Abstract
Purpose
MRI-guided prostate needle biopsy requires compensation for organ motion between target planning and needle placement.
Methods
We propose slice-to-volume registration algorithms for tracking the prostate motion. Three orthogonal intra-operative slices are acquired in the approximate center of the prostate and registered with a high-resolution target planning volume. Both rigid and deformable scenarios were implemented. MRI-guided robotic prostate biopsy cases were analyzed retrospectively.
Results
Average registration errors were 2.55mm for the rigid algorithm and 2.05mm for the deformable algorithm.
Conclusion
Slice-based tracking appears to be promising. Deformable registration does not seem warranted.
Keywords: Biopsy, prostate, motion tracking, slice-to-volume registration
I. Introduction
Prostate cancer continues to be a worldwide health problem and the most common type of cancer among men [1]. Ultrasound guided core needle biopsy is the gold standard in achieving a positive diagnosis. As ultrasound lacks in both sensitivity and specificity, Magnetic Resonance Imaging (MRI) is increasingly becoming the modality of choice in prostate biopsy. During a prostate biopsy procedure, the prostate moves and deforms between target planning and needle insertion and retraction. Confirming clinical observation in a recent longitudinal study of MRI-guided transrectal prostate biopsy cases accrued over several years, Xu et al. found an average biopsy target displacement of 5.4mm [2]. This creates the need for a system to track the prostate position throughout the biopsy procedure. Intra-operative prostate motion tracking in MRI has been achieved before [3, 4], but these depend on custom MR sequences and access to the MR machine to control the timing and pose of tracking scans. Such advanced approaches are not available for average care facilities and cannot be considered as clinically practical. Acquiring intra-operative MR volume would promise the possibility of volumetric registration between the original and moved biopsy locations. Unfortunately, both the acquisition time and processing time for volumetric MR are prohibitively long. We need a workable solution that is sufficiently accurate, available as a standard feature on every MRI scanner, and affordable in terms of OR time and labor.
We propose to position the prostate in the scanner’s isocenter and acquire multiple statically set slices in this position. It is posited that full six degree-of-freedom (DOF) motion of the prostate can be recovered through the registration of a target planning MR volume and multiple MR slices acquired immediately before and after needle insertion. The tracking slices are acquired with ordinary anatomical imaging sequences through the scanner’s console. This approach is affordable, as slice acquisition takes only a few seconds and is available on any commercial MRI scanner without restriction.
In the literature, slice-to-volume registration has been investigated in the contexts of CT guidance [5] and ultrasound [6]. In the context of prostate tracking in MRI, two works in particular inspired our project. Fei, et al. [7] developed a slice-to-volume registration algorithm with application to radio-frequency thermal ablation of prostate cancer, in which 15 actual interventional prostate MRI slices from transverse, sagittal, and coronal orientations were registered to a pre-operative MRI volume, respectively. The slices from each orientation were independently registered to the pre-operative volume, meaning that three independent registrations were performed and the results were compared. Their algorithm featured a multi-resolution approach with an automatic restart. The problem of local extrema traps and the inefficiency of Fei’s optimization were addressed by Gill et al. [8]. They eliminated the need for restarting the routine by performing a multi-resolution registration alone on a volume of interest (VOI), and incorporated transverse and sagittal slices centered at the prostate, which were formed into a simulated intra-operative volume. Tadayyon et al. [9] improved on Gill’s performance in a rigid scheme, which despite its speed advantage did not consider deformations [9].
Our present contributions are threefold. Foremost, we constructed a new deformable two-stage organ tracking scheme; eliminated the need for random restarting that hampered earlier works; and analyzed the performance on clinical MRI-guided prostate biopsy data. The straightforwardness of our approach must not belie the investment of creative efforts needed to make it a workable clinical tool, despite the availability of several underlying algorithmic components developed by others in synergistic problems. We constructed an intuitive, practical, and inexpensive solution for a pressing clinical problem. Our solution blends seamlessly with the current MRI scanner install base and workflow, so it could be translated to patient care in a reasonable timeframe with minimal cost.
