1g). Misalignment of the jaw in the Wwp2Gt/Gt mice resulted in the chronic overgrowth of the mandibular incisors (Fig. 1g). To further quantitate the cranial facial abnormalities, we utilized quantitative computed tomography (μ-QCT) to scan the skulls of the Wwp2Gt/Gt and WT mice. The nasal bones of Wwp2Gt/Gt mice were decreased by 20% in length and were twisted by 10° to 25° in those mice with the more severe asymmetric nasal bone phenotype (Fig. 1h–I).
Figure 1. Craniofacial patterning defects are present in Wwp2GT/GT mice.
a) Schematic depicting the position of B-gal insertion into the Wwp2 locus. b) Analysis of Wwp2 transcript levels by qPCR in WT mice (WT), Wwp2GT/+ (GT/+) and Wwp2GT/GT (GT/GT) mice. Values represent means ± s.d. (n=6 for each genotype) c) Immunoprecipitation-Western blot analysis depicting the absence of Wwp2 protein in the Wwp2GT/GT mice. d) Photograph of four-week old male WT and Wwp2GT/GT mice. e) Body weight of male and female WT and Wwp2GT/GT at four-weeks of age. Values represent means ± s.d. (n=6 for each genotype) f) Presence of shortened snout in Wwp2GT/GT mice as shown grossly (left) as well as by alizarin red staining of WT and Wwp2GT/GT skulls (right). g) Photograph depicting misaligned jaw and overgrown of mandibular incisor of Wwp2GT/GT (GT/GT) mice. h) μCT analysis of skulls from WT mice as well as Wwp2GT/GT mice depicting the shortened (middle) or twisted nasal bone (right) in the Wwp2GT/GT mice. i) Quantitative analysis of the distances between the various landmarks in the skulls of four-week old Wt and Wwp2GT/GT mice. Values represent means ± s.d. (n=8 for each genotype) (*p<0.001). Uncropped images of blots are shown in Supplementary Fig.S7.
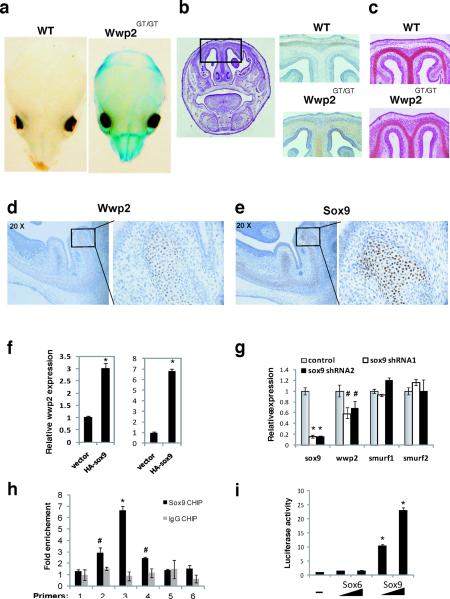
The Wwp2 gene-trap allele allows LacZ to be expressed from the endogenous Wwp2 locus (Fig. 1a). Thus LacZ can be used as a surrogate marker to track Wwp2 expression in vivo. Staining of whole-mount skulls isolated from WT and Wwp2Gt/Gt neonatal mice revealed LacZ to be present in the nasofrontal region of the skull consistent with the type of craniofacial abnormalities observed in Wwp2Gt/Gt mice (Fig.2a). Additional immunostaining analysis of WT and Wwp2Gt/Gt skulls using an anti-X-gal antibody revealed specific LacZ staining patterns in regions of the skull that also positive for Safranin O, a stain used to detect cartilage proteoglycans (Fig. 2b–c). Serial histological sections of Wt skull were analyzed by in situ hybridization for the expression of Wwp2 and immunostaining for Sox9. Wwp2 could be detected in the cartilaginous regions of the skull at various stages of development that were also positive for Sox9 demonstrating that Wwp2 expression in cartilage is necessary for proper patterning of the craniofacial region of the skull (Fig. 2d–e, and Supplementary information, Fig.S1).
Figure 2. Regulation of Wwp2 expression in the skull.
a) Whole mount staining for β-gal in skulls of WT and Wwp2GT/GT mice. b) Detection of β-gal in histological sections of the skull of Wwp2GT/GT mice but not WT mice via immunostaining. Scale bar, 0.5mm c) Safranin O staining identifies cartilaginous regions of the cranial sections. Scale bar, 0.5mm. Analysis of Wwp2 (d) and Sox9 (e) in the skull of WT mice by in situ hybridization and immunostaining, respectively. Scale bar, 0.5mm. Higher magnification of boxed area in both d) and e) is displayed to right. f) Wwp2 transcript levels were measured by qPCR in ATDC5 cells (left) and C3H10T1/2 cells (right) following infection of cells with control or HA-tagged Sox9-expressing lentivirus. Values represent means ± s.d. (n=3) g) Levels of Sox9, Wwp2, Smurf1 and Smurf2 were evaluated by qPCR following infection of ATDC5 cells with a control lenvtivirus or lentivirus containing Sox9-specific shRNAs. Values represent means ± s.d. (n=3) h) Six separate primer sets (1–6) were used in qPCR reactions to scan the Wwp2 intron region that was bound by Sox9 in chromatin immunoprecipitation experiments in C3H10T1/2 cells that utilized anti-Sox9 (black bars) or IgG control (grey bars) antibody. Values represent means ± s.d. (n=3) i) Increasing amounts of Sox9 but not Sox6 lead to induction of luciferase levels in C3H10T1/2 cells transfected with a luciferase reporter construct that contains the region surrounding the Sox9 binding site in the Wwp2 intron. Values represent means ± s.d. (n=3) (*p<0.01, #p<0.05 compared to empty vector control).