II.Methods
A. Workflow
The goal is to develop a rapid and accurate registration of a high-resolution target planning MR volume with static MR slices acquired immediately before and after needle insertion. In the context of this paper, a registration error of less than 2mm is considered to be sufficiently accurate as it is comparable to the diameter of a standard biopsy needle but smaller than the diameter of the clinically significant size of prostate tumor which is about 4mm [10]. The objective of tracking is to ascertain current prostate position prior to insertion of the biopsy needle in order to ascertain the anatomical accuracy of the collected sample. Tracking is requested by the physician and executed by the operator through the console, so the requirement for speed is timely response to the physician’s requests. This timely response may be allowed up to 1 minute. The clinical setup we consider is such that the patient is in prone position, a transrectal robotic probe is inserted into the rectum, and a set of high-resolution transverse MR slices are acquired and compounded into a volume for target planning. This target planning “pre-needle” volume serves as reference for the subsequent slice registration.
The formatting pipeline for the input images is illustrated in Figure 1. The moving image is the pre-needle reference volume and the fixed image is originally input as three orthogonal slices. The pre-processing stage of our algorithm must place the slices in the correct position and orientation in a sparse volume. The slice origin and direction cosines read from the DICOM tags are used to resample the slices into a sparse volume. The bounding box of the prostate is defined as the volume of interest in the sparse volume.
Fig 1.
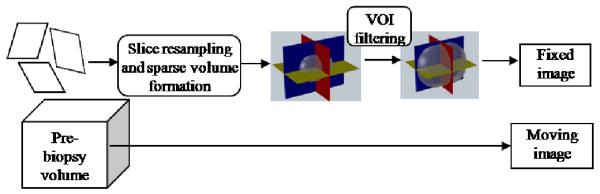
Formatting pipeline for the input images
Figure 2 shows the flow of the registration algorithm. The registration is performed in two stages: A rigid registration is first performed to obtain an initial pose of the pre-needle volume, which is then non-rigidly registered to the fixed, rigid post-needle sparse volume.
Fig. 2.
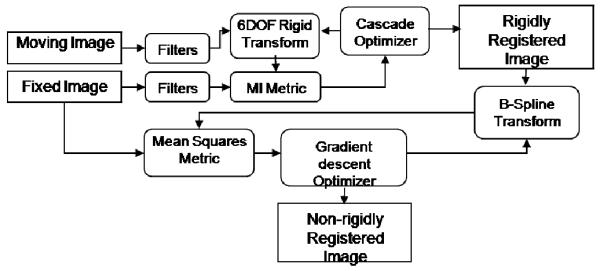
Flow of the registration algorithm
B. Filtering and Similarity Metrics
A histogram matching filter was applied to the moving image to match the intensities of the fixed image. The moving and fixed images were then passed through a Gaussian smoothing filter in order to obtain smooth intensity estimates for our mutual information similarity metric. Random intensity samples were drawn from the fixed image to calculate the joint probability distribution function. The size of the intensity sample population was set to 10% of the total voxel size of the fixed image. The choice of the values of the metric parameters, including smoother variance and population size, was based on trial and error. The smoother variance value was chosen as the optimal balance between smoothness of the density function (low variance) and loss of intensity modes (high variance). The population size was chosen based on the optimal balance between computational speed and representational accuracy of the joint probability distribution function.
C.Transformations and Optimizations
Rigid transformation optimization proceeds in a cascade model, in which the translation parameters are optimized using the CMA Evolutionary Strategy (CMA-ES) [11]. Following translation, rotation is optimized by gradient descent optimization. For our application, the CMA-ES was not able to optimize a 6-DOF search space as it diverged on rotations regardless of scaling. Thus, we decoupled the translation and rotation optimizations and used the CMAES for the parameters that varied the most, i.e. the translation. The gradient descent optimizer converges quickly and accurately for parameters that have a smaller variation range, i.e. the rotations in our case. We feed the rigidly registered pre-needle volume to a deformable registration algorithm, which was implemented as a two level registration pyramid using coarse B-spline grid (5 × 5) followed by a finer grid (15 × 15). We search the parameter space of the B-Spline grid for the minimum mean square of intensity differences using a gradient descent optimizer.
D.Experimental Data
In the pursuit of a more accurate biopsy, Krieger et al. developed robotic assistance under MR image guidance [12]. To date, their system has been used at the U.S. National Cancer Institute in multiple clinical trials. Under ethics board approval, we have obtained five patient data sets from their studies. Each dataset contained a pre-needle volume image used for target planning and a post-needle volume image used for needle placement verification. The high-resolution MRI volumes were acquired from a T2 MRI transverse scan using a 1.5T GE MRI system. The target planning (pre-needle) images had resolutions of 0.78 × 0.78 × 4 mm/pixel for patient 1, 0.625 × 0.625 × 3mm/pixel for patient 2, and 0.55 × 0.55 × 3 mm/pixel for patients 3, 4, and 5. The intra-operative images (post-needle) had resolutions of 0.78 × 0.78 × 4 mm/pixel for patient 1, 0.625 × 0.625 × 3 mm/pixel for patient 2, 0.85 × 0.85 × 3mm/pixel for patients 3 and 4, and 0.94 × 0.94 × 3mm/pixel for patient 5. Their slice dimensions were 256 × 256 pixels for all patients. We extracted three orthogonal slices from each post-needle volume, centered in the prostate. As true sagittal and coronal slices were not available, they were obtained by interpolation between the transverse image slices. We discarded the reformatted sagittal and coronal slices that did not contain the prostate.
III. Results
In order to explore the robustness, capture range and temporal performance of our rigid algorithm on actual clinical data, we applied random perturbations to each patient volume and registered it back to itself. In these tests, the fixed image was a sparse volume of the moving image that was computationally displaced. We applied ±5 mm translation and ±5 degree rotation on all axes and attempted to recover the introduced misalignment. 25 registration tests were performed for each of the 10 volumes (5 volume pairs), totaling the number of registration tests to 250. The results are summed up in Table 1. Registration error was defined as the Euclidean distance between the ground truth voxels and the registered voxels averaged over the prostate sparse volume. We achieved 100% successful registration results where success was defined as a registration error of 2mm or less. The registration error, standard deviation, and registration time are shown in table 1. The higher registration times for patients 1 and 2 can be explained by the fact that the registration tests involved cases where the initial volume displacement was outside of the algorithm’s capture range which caused the optimization to reach the maximum number of iterations. The registration tests revealed that the algorithm could not lock on the solution for initial displacements of more than 10mm for patients 1 and 2.
Table 1.
Registration error (mm) and time (sec) on patient data perturbed around the reference pose
| Registration error | Registration time |
||
|---|---|---|---|
| patient 1 | pre-needle | 0.00 ±0.01 | 6 |
| post-needle | 0.08±0.05 | 66 | |
| patient 2 | pre-needle | 0.09±0.07 | 49 |
| post-needle | 0.04±0.01 | 50 | |
| patient 3 | pre-needle | 0.00±0.00 | 5 |
| post-needle | 0.03±0.01 | 7 | |
| Patient 4 | pre-needle | 0.00±0.00 | 5 |
| post-needle | 0.013±0.00 | 10 | |
| Patient 5 | Pre-needle | 0.00±0.00 | 7 |
| Post-needle | 0.00±0.00 | 7 | |
| Mean | 0.04±0.03 | 21 |
To validate the actual patient registrations, we segmented the prostate in both volumes and computed the mean surface misalignment after registration. We term this measure as surface distance (SD). This result may seem surprising at first, but actually coincides with latest results in the literature contributed by Xu [2] and Karnik [13].
The average rigid registration time was 70 s for the rigid algorithm and 1000 s for the deformable algorithm (includes initial rigid alignment time). The pre-needle insertion and post-needle insertion positions of the first biopsy target for patient 1 are shown in Figure 3. The pre-insertion position was obtained from biopsy data and the post needle position was obtained by our tracking algorithm. The prostate surfaces before and after rigid and deformable registrations for patient 1 are shown in Figure 4. Note that the prostate was segmented for the purpose of validation. No segmentation was involved in the algorithm before or during registration.
Fig. 3.