Based on the restricted pattern of Wwp2 expression in cartilage, we hypothesized that Wwp2 might be regulated transcriptionally by Sox9. We therefore analyzed Wwp2 transcript levels following ectopic expression of Sox9 in two murine cell lines. A marked elevation in Wwp2 expression was observed in the chondrogenic ATDC5 cell line as well as the mesenchymal C3H10T1/2 cell line following Sox9 over expression (Fig. 2f). We observed that reduction in endogenous Sox9 levels in ATDC5 cells by Sox9-specific shRNAs also led to reductions in Wwp2 levels (Fig. 2g). No changes in the expression of the Nedd4-like E3 ligases, Smurf1 and Smurf2, were observed in cells containing the Sox9-specific shRNA constructs (Fig., 2g). We next performed chromatin immunoprecipitation using an anti-Sox9 antibody to determine if Sox9 could bind to the Wwp2 promoter. Scanning both the promoter and intronic regions of the Wwp2 gene for Sox9-binding sites, we identified a peak representing Sox9 binding an intronic region of Wwp2 between exons 4 and 5 (Supplementary Information, Fig. S2). A control IgG antibody showed no significant enrichment of this segment indicating that Sox9 is specifically bound at this genomic region (Fig. 2h). We next generated a luciferase reporter construct in which the promoter contains the 800 base pair fragment of the Wwp2 intron containing the Sox9-binding region. Co-transfection of Sox9 with this reporter construct resulted in marked elevation in luciferase activity (Fig. 2i). Interestingly, the regulation of Wwp2 via this intronic element appears to be specific for Sox9 as ectopic expression of Sox6, another Sry-box containing protein, did not activate this reporter construct (Fig.2i). Collectively, these data suggest that Sox9 regulates the cartilaginous-expression of Wwp2.
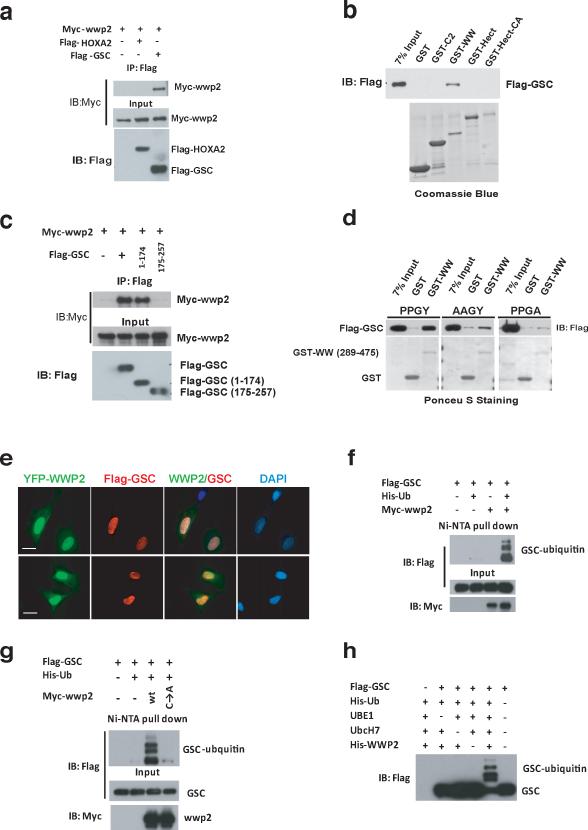
Since Wwp2 is an E3 ligase it likely regulates craniofacial patterning through ubiquitination of specific target proteins. The WW domain of Wwp2 mediates substrate interaction through the binding of a PPxY motif in target proteins. We therefore sought to determine if proteins known to influence craniofacial patterning contain a PPxY motif. From this analysis, we identified this motif in three proteins reported to regulate craniofacial development: Gsc7–10, Hand2 and HoxA211, 12. We questioned if Wwp2 could interact with and promote the ubiquitination of these proteins. Immunoprecipitation of myc-epitope tagged Wwp2 followed by Western blot revealed that Wwp2 interacted with the homeobox protein Gsc but not another homeobox protein HoxA2 (Fig. 3a and Supplementary Information, Fig. S3). We further established that Gsc and Wwp2 could physically interact through in vitro interaction assays utilizing recombinant fragments of these proteins. As shown in Fig. 3b, we could detected a specific interaction between Gsc and the WW domain of Wwp2. Additional analyses revealed that Wwp2 associated with Gsc through an N-terminal fragment of Gsc that contains the PPxY motif (Fig. 3c). Given that the PPxY motif in Gsc is conserved across multiple species (Supplementary Information, Fig. S3), we investigated if this motif is required for its interaction with Wwp2. Mutation of the PPGY motif in Gsc to AAGY or PPGA abolished its interaction with the WW domain of Wwp2 (Fig. 3d). Finally, analysis of the subcellular distribution of Wwp2 and Gsc via immunofluoresence in ATDC5 cells revealed a nuclear co-localization of these proteins suggesting that Wwp2 could control craniofacial development in part through its ubiquitination of Gsc (Fig.3e).