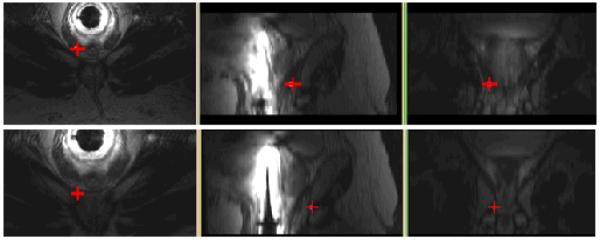
Transverse, sagittal, and coronal views of biopsy target position before needle insertion (top) and after needle insertion (bottom)
Fig. 4.
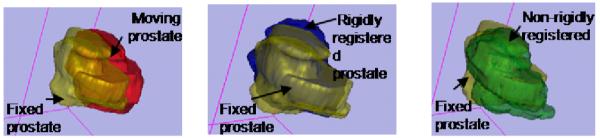
Prostate surfaces before registration (left), after rigid registration (middle), and after deformable registration (right). Just as in Table 2, deformable registration shows no substantial benefits.
IV. Discussion
One major challenge of our evaluation is the fact that in clinical patient data there is no strong ground truth. As opposed to many computer assisted interventions that use fiducials to perform or evaluate registrations, there are no fiducials associated with the in-vivo prostate during a biopsy procedure. Implanting such markers for our study would involve discomfort and risk to the patient. As such, the measure of registration error for actual patient registrations for this study is based on surface misalignment. In the case of this study where an accurate ground truth is not present, point transformations cannot be used to compute error as performed by previous groups [7] since there is no reference transformation to base the error on.
Results of the deformable registration tests show that deformation of the prostate is not significant, which is fully in step with the latest findings of Xu [2] and Karnik [13] who suggest that their results from rigid and non-rigid intra-operative prostate biopsy registrations were not statistically different. An accuracy gain of 1.25 fold at the cost of 14 fold loss in temporal performance does not favour the deformable algorithm for clinical applications.
Another source of tracking error is the needle artifact, but its contribution is probably small since it only occupies a few voxels. Table 1 also suggests that tracking in the pre-needle volume is consistently better than post-needle volumes, indicating that imaging quality deteriorates during the procedure.
We must stress again that our experiments were conducted with reformatted sagittal and coronal images. Under true clinical circumstances, the resolution of the sagittal and coronal tracking images will be several times higher, which will undoubtedly improve registration performance. In essence, the current tests with low-resolution non-axial slices underestimate the true power of our prostate tracking method. Also, in practice tracking slices will be acquired more often than in the datasets we received for the analysis. In real cases, we will run an additional tracking sequence immediately before needle insertion, which will divide current prostate motion and deformation errors in about half - a major improvement in target tracking performance. In addition, because slice imaging is fast, we will probably acquire more than just three canonical slices and the use of three transverse, one coronal and one sagittal slice will surely surpass present accuracy and robustness.
Our registration time for the rigid algorithm is clinically acceptable. Deformable registration times could raise feasibility concerns, but in all we are not particularly concerned about time, because for clinical trials the registration should be ported to the GPU which then obsoletes all prior considerations.
In conclusion, results on limited clinical patient data indicate that the rigid registration is sufficient for tracking of the prostate in MRI-guided robotic biopsy. Our rigid algorithm has demonstrated convergent results for initial prostate displacements up to 8mm. Work continues with performing target registration error studies in typical sextant and octant biopsy locations and, most importantly, moving toward a prospective clinical trial with the use of true sagittal and coronal slices.
Table 2.
Mean surface distances (mm) before and after registration of actual patient datasets
| Initial SD | Rigid SD | Non-rigid SD | |
|---|---|---|---|
| Patient 1 | 4.51 | 1.82 | 1.32 |
| Patient 2 | 2.23 | 2.38 | 1.93 |
| Patient 3 | 7.76 | 1.65 | 1.62 |
| Patient 4 | 4.13 | 1.88 | 1.83 |
| Patient 5 | 7.67 | 5.01 | 3.56 |
| Overall | 5.26 | 2.55 | 2.05 |
Acknowledgments
This work has been supported by the National Institute of Health under grants 5R01CA111288-04 and 5R01EB002963-05.