Figure 3. Gsc interacts with and is ubiquitinated by Wwp2.
a) Co-immunoprecipitation experiments were conducted in 293T cells transfected with myc-Wwp2 and flag-Gsc or flag-HoxA2 expression constructs. Wwp2 was immunoprecipitated from cell lysates with anti-flag antibody, followed by Western blot analysis with anti-myc antibody. b) Purified recombinant fragments of Wwp2 were used for in vitro interaction analysis. Western blot analysis with an anti-flag antibody was used to detect Gsc interaction with the various fragments of Wwp2. c) Co-immunoprecipitation experiments were conducted in 293T cells with flag-epitope tagged Gsc deletion mutants and myc-Wwp2 as described in (a). d) Interaction analysis between recombinant WW domain fragments of Wwp2 and wild-type Gsc (PPGY) or Gsc bearing a AAGY or PPGA mutation were analyzed via GST-pull down followed by Western blot analysis. e) Immunofluoresence analysis of ATDC5 chondrocyte cell line transfected with YFP-tagged Wwp2 expression construct and a flag-epitope tagged Gsc expression construct reveals colocalization of these proteins in the nucleus. Scale bar, 10μm f) Analysis of Gsc ubiquitination was performed in 293T cells transfected with flag-Gsc, myc-Wwp2 and His-epitope-tagged ubiquitin. Ubiquitinated flag-Gsc proteins were detected in cells by precipitating ubiquitinated proteins from denatured lysates with Ni-NTA-agarose, followed by Western blot analysis with anti-flag antibody. g) Additional Gsc ubiquitination experiments were conducted as described in f) that utilized either WT Wwp2 or functionally inert Wwp2 harboring a cysteine to alanine mutation. h) In vitro ubiquitination of Gsc was performed using flag-Gsc purified from 293T cells transfected with flag-Gsc. Flag-Gsc was incubated with recombinant Wwp2, His-ubiquitin, UBE1, Ubch7. Reactions were then subjected to Western blot analysis with anti-Flag antibody to detect Gsc proteins. Uncropped images of blots are shown in Supplementary Fig.S7.
To determine if the interaction between these two proteins results in the ubiquitination of Gsc, we transfected 293T cells with His-ubiquitin, Flag-Gsc and Myc-Wwp2. Western blot analysis of the purified ubiquitinated proteins using an anti-Flag antibody revealed that co-transfection of Wwp2 resulted in ubiquitination of Gsc whereas no ubiquitination of Gsc was observed in the absence of Wwp2 (Fig. 3f). A functionally inert Wwp2, in which the active cysteine residue was mutated to alanine, was unable to promote ubiquitination of Gsc. (Fig. 3g). To establish that Wwp2 directly ubiquitinates Gsc via in vitro ubiquitination reactions, we utilized purified Gsc and recombinant Wwp2 as well as the recombinant ubiquitin-activating E1 enzyme UBE1 and recombinant E2 ubiquitin ligase UbcH7. Significant ubiquitination of Gsc was observed during in vitro reactions containing Wwp2, E1 and E2 enzymes (Fig. 3h). Conversely, Gsc was unable to be ubiquitinated in these in vitro reactions lacking any one component (Fig. 3h). These data demonstrate that Wwp2 ubquitinates Gsc through a direct association with this protein.
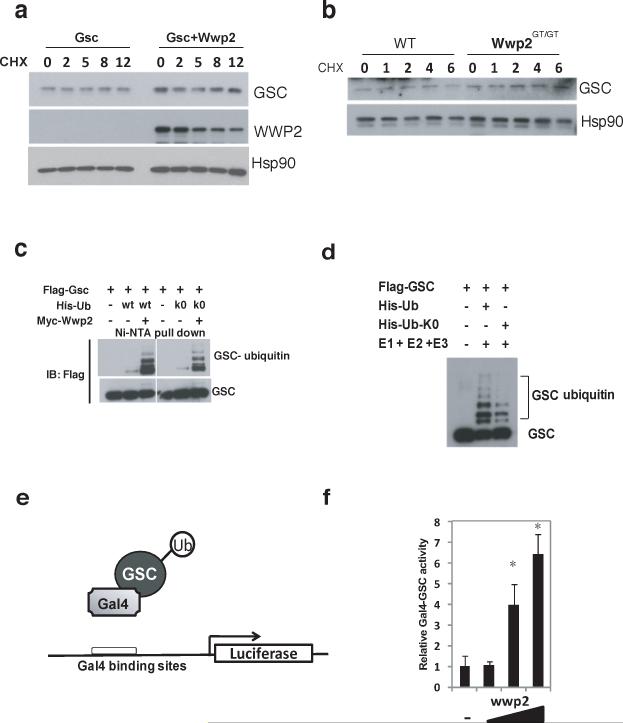
Ubiquitination of a protein generally results in proteasome-mediated degradation. We therefore evaluated Gsc protein levels following cycloheximide treatment to determine if Wwp2 augments the turnover of Gsc. Interestingly, co-expression of Wwp2 with Gsc in 293T cells did not alter Gsc protein levels (Fig. 4a). Furthermore, the levels of Gsc protein were similar in cells isolated from the nasal cartilage of Wt and Wwp2Gt/Gt mice (Fig. 4b). These findings led us to pursue the possibility that Gsc was being mono-ubiquitinated by Wwp2, a posttranslational modification that does not change protein levels. To address this, we analyzed Wwp2-mediated ubiquitination of Gsc in the presence of a His-ubiquitin construct in which all seven lysines were mutated to arginines (Ub-K0). The conjugation of Ub-K0 to a target protein prevents the formation of poly-ubiquitin chains allowing for proteins to only be mono-ubiquitinated. Interestingly, the ubiquitination pattern of Gsc was identical whether cells were transfected with Wwp2 and the K0-ubiquitin expressing construct or the Wt ubiquitin construct demonstrating that Gsc is monoubiquitinated at multiple sites (Fig. 4c). We confirmed that Gsc was being mono-ubiquitinated by Wwp2 via in vitro ubiquitination assays that utilized purified Gsc, recombinant Wwp2 and ubiquitin-K0 proteins that resulted in a pattern of Gsc ubiquitination similar to WT ubiquitin (Fig 4d).