Contributor Information
Hadi Tadayyon, Department of Electrical and Computer Engineering, Queen’s University, Kingston, Canada K7L3N6 (hadi.tadayyon@queensu.ca).
Andras Lasso, School of Computing, Queen’s University, Kingston, Canada K7L3N6 (lasso@cs.queensu.ca).
Sean Gill, School of Computing, Queen’s University, Kingston, Canada K7L3N6 (gill@cs.queensu.ca).
Aradhana Kaushal, National Institutes of Health, Bethesda, USA.
Peter Guion, National Institutes of Health, Bethesda, USA.
Gabor Fichtinger, School of Computing, Queen’s University, Kingston, Canada K7L3N6 (gabor@cs.queensu.ca).
References
- [1].Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59(4):225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- [2].Xu H, Lasso A, Vikal S, Guion P, et al. SPIE Medical Imaging, Visualization 2010, Image-guided Procedures and Modeling. Vol. 7625. 2010. Accuracy validation for MRI-guided robotic prostate biopsy. to appear in. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].DiMaio SP, Pieper S, Chinzei K, et al. Robot-assisted Needle Placement in Open-MRI: System Architecture, Integration and Validation, Computer Aided Surgery. 2007;12(1):15–24. doi: 10.3109/10929080601168254. [DOI] [PubMed] [Google Scholar]
- [4].Hata N, Tokuda J, Morikawa S, Dohi T. Projection Profile Matching for Intraoperative MRI Registration Embedded in MR Imaging Sequence. MICCAI 2002. 2002:164–169. [Google Scholar]
- [5].Xu S, Fichtinger G, Lindisch D, Cleary K. SPIE Medical Imaging 2005: Visualization, Image-Guided Procedures, and Display. Vol. 5744. 2005. Validation of 3D Motion Tracking of Pulmonary Lesions for Image-guided Lung Biopsy; pp. 60–68. [Google Scholar]
- [6].Blackall JM, Penney GP, King AP, Hawkes DJ. Alignment of sparse freehand 3-D ultrasound with preoperative images of the liver using models of respiratory motion and deformation. IEEE Trans Med Imaging. 2005;24(11):1405–16. doi: 10.1109/TMI.2005.856751. [DOI] [PubMed] [Google Scholar]
- [7].Fei B, Duerk JL, Boll DT, et al. Slice-to-Volume Registration and its Potential Application to Interventional MRI-Guided Radio-Frequency Thermal Ablation of Prostate Cancer. IEEE Trans Med Imaging. 2003;22(4) doi: 10.1109/TMI.2003.809078. [DOI] [PubMed] [Google Scholar]
- [8].Gill S, Abolmaesumi P, Vikal S, et al. Intraoperative Prostate Tracking with Slice-to-Volume Registration in MR; International Conference of the Society for Medical Innovation and Technology; Vienna, Austria. 2008; 2008. pp. 154–158. Electronic Proceedings, ISBN 3-902087-25-0. [Google Scholar]
- [9].Tadayyon H, Vikal S, Gill S, Lasso A, Fichtinger G. MRI-guided prostate motion tracking by means of multislice-to-volume registration. Proc. SPIE. 2010;7625 doi: 10.1117/12.844454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Canto EL, Singh H, Shariat SF, et al. Effects of systematic 12-core biopsy on the performance of percent free prostate specific antigen for prostate cancer detection. Journal of Urology. 2004;172:900–904. doi: 10.1097/01.ju.0000134619.72675.8d. [DOI] [PubMed] [Google Scholar]
- [11].Hansen N. The CMA Evolution Strategy: A Comparing. Springer; 2006. pp. 75–102. [Google Scholar]
- [12].Krieger A, Susil R, Menard C, et al. Design of novel MRI compatible manipulator for image guided prostate interventions. IEEE Trans. Biomed. Eng. 2008;52(2):295–304. doi: 10.1109/TBME.2004.840497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Karnik V, Fenster A, Bax J, et al. Assessment of registration accuracy in three-dimensional transrectal ultrasound images of prostates. SPIE Medical Imaging, Visualization, Image-guided Procedures and Modeling. 2010:7625. doi: 10.1118/1.3298010. [DOI] [PubMed] [Google Scholar]