Figure 4. Monoubiquitination augments transcriptional activity of Gsc.
a) Western blot analysis of Gsc and Wwp2 protein levels in cell lysates generated from 293T cells that were transfected with either a Gsc-expression construct or a Gsc-expression constuct and a Wwp2-expression construct. Transfected cells were treated with cyclohexamide (CHX) for 2, 5, 8 and 12 hours prior to cell lysis. b) Endogenous Gsc protein levels were assessed in lysates generated from nasal cartilage cells isolated from WT and Wwp2GT/GT mice by Western blot analysis using anti-Gsc antibody. c) Ubiquitination of Gsc was evaluated in 293T cells transfected with expression constructs of flag-Gsc, myc-Wwp2 and either His-wild type ubiquitin or His-K0-ubiquitin. Ubiquitinated flag-Gsc proteins were detected as described above d) Analysis of Gsc ubiquitination was also assessed using recombinant his-wild type ubiquitin or his-K0-ubiquitin through in vitro ubiquitination assays similar to those described above. e) Schema depicting the Gsc-gal4 fusion protein and reporter construct used in subsequent luciferase experiments. f) Luciferase levels were evaluated following transfection of Gsc-gal4 expresson construct, Gal4-luciferase reporter and increasing amounts of Wwp2 expression construct. Values represent means ± s.d. (n=3) (*p<0.01 compared to empty vector control). Uncropped images of blots are shown in Supplementary Fig.S7.
Mono-ubiquitination of transcription factors has been demonstrated to alter their ability to activate gene transcription. We investigated if Wwp2-mediated mono-ubiquitination alters the transcriptional activity of Gsc. Regulation of transcription factor activity by ubiquitination has been suggested to be promoter-specific. Therefore, we initially performed gene reporter assays where Wwp2 was co-transfected in cells with Gsc fused to a GAL4 DNA-binding domain along with a GAL3-repsonsive luciferase reporter (Fig. 4e). As shown in Fig 4f, co-transfection of increasing amounts of Wwp2 led to increased Gsc-driven luciferase reporter expression in a dose-dependent manner. These results indicated that Wwp2 augments the transcription activity of Gsc.
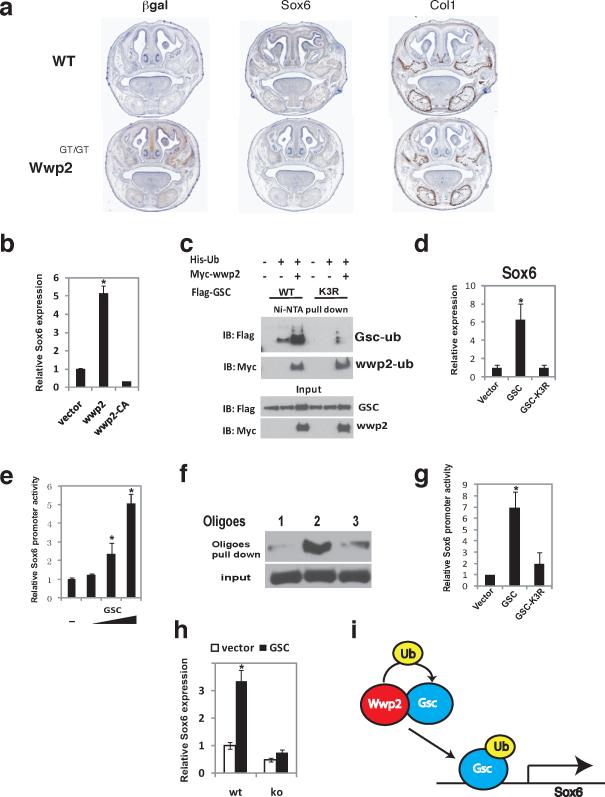
Analysis of Wwp2 and Gsc expression via in situ hybridization of E18.5 embryos revealed that these two genes were co-expressed in several tissues including the nasal cartilage (Supplementary information, Fig.S4). To identify genes regulated by Wwp2-monoubiquitinated Gsc that may influence craniofacial patterning, we performed gene expression analysis of cells isolated from the nasal cartilage of Wt and Wwp2GT/GT neonatal mice. A number of candidate genes implicated in craniofacial patterning were found to be dysregulated in Wwp2-null cells. To establish which of these genes is directly regulated by Wwp2, we examined their expression in nasal cartilage cells infected with control or Wwp2-expressing lentivirus (Supplementary Information, Fig. S4). One gene of particular interest identified was Sox6, a factor involved in cartilage biology13, 14. Evaluation of Sox6 levels via in situ hybridization of Wt and Wwp2GT/GT skulls revealed decreased Sox6 levels in Wwp2GT/GT mice, confirming our in vitro observations (Fig. 5a). As control, the levels of the osteoblast marker Col1a1 were similar between the Wt and Wwp2GT/GT sections (Fig. 5a). Infection of both Wt and Wwp2-null cells with Wwp2-expressing lentivirus resulted in elevated Sox6 levels indicating that Wwp2 can influence Sox6 expression (Fig 5b). Interestingly, Sox6 expression was not induced when cells were infected with a lentivirus expressing a mutant of Wwp2 that is unable to ubiquitinate target proteins (Fig. 5b).
Figure 5. Ubiquitination of Gsc by Wwp2 is required for optimal expression of Sox6.
a) Coronal sections of WT and Wwp2GT/GT skulls were evaluated for expression of β-gal via immunostaining and Sox6 and Col1a1 via in situ hybridization. Scale bar, 1mm b) Sox6 mRNA levels were analyzed by qPCR in WT nasal cartilage cells transduced with a control lentivirus, Wwp2-expressing lentivirus or lentivirus expressing Wwp2 with a non-functional Hect domain (Wwp2-CA). Values represent means ± s.d. (n=3, *p<0.01). c) 293T cells were transfected with expression constructs for flag-myc-Wwp2, His-ubiquitin and flag-tagged Gsc or flag-tagged Gsc-K3R, a Gsc protein with the three lysines mutated to arginine. Ubiquitination of the wild-type Gsc and mutant Gsc-K3R were analysed by immunoprecipitation and Western blot analysis d) wild-type nasal cartilage cells were infected with lentivirus expressing wild-type Gsc or mutant Gsc-K3R or control lentivirus. Transcript levels of Sox6 in these cell populations were then evaluated by qPCR Values represent means ± s.d. (n=3, *p<0.01). e) 293T cells were transfected with Sox6pro-luc reporter construct and increasing amounts of a Gsc-expression construct. Results were normalized to the expression of the pRL-Tk plasmid Values represent means ± s.d. (n=3, (*p<0.01). f) Gsc DNA binding to the Sox6 promoter was determined through nuclear extracts generated from nasal cartilage cells. Gsc binding to region −235 to −185 (probe #1), −187 to −136 (probe #2) and −136 to −87 (probe #3) was detected by Western blotting following oligo pulldown experiments. g) Analysis of Gsc-K3R mutant's ability to transactivate Sox6pro-luc was evaluated posttransfection of 293T cells and was compared to cells transfected with the Sox6pro-luc and construct expressing wild-type Gsc. Levels of luciferase in these experiments were normalized to the expression of the pRL-Tk plasmid. Values represent means ± s.d. (n=3, *p<0.01). h) Sox6 transcript levels were analyzed by qPCR in nasal cartilage cells from WT or Wwp2GT/GT mice infected with control or Gsc-expressing lentivirus. Values represent means ± s.d. (n=3, *p<0.01) i) Model depicting the mechanism through which Wwp2-mediated monoubiquitination of Gsc leads to augmented Sox6 expression. Uncropped images of blots are shown in Supplementary Fig.S7.
The above results suggested that Wwp2 augments expression of Sox6 through its function as an E3 ligase. We sought to determine if Wwp2-mediated mono-ubiquitination of Gsc is required for optimal expression of Sox6. We explored if Gsc could regulate the expression of Sox6. Indeed, infection of both the ATDC5 and C3H10T1/2 cell with Gsc-expressing lentivirus led to increased Sox6 transcript levels (Supplementary Information, Fig. S5). To determine if ubiquitination of Gsc is important for its transcriptional control of Sox6, we generated a series of Gsc protein mutants where each individual lysine residue as well as different combination of lysine residues was mutated to arginine. Ubiquitin is conjugated to the lysine ε-amino group of the targeted protein, therefore the mutation of the critical lysine residues in Gsc should result in hypo-ubiquitination of this proteins. From this analysis, we identified three individual lysine residues within Gsc (K219R, K231R, K234R) that when mutated to arginine (Gsc-K3R) led to a dramatic reduction in the level of Wwp2-mediated ubiquitination (Fig. 5c). While these mutations in Gsc affected its ability to be ubiquitinated, they did not alter protein levels (Fig. 5c). We next examined if the reduced ubiquitination of Gsc-K3R correlated with a decrease in transcriptional activity. For this, we infected primary cartilage cells with lentivirus expressing Wt-Gsc or the Gsc-K3R mutant. A marked reduction in the expression of Sox6 levels was observed in those cells transduced with the Gsc-K3R lentivirus when compared to cells infected with the Wt-Gsc (Fig. 5d).
To confirm that Gsc directly regulates Sox6 expression, we attempted to identify the region of the Sox6 promoter that is bound by Gsc. Transfection of a Gsc-expression construct led to a dose-dependent increase in the expression of reporter construct that contained a 2.4kb region of the Sox6 promoter (Fig. 5e). To identify Gsc binding sites in the Sox6 promoter, a series of luciferase-reporters containing truncated fragments of the Sox6 promoter were generated and retested. We found that Gsc expression had no effect on the (−136)Sox6pro-luc whereas Gsc could induce expression of the (−187)Sox6pro-luc construct (Supplementary Information, Fig. S6). We confirmed that Gsc could interact with this region of DNA through oligonucletide binding assays utilizing the −187 to −136 region of the Sox6 promoter (Fig. 5f). As a control, the region (−235 to −185) and (−136 to −87) of the Sox6 promoter showed a marked reduction in the ability to bind Gsc (Fig. 5f). The Gsc-K3R mutant was significantly impaired in its ability to induce the expression of these reporter constructs (Fig. 5g). Lastly, we asked if Gsc could induce Sox6 expression in the absence of Wwp2 by analyzing Sox6 expression levels in nasal cartilage cells isolated from WT or Wwp2GT/GT mice transduced with control lentivirus or a Gsc-expressing lentivirus. Infection of WT nasal cartilage cells with Gsc-expressing lentivirus led to a marked increased in Sox6 expression (Fig. 5h). Strikingly, infection of Wwp2GT/GT nasal cartilage cells with Gsc-expressing lentivirus failed to induce Sox6 expression (Fig. 5h).
Previous reports demonstrate that Sox9 is required for Sox6 expression in vivo and that the co-expression of these two proteins function cooperatively to regulate target genes within chondrocytes15. We sought to address how Wwp2's regulation of Sox6 downstream of Sox9 fit into this cooperative model. Knockdown of endogenous Sox9 via Sox9-sepcific shRNA resulted in significant reductions in Sox6 mRNA levels in WT and Wwp2-deficient cells (Supplementary information, Fig.S6). These data suggests that Sox9 regulates Sox6 expression in part through a Wwp2-indepent pathway and may explain why the absence of Wwp2 in vivo results in levels of Sox6 that are diminished but not abolished. However, we also observed that the induction of Sox6 in Wwp2-deficient cells following ectopic expression of Sox9 is markedly reduced compared to the Sox6 levels that are induced following ectopic expression of Sox9 in Wt cells (Supplementary information, Fig.S6). Collectively, we interpret these data to suggest that Wwp2 functions downstream of Sox9 during patterning of the craniofacial region to “fine-tune” the temporo-spacial expression of Sox6. Wwp2 achieves this regulation through a unique non-proteolytic mechanism that is required for optimal activity of Gsc transcription factor during craniofacial development.
MATERIALS AND METHODS
Generation of Wwp2GT/GT mice
ES cell clone HMA140 containing the bacterial β-galactosidase (LacZ) gene between Intron 3 and 4 of the Wwp2 gene locus was obtained from BayGenomics. Confirmed ES cells were injected into blastocysts to achieve initial germ-line transmission. Genotyping of founders as well as subsequent offspring was performed on tail clippings using primers flanking the position of the LacZ insertion coupled with primers that amplified a portion of the LacZ insertion. Wwp2GT/+ pups were backcrossed to C57BL/6 mice for 5 generations prior to analysis. Mice analyzed in all studies were sex and age matched. All mice were bred and housed in the pathogen-free facility at Harvard School of Public Health and were handled in accordance with guidelines from the Center for Animal Resources and Comparative Medicine (ARCM) at Harvard Medical School.
Cell culture, transfection and infection
HEK293T were maintained in DMEM supplemented with glutamine, penicillin/streptomycin and 10% FBS. HEK293T cells were transiently transfected using Effectene reagent according to the manufacturer's instructions (Roche, Basel, Switzerland). The murine mesenchymal stem cell line C3H10T1/2 was maintained in DMEM (Mediatech, Inc.) and 10%FBS. Nasal cartilage cells were enzymatically released from nasal cartilaginous nodules of neonatal WT and Wwp2GT/GT littermates and were plated in αMEM+10% FBS in 6-well dish. Cells were harvested at a subconfluent stage and replated in a 6-well dish for lentivirus infection. Forty-eight hours before harvest, 2μg/ml puromycin was added to the cultures to select infected cells. Lentiviral vectors harboring Sox9 specific shRNAs were obtained from the TRC shRNA library (http://www.broadinstitute.org/rnai/trc/lib) and targeted the following target sequences: Sox9 shRNA1: CGTCAATGAGTTTGACCAATA, Sox9 shRNA2: CAGACTCACATCTCTCCTAAT
Plasmids, oligonucleotides and recombinant proteins
p6xMyc-Wwp2 was made by inserting a EcoRI/XbaI (5'-ttttgaattcaagcttatggcatctgccagctctagccgggcag 3' 5'-ctatdtctagaggcagggaggactcaggactacatg 3') digested polymerase chain reaction product into digested p6xMyc vector. pFlag-CMV2-Gsc was made by inserting a hindIII/XbaI (5'-atagtctaagcttatgcccgccagcatgttcagcatcga 3', 5'-ctattctagactgtgcaagtccttcgagttagg 3') digested PCR products into pFlag-CMV2 vector. Hect domain mutation of Wwp2 and lysine mutations of Gsc were made by using a quickChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. His-ubiquitin (Ub) expression vector was kindly provided by Dr. Dong-Er Zhang (Scripps Institute), His-Ub K0 mutation expression vector, which expresses ubiquitin with all lysine residues mutated to arginine residues, was provided by Dr. Wei Gu. The Sox9 response element in the Wwp2 intron was cloned from mouse genomic DNA using the primers, (5'-tttggtaccttccataagagagctcctgaagc 3'and 5'-ttttaagcttcctagtgctggagagagaagc 3'). PCR products were digested by using KpnI/HindIII enzyme and cloned into the KpnI/HindIII digested luciferase report vector pGL3-Basic (Promega).
Statistical methods
For statistcal analysis, mean values with standard deviation (s.d.) are depicted for most graphs that were generated from multiple independently obtained data sets. P-values were obtained from t-test with paired or unpaired samples with significance set at P-values <0.05.
Chromatin Immunoprecipitation (ChIP)
The ChIP assay was performed as described previously16. Briefly, cells were crosslinked with 1% formaldehyde (vol/vol) for 10 min at room temperature, and formaldehyde was then inactivated by the addition of glycine to the final concentration of 125 mM. Chromatin extracts containing DNA fragments with an average size of 500 bp were immunoprecipitated using rabbit anti-Sox9 polyclonal antibody (Millipore, AB5535) or rabbit IgG polyclonal antibody (PP64B). 5μg of antibody was utilized for each immunoprecipitation. Relative occupancy values were calculated by determining the apparent immunoprecipitation efficiency (ratios of the amount of immunoprecipitated DNA to that of the input sample) and normalized to the level observed at a control region, which was defined as 1. ChIP experiments were repeated three times. For all the primers used, each gave a single product of the right size, as confirmed by dissociation curve analysis. These primers also gave no DNA product in the no-template control. The primers sets used for real-time PCR to quantitate the ChIP enriched DNA are listed in the Supplementary information, Table 1 online.
Luciferase assays
Cells were transfected with a luciferase reporter gene plasmid and different combinations of expression constructs, as indicated, using the Effectene transfection reagent (Qiagen). Total amounts of transfected DNA were kept constant by supplementing with control empty expression vector plasmids as needed. All cells were cotransfected with pRL-TK (Promega) as a normalization control for transfection efficiency. Forty-eight hours after transfection, cells were harvested and lysed in 1×Passive Lysis Buffer (Promega). Luciferase assays were performed using the Dual-Luciferase Reporter Assay System (Promega).
Ni-NTA-agarose purification
Forty-eight hours post transfection, cells were washed with PBS and were lysed in 8 M urea containing 0.5% triton and 10 mM imidazole. Ni-NTA agarose beads (20 μl) (Qiagen) were then added to cell extracts (500 mg) and rotated at room temperature (RT) for 4 h. Precipitates were washed three times with lysis buffer and then boiled in 1× Laemmli sample buffer.
Immunoprecipitation (IP) and western blot analyses
Forty-eight hours posttransfection, cells were lysed in modified RIPA buffer (50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1% NP-40, 0.25% Deoxycholate, 0.1% SDS). Immunocomplexes were precipitated with a mixture of Protein-A and -G-agarose (Amersham Biosciences, Piscataway, NJ). Immunoprecipitates were washed with the same buffer three times and boiled in SDS-PAGE sample buffer. Antibodies against Flag (Sigma) and Myc (Santa cruz, CA) were purchased from the respective manufacturers and used at a 1:2000 dilution for Western blot. Rabbit anti-mouse wwp2 polyclonal antibodies were produced by immunizing rabbits with wwp2 peptides. For Western blot analysis, antibody for Wwp2 was used at 1:1000 dilution. Western blot analysis was performed under standard conditions using the indicated antibodies.
Histology and Immunostaining
Generation and preparation of skeletal tissue for histological analysis and in situ hybridization were performed as described previously17. Immunohistochemistry was performed using TSA-biotin amplification system (Perkin Elmer Life Sciences) according to manufacture's instructions utilizing antibodies against Sox9 (Millipore) and β-gal (Abcam). Antibodies were used at a 1:2000 dilution for immunostaining. In situ probes for Wwp2, Gsc and Sox6 were generated by PCR utilizing the following primer sets: Wwp2-5:TACGCCATCGAGGAGACTGAGG, Wwp2-3-T7: TTGTAATACGACTCACTATAGGGAGCTGGCGCAAGTGTTAAGCAG, Sox6-5: CATGTCCAACCAGGAGAAGCAACC, Sox6-3-T7: TTGTAATACGACTCACTATAGGGGACTCATAAGACGGCGATGG, GSC-5: ACCACTTGGAGACTTCTTCTTCTTCG, mGSC-3-T7: TTGTAATACGACTCACTATAGGGTTTTTCATGGCTGGTCTACATTGC. Collagen I was kindly provided by Dr. Beate Lanske. Digoxigenine (DIG)-labelled RNA probes were then generated by T7 or T3 RNA polymerase according to manufacturer's protocol.
Oligonucleotide Binding Assays
HEK 293T cells were pelted and nuclear extracts generated 48hrs after transfection with a Flag-Gsc expression construct. Nuclear protein was incubated with biotinylated double-stranded oligoes plus streptavidin-agarose for 1 hour at 4 degrees C in Binding buffer (100 mM NaCl, 10 mM Tris-Hcl pH 7.6, 0.1 mM EDTA, 1mM DTT, 5% glycerol, 1 mg/ml BSA, 20 μg/ml poly dI/dC plus protease inhibitors. Beads were washed 3 times in Binding buffer, and bound proteins were resolved by SDSPAGE followed by immunoblotting for overexpressed Flag-Gsc. The oligoes utilized in these experiments were synthesized from Sigma: Biotin-235-185: gcccagccagcaacccagtggagagaatgccccttccccccgcccaccag, 235-185-R: ctggtgggcggggggaaggggcattctctccactgggttgctggctgggc, Biotin-2 (187-136): agggtctcctctcttctgtcctttgtagtcacatgtccgggttttccccctg, 187-136: cagggggaaaacccggacatgtgactacaaaggacagaagagaggagaccct, Biotin-136-87: ggtccgagaacagacacagccccgcccagtctatttctagaaggtgggtt, 136-87-R: aacccaccttctagaaatagactgggcggggctgtgtctgttctcggacc.
μCT Analysis of the skull
Skulls isolated from age and sex-matched WT and Wwp2GT/GT mice were fixed in 70% ethanol and scanned using a Scanco μCT-35 (Scanco Medical AG) with a spatial resolution of 20 μm. The scans were reconstructed into two-dimensional (2D) coronal slices. From these slices, the following 8 different landmarks were identified by examining the slices individually: a) most distal point on the nasal bone, b) midpoint of the frontal-nasal suture, c) midpoint of the frontal-parietal suture, d) midpoint of the parietal-intraparietal suture, e) midpoint of the intraparietal-occipital suture, f) most distal point on right parietal bone, g) most distal point on left parietal bone, h) end of palatine process. These landmarks were further defined by three-dimensional (3D) coordinates. The straight-line distance between the points was calculated using the three-dimensional distance formula. To quantify the curvature of the nasal bone, the angle between point A and the plane defined by points B, C, and H was calculated.
Supplementary Material
Acknowledgements
The authors thank Dr. Dong-Er Zhang, Dr. Wei Gu, Dr. Tatsuya Kobayashi, Dr. Nakamura Yukio and Dr. Jian Xu for kindly providing reagents. We also thank Dr. Marc Wein for thoughtful comments on the manuscript. Supported by NIH grants HD055601 (LHG), K99AR055668 (DJ) and a grant from Merck Pharmaceuticals. WGZ is a Yerby Fellow at the Harvard School of Public Health.
Footnotes
Competing Financial Interest: L.H.G. holds equity in and is on the corporate board of directors of Bristol-Myers Squibb.
References
- 1.Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 2.Dhananjayan SC, Ismail A, Nawaz Z. Ubiquitin and control of transcription. Essays Biochem. 2005;41:69–80. doi: 10.1042/EB0410069. [DOI] [PubMed] [Google Scholar]
- 3.Mackie EJ, Ahmed YA, Tatarczuch L, Chen KS, Mirams M. Endochondral ossification: how cartilage is converted into bone in the developing skeleton. Int J Biochem Cell Biol. 2008;40:46–62. doi: 10.1016/j.biocel.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 4.Li H, et al. Wwp2-mediated ubiquitination of the RNA polymerase II large subunit in mouse embryonic pluripotent stem cells. Mol Cell Biol. 2007;27:5296–5305. doi: 10.1128/MCB.01667-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McDonald FJ, et al. Ubiquitin-protein ligase WWP2 binds to and downregulates the epithelial Na(+) channel. Am J Physiol Renal Physiol. 2002;283:F431–436. doi: 10.1152/ajprenal.00080.2002. [DOI] [PubMed] [Google Scholar]
- 6.Xu HM, et al. Wwp2, an E3 ubiquitin ligase that targets transcription factor Oct-4 for ubiquitination. J Biol Chem. 2004;279:23495–23503. doi: 10.1074/jbc.M400516200. [DOI] [PubMed] [Google Scholar]
- 7.Latinkic BV, Smith JC. Goosecoid and mix.1 repress Brachyury expression and are required for head formation in Xenopus. Development. 1999;126:1769–1779. doi: 10.1242/dev.126.8.1769. [DOI] [PubMed] [Google Scholar]
- 8.Rivera-Perez JA, Mallo M, Gendron-Maguire M, Gridley T, Behringer RR. Goosecoid is not an essential component of the mouse gastrula organizer but is required for craniofacial and rib development. Development. 1995;121:3005–3012. doi: 10.1242/dev.121.9.3005. [DOI] [PubMed] [Google Scholar]
- 9.Rivera-Perez JA, Wakamiya M, Behringer RR. Goosecoid acts cell autonomously in mesenchyme-derived tissues during craniofacial development. Development. 1999;126:3811–3821. doi: 10.1242/dev.126.17.3811. [DOI] [PubMed] [Google Scholar]
- 10.Yamada G, et al. Targeted mutation of the murine goosecoid gene results in craniofacial defects and neonatal death. Development. 1995;121:2917–2922. doi: 10.1242/dev.121.9.2917. [DOI] [PubMed] [Google Scholar]
- 11.Gendron-Maguire M, Mallo M, Zhang M, Gridley T. Hoxa-2 mutant mice exhibit homeotic transformation of skeletal elements derived from cranial neural crest. Cell. 1993;75:1317–1331. doi: 10.1016/0092-8674(93)90619-2. [DOI] [PubMed] [Google Scholar]
- 12.Xiong W, et al. Hand2 is required in the epithelium for palatogenesis in mice. Dev Biol. 2009;330:131–141. doi: 10.1016/j.ydbio.2009.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lefebvre V. Toward understanding the functions of the two highly related Sox5 and Sox6 genes. J Bone Miner Metab. 2002;20:121–130. doi: 10.1007/s007740200017. [DOI] [PubMed] [Google Scholar]
- 14.Smits P, et al. The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev Cell. 2001;1:277–290. doi: 10.1016/s1534-5807(01)00003-x. [DOI] [PubMed] [Google Scholar]
- 15.Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;16:2813–2828. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 17.Shim JH, et al. TAK1 is an essential regulator of BMP signalling in cartilage. EMBO J. 2009;28:2028–2041. doi: 10.1038/emboj.2009.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.